Abstract
Complex I is the first and largest enzyme of the respiratory chain, playing a central role in cellular energy production by coupling electron transfer between NADH and ubiquinone to proton translocation. It is implicated in many common human neurodegenerative diseases. Here we report the first crystal structure of the entire, intact complex I (from T. thermophilus) at 3.3 Å resolution. The structure of the 536 kDa complex comprises 16 different subunits with 64 transmembrane helices and 9 Fe-S clusters. The core fold of subunit Nqo8 (NuoH/ND1) is, unexpectedly, similar to a half-channel of the antiporter-like subunits. Small subunits nearby form a linked second half-channel, thus completing the fourth proton translocation pathway, in addition to the channels in three antiporter-like subunits. The quinone-binding site is unusually long, narrow and enclosed. The quinone headgroup binds at the deep end of this chamber, near cluster N2. Strikingly, the chamber is linked to the fourth channel by a “funnel” of charged residues. The link continues over the entire membrane domain as a remarkable flexible central axis of charged and polar residues. It likely plays a leading role in the propagation of conformational changes, aided by coupling elements. The structure suggests that a unique, out-of-the-membrane quinone reaction chamber allows the redox energy to drive concerted long-range conformational changes in the four antiporter-like domains, resulting in translocation of four protons per cycle.
Complex I (NADH:ubiquinone oxidoreductase, EC1.6.5.3) plays a central role in the respiratory chain in mitochondria and many bacteria1-7. It catalyses the transfer of two electrons from NADH to ubiquinone, coupled to the translocation of four protons (current consensus value8,9) across the bacterial or inner mitochondrial membrane:
The transfer of two electrons from NADH to oxygen, through complexes I, III (bc1) and IV (cytochrome c oxidase), results in the translocation of 10 protons across the membrane10, creating the proton-motive force (pmf) for the synthesis of ATP by ATP synthase11. Complex I is a reversible machine12, able to utilize pmf and ubiquinol to reduce NAD+.
Mutations in complex I subunits lead to the most common human neurodegenerative diseases5,13. The enzyme is also a major source of reactive oxygen species in mitochondria14, which can lead to mitochondrial DNA damage and are implicated in Parkinson’s disease15 and aging16.
Complex I is one of the largest known membrane proteins. The mitochondrial enzyme consists of 44 different subunits (~980 kDa in total)17,18. The simpler prokaryotic version normally comprises 14 “core” subunits (~550 kDa total), highly conserved from bacteria to humans1,2,5,19,20, suggesting that the mechanism is also conserved. Both enzymes contain equivalent redox components and have a similar L-shaped structure, formed by the hydrophilic and membrane domains2,5,21. The ~30 “accessory” subunits of the mitochondrial enzyme mostly form a protective shell around the core3,22,23, although some may have a specialized functional role23-25.
We determined the first structures of the eight-subunit hydrophilic domain of Thermus thermophilus complex I at up to 3.1 Å resolution26,27. It contains all the redox centres of the enzyme – non-covalently bound flavin mononucleotide (FMN) and nine iron-sulphur (Fe-S) clusters. NADH transfers two electrons to FMN as a hydride ion and then electrons are transferred one by one, along the uniquely long (95 Å) chain of seven conserved Fe-S clusters, to the quinone-binding site at the interface with the membrane domain.
We subsequently determined the architecture of the entire T. thermophilus complex I at 4.5 Å resolution, with the membrane domain resolved at the level of arrangement of subunits and α-helices21. X-ray analysis at 6.3 Å resolution of the Yarrowia lipolytica mitochondrial enzyme was published later28, but no subunits were identified and no models were deposited from this work, due to limited resolution. Currently all information on atomic structures of complex I is from our studies.
Recently we have determined the 3.0 Å resolution structure of the membrane domain from E. coli complex I29. The three largest subunits, Nqo12/NuoL, Nqo13/NuoM and Nqo14/NuoN (T. thermophilus/E. coli nomenclature, Supplementary Table 3), are homologous to each other and to Na+/H+ antiporter complex (Mrp) subunits30,31. Each contains 14 conserved TM helices and a putative proton translocation channel29. However, the crystals lacked Nqo8/NuoH/ND1, the only core subunit of unknown structure.
The overall architecture of complex I suggests that the coupling mechanism involves long-range conformational changes: there are no cofactors in the membrane and the antiporter-like subunits (which we will call ‘antiporters’ for brevity) are distant from the interface with the hydrophilic domain21,29. Exactly how these changes are coupled to redox reactions remains unclear, although mechanical coupling elements have been suggested21,28,29. Surprisingly, Fe-S cluster N2, which donates electrons to the quinone, is ~25-30 Å away from the membrane surface21, suggesting that the quinone has to move out of the membrane to accept electrons. Nqo8 is the most conserved membrane subunit of complex I (Supplementary Fig. 1), forming the interface with the hydrophilic domain and contributing to the quinone (Q) binding site32. This subunit emerged only once during evolution, joining hydrogenase and antiporter modules of complex I-related enzymes20. Clearly, the atomic structure of the entire complex, including Nqo8, holds the key to understanding the enigmatic coupling mechanism.
Determination of structures
The diffraction of crystals of the entire T. thermophilus complex has been improved to 3.3 Å resolution (Methods). Crystals are, however, twinned and so to overcome the problem of model bias, we crystallised the isolated T. thermophilus membrane domain. These crystals were non-twinned and contained subunit Nqo8. The structure was solved at 3.3 Å resolution by molecular replacement with our E. coli model (PDB 3RKO) (Supplementary Tables 1 and 2, Supplementary Fig. 2). It contains seven subunits (Nqo12 with 16 TM helices, Nqo13 - 14, Nqo14 – 14, Nqo10 – 5, Nqo11 – 3, Nqo7 – 3 and Nqo8 – 9). Antiporters Nqo12-14 show an arrangement of helices (Supplementary Fig. 3) and key residues similar to the E. coli structure - each subunit contains two inverted symmetry-related half-channels. The cytoplasm-linked TM4-8 half-channel contains a central lysine on the discontinuous, thus flexible, TM7 and its pKa-modulating glutamate on TM5, while the periplasm-linked TM9-13 half-channel contains a central lysine (Glu in Nqo13) on discontinuous TM12 (Fig. 1b). The half-channels are linked into a single full proton translocation channel by charged residues, including a lysine from the broken (partly unwound in the middle) TM8 (His in Nqo12). The long connecting helix HL, from the C-terminal extension of Nqo12, is straighter in T. thermophilus than in E. coli (Supplementary Fig. 3). On the opposite side of the domain, the β-hairpin-helix connecting element (βH) shows a very similar arrangement in both species. Thus, both previously proposed coupling elements29 appear to be a common complex I feature.
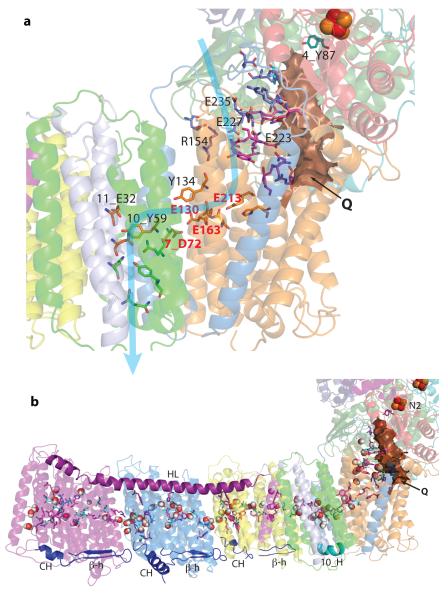
Fig. 1.
a) Structure of the entire complex I from T. thermophilus. FMN and Fe-S clusters are shown as magenta and red-orange spheres, respectively, with cluster N2 labelled. Key helices around the entry point (Q) into the quinone reaction chamber, and approximate membrane position are indicated. b) Putative proton translocation channels in the antiporter-like subunits. Polar residues lining the channels are shown as sticks with carbon in dark blue for the first (N-terminal) half-channel, in green for the second (C-terminal) half-channel and in orange for connecting residues. Key residues, GluTM5 and LysTM7 from the first half-channel, Lys/HisTM8 from the connection and Lys/GluTM12 from the second half-channel, are labelled. Approximate proton translocation paths are indicated by blue arrows.
T. thermophilus Nqo8 contains eight conserved TM helices and an additional C-terminal TM helix. Unexpectedly, TM1 of Nqo7/NuoA is in a different position compared to the E. coli structure, forming a part of the Nqo8 helical bundle (Fig. 1a and Supplementary Fig. 3). Presumably, when NuoH dissociates in E. coli, this helix moves closer to the remaining subunits.
The T. thermophilus membrane domain structure was then used with the hydrophilic domain structure (PDB 3I9V) to solve the structure of the entire complex by molecular replacement. The 3.3 Å resolution structure (Fig. 1a) of the 536 kDa complex contains nine hydrophilic subunits with 9 Fe-S clusters and one FMN molecule, as well as seven membrane subunits with 64 TM helices (Supplementary Tables 1 and 2). This includes the novel assembly factor-like hydrophilic subunit Nqo16, essential for crystallization but not necessary for activity (Supplementary Discussion). Although many assembly factors for mitochondrial complex I are known33, this is the first example for bacterial complex I.
Features at the interface between the two main domains, missing in the individual structures, were resolved in that of the entire complex, including loops from Nqo4/7/8 (Supplementary Figs. 4 and 5). The quinone-binding site is found, as expected21,34, at the interface of Nqo4, 6 and 8 and is described below. Most of the interactions between the two domains involve Nqo8 (Supplementary Discussion and Table 4), including the highly conserved first cytoplasmic loop. Another highly conserved Nqo8 loop (third cytoplasmic) lines the Q cavity. The first cytoplasmic loop from Nqo7/NuoA wraps around Nqo8, stabilizing the domains’ interface.
Subunit Nqo8/NuoH/ND1 forms part of a proton channel
The fold of Nqo8 is unusual, with some TM helices very short (TM5, 14 residues), others very long (TM1, 35 residues) and with nearly all helices highly tilted relative to membrane normal (Fig. 2). Unexpectedly, TM helices 2-6 can be superimposed (RMSD 2.1-2.6 Å over ~140 residues, PDBeFOLD) to the antiporters’ “half-channel” TM helices 4-8 or 9-13 (Fig. 2c). This similarity is not apparent from the sequence (~ 11-18% identity). Although in the antiporters most helices are roughly normal to the membrane, TMs 2-6 from Nqo8 are tilted dramatically, up to 45°. In contrast, TM1 is tilted in the opposite direction, so that it crosses TM6 at nearly 90°. TM9 is peripheral, consistent with its absence in most species.
Fig. 2. Fold of subunit Nqo8.

Coloured blue to red from N to C terminus. Neighbouring subunits Nqo7/10 are shown in light/dark grey. a) View from the cytoplasm. TM helices are numbered, with helices corresponding to the antiporter half-channel in bold. The conserved salt bridge Arg36-Asp62, supporting amphipathic helix AH1, is shown. b) Side view. Charged residues from the conserved 3rd cytoplasmic loop, mainly lining the Q cavity, are shown as sticks. c) Alignment of TM helices 2-6 of Nqo8 (orange) with TM helices 4-8 of Nqo13 (blue). LysTM7 from Nqo13 and Glu213 from Nqo8 TM5 are shown as sticks.
In Nqo8, charged residues are found in similar positions to key antiporters’ residues: Glu130/Glu163 in the GluTM5 position, Glu213/Glu248 - near LysTM7 (Fig. 2c). Overall, Nqo8 contains many more charged residues in the membrane (Fig. 3a) than the antiporters, and many of these residues (including Glu163 and Glu213) are conserved in complex I and in membrane-bound hydrogenases (Supplementary Figs. 1 and 6). They form an unusual chain (or “funnel”) of charged residues leading from the Q-binding site to a remarkable network of four interacting carboxylates deep in the membrane (Glu130, Glu163 and Glu213 from Nqo8 plus conserved 7_Asp72 (prefix indicates subunit)). The charged network congregates around the highly conserved broken 10_TM3, a hotspot for human disease mutations29. On the other side of the break, conserved 10_Tyr59 interacts with essential 11_Glu32, part of a fourth proton translocation channel proposed previously29. Nqo11 superimposes with helices 4-6 from the antiporter half-channels, overlaying Glu32 with GluTM529.
Fig. 3.
a) E-channel (fourth proton translocation channel). Charged and polar residues constituting the channel are shown as sticks. Central residues are shown with carbon in yellow, those forming a link to the Q site in magenta, link to the cytoplasm in blue, link to the periplasm in green and those interacting with quinone headgroup in cyan. Key residues are labelled, with Glu/Asp quartet in red. Approximate proton translocation path is indicated by blue arrow. Quinone cavity is shown with surface in brown. b) Central axis of charged and polar residues. Residues shown are either central to half-channels or are forming the connection between them (charged residues have carbon in magenta, polar in cyan). Most of them are located near the breaks in key helices TM7/8/12 (antiporters), 10_TM3 and 8_TM5. Predicted waters nearby, modelled using Dowser software44, are shown as spheres. Connecting elements are shown in solid colours: helix HL in magenta and the βH element in blue, with the C-terminal helix CH29 and the β-hairpin from each antiporter labelled. The contacting Nqo10 helix is labelled 10_H. Subunits are coloured as in Fig. 1.
Thus, the first half-channel formed by Nqo8 is linked to the second half-channel in Nqo10/11, with 8_TM5 and 10_TM3 playing the roles of the antiporters’ key discontinuous helices TM7/12. This strongly suggests that input from the cytoplasm into the fourth channel is not, as previously proposed29, at the interface between Nqo14 and Nqo11 (which in T. thermophilus is more clearly closed from the cytoplasm than in E. coli), but is via Nqo8 instead. Similarly to the antiporters’ N-terminal half-channel, the Nqo8 half-channel is closed from the periplasm by large hydrophobic residues but is connected through an extensive network of polar residues both to the cytoplasm and to the Q site. Furthermore, similarly to the antiporters’ C-terminal half-channel, the Nqo10/11 half-channel is blocked from the cytoplasm, but is connected to the periplasm by polar residues. The two half-channels are linked into a single channel by the Glu/Asp quartet and putative water molecules (Fig. 3). We dub it the E-channel due to abundance of glutamates in its centre. Many residues in the E-channel are conserved and essential for activity (Supplementary Table 7). Therefore it is, unexpectedly, arranged similarly to the three channels within the antiporters. This implies that proton-pumping stoichiometry in complex I is indeed 4 (not 3, as proposed recently35).
Unique quinone reaction chamber
To determine exactly where quinone binds, intact complex I was co-crystallised, or crystals soaked with quinone analogues: inhibitor Piericidin A and decyl-ubiquinone (DQ). Although in vivo T. thermophilus complex I uses menaquinone-8, it is also fully active with DQ21, and, in contrast to E. coli complex I36, the T. thermophilus enzyme does not contain any bound endogenous quinone after purification. X-ray data (Supplementary Table 1) clearly show (Fig. 4ab) that Piericidin A and DQ bind in a very similar manner, ~15 Å away from the membrane surface, at the deep end of long narrow cavity. In this position, the quinone headgroup is ~12 Å (centre-to-centre) from the Fe-S cluster N2, appropriate for efficient electron transfer37. One of the DQ ketone groups is, as predicted26,34, hydrogen-bonded to 4_Tyr87, while another interacts, unexpectedly, with 4_His38. Both residues are invariant and essential for activity34,38.
Fig. 4. Quinone reaction chamber.
Subunits are coloured as in Fig. 1. Iron-sulphur cluster N2 is shown as red-orange spheres. a-b) Experimental electron density (2mFo-DFc in blue, contoured at 1σ, and mFo-DFc in green, contoured at 3σ) and models obtained from crystals with bound Piericidin A (a) and Decyl-ubiquinone (b). Difference electron density was calculated before ligand modelling. Nqo4 residues interacting with the headgroup are indicated. Potential polar interactions are shown labelled with distances in Å. c) Surface (solvent-accessible) representation of the interface between two main domains. The empty crevice (C, circled, Supplementary Discussion) between Nqo10 and 7_TM1/Nqo8, as well as helices framing the entry point to the quinone site (Q) are indicated. d) Quinone reaction chamber, with its internal solvent-accessible surface coloured red for negative, white for neutral, and blue for positive surface charges. Charged residues lining the cavity are shown with carbon in magenta and hydrophobic residues in yellow. Residues are labelled with prefix indicating subunit (omitted for Nqo8). Ala63, the site of the primary LHON disease mutation45, is labelled in red. e) Theoretical model of bound ubiquinone-10. Carbon atoms in cyan indicates the 8th isoprenoid unit. The quinone chamber is shown with surface in brown and helices framing its entry point are indicated. Movable helix 6_H127, interacting with 8_AH1, is also labelled.
One of the most surprising structural features is that this 30 Å long chamber is completely enclosed from the solvent, with only a narrow (about 2-3 × 4-5 Å) apparent entry point for the quinone, framed by helices TM1, TM6 and amphipathic AH1 from Nqo8 (Figs. 1a and 4), as well as 7_TM1. All residues facing the lipid bilayer here are hydrophobic, but the inside of the chamber is lined, unexpectedly, mostly by hydrophilic residues, especially in the area (“front”) facing the tip of the membrane domain (to the left in Fig. 4d). However, at the opposite side (“back”), a hydrophobic patch formed, surprisingly, mainly by residues from hydrophilic subunits Nqo4/6, extends towards the entrance, sufficient to accommodate the quinone tail. The cavity “front” is mostly negatively charged, whilst the “back” is neutral and the “top” (near cluster N2) is positively charged (Fig. 4d). The ionisable residues lining the chamber are all highly conserved (Supplementary Fig. 6 and Table 8) and mutations to many of these residues lead to human diseases and loss of complex I activity (ref5, Supplementary Tables 5 and 6).
Since the Q chamber is long and narrow, it will restrict the quinone tail to an extended conformation. Thus, knowing the headgroup position, we can model the mode of binding of native ubiquinone with 8 or 10 isoprenoid units (Fig. 4e). Remarkably, only the last 1-3 isoprenoid units will protrude out of the cavity into the lipid. The passage around the entrance is narrower than the rest of the cavity, so that the quinone tail will block solvent access to the cavity, sealing the reaction chamber. That is a surprising feature and suggests that slight structural re-arrangements are necessary to allow the Q headgroup to move in and out of the cavity.
Thus, complex I contains a unique long enclosed reaction chamber, where nearly the entire quinone molecule can be accommodated, in contrast to other membrane proteins, where Q binding sites are usually open (Supplementary Discussion). It is possible that sealing off the cavity has functional importance, although it is not necessary for proton pumping per se, as pumping proceeds with short-tailed Q1 and DQ8,12. However, even with short-tailed quinones, steric restrictions in the cavity will not allow solvent access to the bound headgroup. Previously, a second quinone binding site in complex I has been discussed39, but structural data argues strongly against its existence (Supplementary Discussion).
Coupling between electron transfer and proton translocation
The structure provides clear novel implications for the coupling mechanism of complex I, described in the Supplementary Discussion and summarised briefly here. Whilst a part of the redox energy in complex I is utilised upon reduction of cluster N227, most of it is released during quinone chemistry20. Upon Q reduction, either Q2− or key charged residues nearby are likely to remain unprotonated in order to drive conformational changes. The enclosed Q reaction chamber is well suited for this purpose: due to tight protein packing near the bound headgroup, the quinone can be protonated only via the coordinating 4_Tyr87 and 4_His38. Importantly, the charged species can exist in the chamber because it is relatively hydrophilic and distal from the membrane. The Q site is linked to the Glu/Asp quartet in the centre of the E-channel by a hydrophilic “funnel”, so these negatively charged species can interact electrostatically, driving conformational changes in this channel. Additional driving force is likely provided by moving upon N2 reduction Nqo4/6 helices40, which directly interact with flexible parts of Nqo8.
One of the most fascinating features of the structure is that the hydrophilic “funnel” is then continued, through a series of conserved residues, all the way to the tip of Nqo12. Interacting charged and polar residues, surrounded by a “river” of water molecules, form a remarkable continuous hydrophilic axis in the middle of membrane, spanning the entire length of the membrane domain (Fig. 3b). Its residues are found in the half-channels and in the connections between them and most are on or near the breaks in discontinuous helices, allowing for the flexibility along this axis, linking membrane subunits in an overall conformational cycle. Tight coupling observed between proton translocation and quinone chemistry29 can be explained if opening of the Q site entrance to allow quinone in and out forms a part of this cycle.
Conformational changes likely involve both the previously described coupling elements (helix HL and βH element29) and the central hydrophilic axis. The striking architecture of this axis, discussed previously29 but visible only now in its full extent suggests that it probably plays a prominent role in the cycle. The most plausible scenario is that the electrostatically (and N2) -driven E-channel drives conformational changes first in the neighbouring antiporter Nqo14/NuoN, which in its turn drives distal subunits Nqo13/NuoN and Nqo12/NuoL, all via the central axis. The extent of the two coupling elements’ movement during the catalytic cycle is currently unclear, but they likely contribute at least to the coordination between the three antiporters. The concerted conformational changes (conformational switch41) would lead to changes in pKa (Supplementary Table 9) and solvent exposure of key residues, resulting in proton translocation (Fig. 5).
Fig. 5. Proposed coupling mechanism of complex I.
a) Overview showing key helices and residues. Upon electron transfer from cluster N2, negatively charged quinone initiates a cascade of conformational changes, propagating from the E-channel (Nqo8/10/11) to the antiporters via the central axis (red arrows) of charged and polar residues located around flexible breaks in key TM helices. Cluster N2-driven shifts of Nqo4/6 helices27 (blue arrows) likely assist overall conformational changes. Helix HL and the βH element help coordinate conformational changes by linking discontinuous TM helices between the antiporters. In the antiporters, LysTM7 from the first half-channel is assumed to be protonated (via the link to cytoplasm) in the oxidised state29. Upon reduction of quinone and subsequent conformational change, the first half-channel closes to the cytoplasm, GluTM5 moves out and LysTM7 donates its proton to the connecting Lys/HisTM8 and then onto Lys/GluTM12 from the second half-channel. Lys/GluTM12 ejects its proton into periplasm upon return from reduced to oxidised state. A fourth proton per cycle is translocated in the E-channel in a similar manner. TM helices are numbered and key charged residues (GluTM5, LysTM7, Lys/GluTM12, Lys/HisTM8 from Nqo12-14, 11_Glu67, 11_Glu32, interacting with 10_Tyr59, 8_Glu213 and some residues from the connection to Q cavity) are indicated by red circles for Glu and blue circles for Lys/His. b) Schematic drawing illustrating conformational changes between the two main (low energy) conformations. Analysis of networks of polar residues and modelled waters in the structure suggests that in the oxidised state (as crystallised) periplasmic half-channels are likely to be open. Residues shown as black circles indicate conserved prolines from the break in TM12.
The view that anionic ubiquinol is the main driving force for the conformational changes is supported by evolutionary considerations, now including the antiporter fold of Nqo8: membrane-bound proton-translocating hydrogenases, ancestors of complex I20, do not use quinone and likely evolved when soluble hydrogenase attached to the antiporter complex (which is driven essentially by membrane polarization). Sealing of the interface between the two complexes, containing charged NiFe centre reaction intermediates and thus mimicking membrane polarization, would immediately result in coupling of the oxidoreductase and proton translocation activities.
We can now finally begin to make sense of the enormous complexity of this molecular machine and understand how it evolved, with all pieces of the puzzle falling into place. The three novel and unexpected features of the structure - a sealed Q chamber, an antiporter-like Nqo8 fold and a hydrophilic “funnel” connecting Q site to the E-channel - combine to suggest that redox-driven conformational changes propagate to four proton channels via the unique central hydrophilic axis, aided by coupling elements. Mutations in any core complex I subunit, whether known to cause human disease or introduced in model studies, can now be understood on a structural basis: observed effects are consistent with the proposed mechanism (Supplementary Discussion, Tables 5, 6 and Fig. 7). Further details of conformational changes and how exactly they lead to proton translocation will need clarification from structures of different redox states of the enzyme and time-resolved studies.
Methods
Protein purification
Intact complex I from Thermus thermophilus was purified as described previously21, except that the DEAE column was replaced by a Mono-S cation-exchange column. Fractions from the ANX column were diluted to ~5 mM NaCl with buffer A (20 mM Bis-Tris pH 6.0, 0.002% PMSF, 2 mM CaCl2, 10% (v/v) glycerol and 0.05% n-tridecyl-β-maltoside (TDM)) and applied to a Mono-S HR 16/10 column, equilibrated with buffer A. The protein was eluted with a linear gradient of buffer B (0.5 M NaCl in buffer A), using a Gilson HPLC system at room temperature, and the fractions were assessed by SDS-PAGE. Two peaks of NADH:FeCy activity contained complex I lacking Nqo16 protein in the first peak, eluting at ~80 mM NaCl, and complex I with Nqo16 protein in the second peak, eluting at ~100 mM NaCl. These two peak fractions were pooled separately, concentrated to about 1 ml using 100 kDa MWCO concentrators and each applied to a HiLoad 16/60 Superdex 200 gel-filtration column, equilibrated with GF buffer (buffer A with 100 mM NaCl). Fractions from each run were pooled on the basis of purity (assessed by SDS-PAGE), concentrated to about 25 mg ml−1 and an additional 15% glycerol added for storage in liquid nitrogen. Only the protein containing Nqo16 was crystallisable.
The membrane and hydrophilic subunits were separated by treatment of intact complex I at pH 4.0. Purified complex I (2 mg) was incubated in 300 mM sodium acetate pH 4.0, 100 mM NaCl, 50 mM EDTA, 25% (v/v) ethylene glycol (EG) and 0.03% (w/v) n-undecyl-β-maltoside (UDM) for 3 h at room temperature. The protein solution was loaded into a HiLoad 16/60 Superdex 200 gel-filtration column equilibrated with buffer M (20 mM Bis-Tris pH 6.0, 2 mM CaCl2, 100 mM NaCl, 10% EG and 0.03% UDM). Fractions containing the membrane domain were pooled on the basis of purity (assessed by SDS-PAGE), concentrated to about 10 mg ml−1 using 100 kDa MWCO concentrators and used immediately for crystallisation trials. In addition to membrane domain subunits, the preparation contained the hydrophilic subunits Nqo4, 5, 6 and 9, which dissociated upon crystallisation.
Crystallisation
Crystals of intact T. thermophilus complex I were grown at 23 °C using sitting drop crystallisation. First, purified complex I (18 mg ml−1 in 20 mM Bis-Tris pH 6.0, 2 mM CaCl2, 100 mM NaCl, 0.002% PMSF, 25% glycerol and about 2% TDM) was incubated with additional TDM detergent (final concentration of about 4% (w/v)) at room temperature, and then mixed at 2:1 (v/v) ratio with crystallisation reagent comprising 100 mM Bis-Tris pH 6.0, 19-24% (w/v) polyethylene glycol (PEG) 4000, 100 mM KCl, 100 mM glutaric acid pH 6.0 and 2.2 mM FOS-CHOLINE-8 fluorinated. The addition of extra TDM prior to crystallisation trials improved the size of the rod crystals (to about 50 × 50 × 500-700 μm). Crystals used for the iron peak data collection were grown in similar conditions, except that the FOS-CHOLINE-8 additive in the crystallisation reagent was replaced by either 0.6% (w/v) n-dodecyl-β-maltoside (DDM) or 7.6 mM CYMAL-4. Different detergent additives promoted crystal growth to a variable extent but they did not affect diffraction properties. Crystals were fully grown within 1 week. They did not tolerate dehydration and were cryo-protected in 100 mM Bis-Tris pH 6.0, 9% PEG4000, 50 mM KCl, 50 mM glutaric acid pH 6.0, 25% EG and 0.01% TDM prior to plunging into liquid nitrogen.
Crystals of the membrane domain of T. thermophilus complex I were grown at 17 °C using hanging drop crystallisation. Purified membrane domain (10 mg ml−1 in 20 mM Bis-Tris pH 6.0, 2 mM CaCl2, 100 mM NaCl, 10% EG and about 1% UDM) was mixed with crystallisation reagent comprising 100 mM phosphate-citrate pH 4.5, 26% (v/v) PEG300 and 5 mM CHAPS at 1:1 (v/v) ratio. Crystals grew for about 2 weeks and were rectangular-shaped (about 100 × 100 × 500-700 μm). Crystals were dehydrated and cryo-protected in 100mM phosphate citrate pH4.5, 26% PEG 300, 30% PEG 3350, 0.03% UDM, 5mM CHAPS and 7% EG for 6 hours prior to plunging into liquid nitrogen.
Data collection and processing
Data from crystals of the entire complex were collected (using several points along the crystal to minimize radiation damage) at 100 K with a Pilatus 6M detector at beamline ID29 at the European Synchrotron Radiation Facility (ESRF, Grenoble). Image data were processed with XDS and XSCALE46, which helped to deal with the problem of reflections overlap due to a large unit cell. Data from crystals of the membrane domain were collected at 100 K with an ADSC Q210 detector at ESRF beamline ID29. Image data were processed with iMosflm47 and SCALA from the CCP4 suite48.
Crystals of the entire complex I belong to P21 space group with 2 molecules per asymmetric unit (ASU) and 69% solvent content. Crystals are pseudo-merohedrally twinned with twin fractions approaching 0.521. Data from three isomorphous crystals were combined to achieve maximal resolution and completeness (Supplementary Table 1). Overall resolution extends to about 3.3 Å by CC0.5 criterion49, although diffraction along axis b is weaker. Using the Diffraction Anisotropy Server (http://services.mbi.ucla.edu/anisoscale/)50, the data set was anisotropically scaled and truncated to 3.3 Å, 3.5 Å and 3.3 Å resolution, where the F/σ ratio drops to ~2.6–2.8 along the a*, b* and c* axes, respectively. Due to twinning, model bias presented a problem when using the relatively diverse (~30-35% identity) model of the E. coli membrane domain for molecular replacement. To overcome this, we crystallized the isolated T. thermophilus membrane domain. Crystals were in P1 space group and not twinned. Packing resembles that of the E. coli membrane domain29, with 2 molecules per ASU and 66% solvent content. Crystals contained subunit Nqo8, in contrast to the E. coli case. The diffraction was also anisotropic and so the best data set, of about 3.3 Å overall resolution, was anisotropically scaled and truncated to 3.6 Å, 3.4 Å and 3.3 Å resolution along the a*, b* and c* axes, respectively.
Structure solution and refinement
From trials with many heavy atoms for experimental phasing, tungsten derivatives of the membrane domain crystals were obtained. However, W atoms were wedged between the two domains and so disrupted the packing, resulting in lower resolution. Therefore, the structure of the membrane domain was solved by molecular replacement with our E. coli model (PDB 3RKO)29. Initially, individual subunits were refined as rigid bodies in Phaser51, sequence replaced for T. thermophilus with Chainsaw48 and side-chains re-built with SCWRL452. This model was re-built by one cycle of Rosetta_MR protocol in PHENIX53, followed by DEN refinement in CNS54, resulting in R/Rfree=31.5/36.6%. Improved electron density (both calculated and density-modified) allowed for cycles of manual building of subunit Nqo8 and re-building of some regions in other subunits in COOT55, followed by refinement in PHENIX with secondary structure/NCS restraints and using TLS. The final model was refined to R/Rfree=20.9/26.3% (Supplementary Table 1). It contains 2144 residues (Supplementary Table 2) and two bound UDM molecules per domain. All nine TM helices of Nqo8 are resolved in the structure, but the long highly conserved cytoplasmic loops 1 (TM1-2) and 3 (TM5-6) were disordered.
The structure of the entire complex in twinned P21 crystals was solved by molecular replacement (using Phaser) with the new structure of T. thermophilus membrane domain and the previously described structure of the hydrophilic domain (PDB 3I9V). First, to exploit anomalous signal from intrinsic Fe-S clusters, we used combined data from two rare crystals with relatively low twin fraction (~0.4), collected at Fe peak wavelength (Supplementary Table 1). Jelly body refinement of the initial model with SAD target function in Refmac56 resulted in improved electron density for novel features at the interface of the two domains, missing in either starting structure (including Nqo16). Then, manual model building was followed with cycles of refinement in PHENIX against the best native data set, with secondary structure/NCS restraints and using TLS. The structure was refined at 3.3 Å resolution to R/Rfree =20.2%/23.9% (R factors are lower than usual due to twinning). One complex contains 4780 residues, nine Fe-S clusters and one FMN molecule (Supplementary Table 2). Both the membrane domain and the entire complex structures were validated in MOLPROBITY57 and were found to be of better than average quality for the resolution (Supplementary Table 1).
Co-crystals of intact complex I with Piericidin A were obtained by mixing protein and inhibitor at a 1:1 molar ratio prior to crystallisation. The co-crystal was further soaked in 100 μM Piericidin A (added from 10 mM stock in 100% DMSO) for approximately 4 hours, prior to cryo-cooling. Decyl-ubiquinone soaks were performed by soaking native complex I crystals overnight in solution containing 500 μM decyl-ubiquinone (added from 50 mM stock in 100% ethanol). Crystals were otherwise treated and data processed as for native crystals. MR with the intact complex structure in Phaser was followed by refinement in Refmac for 40 cycles with Jelly body restraints (sigma 0.02). This resulted in a significant drop of Rfree, however, the models were not re-built manually and should be considered preliminary. A strong positive difference electron density (shown in green in Fig. 4ab) identified the position of the aromatic ring of the compounds; they were modelled into the electron density, added to the initial structure and Refmac refinement repeated, resulting in the models shown in Fig. 4ab.
Bioinformatics
Structure-based multiple sequence alignment was performed in CLUSTALW v1.8358 with the profile alignment option. Water molecules were modelled using Dowser software59. When applied to the E. coli membrane domain structure (PDB 3RKO), a similar “river” of waters was predicted, with crystallographic waters, where observed, coinciding with those modelled. Conservation scores for Supplementary Fig. 6 were calculated with ConSurf server60, with sequences for alignment selected to be between 90% and 30% similar, resulting in about 300-400 sequences per subunit. Figures were prepared in PyMol. Surface charges were calculated with APBS plug-in in PyMol.
Supplementary Material
Acknowledgements
This work was funded by the Medical Research Council. We thank the European Synchrotron Radiation Facility and the Swiss Light Source for provision of synchrotron radiation facilities. We are grateful to the staff of beamlines ID29, ID23-2 (ESRF, Grenoble) and X06SA (Swiss Light Source, Villigen) for assistance.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information: The coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 4HE8 (membrane domain) and 4HEA (entire complex).
References
- 1.Walker JE. The NADH - ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 2.Yagi T, Matsuno-Yagi A. The proton-translocating NADH-Quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry. 2003;42:2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 3.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu. Rev. Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 4.Ohnishi T. Iron-sulfur clusters/semiquinones in complex I. Biochim. Biophys. Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 5.Sazanov LA. Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 6.Hirst J. Towards the molecular mechanism of respiratory complex I. Biochem. J. 2010;425:327–339. doi: 10.1042/BJ20091382. [DOI] [PubMed] [Google Scholar]
- 7.Sazanov LA, editor. A structural perspective on respiratory complex I : structure and function of NADH:ubiquinone oxidoreductase. Springer; 2012. [Google Scholar]
- 8.Galkin AS, Grivennikova VG, Vinogradov AD. H+/2e− stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 1999;451:157–161. doi: 10.1016/s0014-5793(99)00575-x. [DOI] [PubMed] [Google Scholar]
- 9.Galkin A, Drose S, Brandt U. The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim. Biophys. Acta. 2006;1757:1575–1581. doi: 10.1016/j.bbabio.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim. Biophys. Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U S A. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinogradov AD. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim. Biophys. Acta. 1998;1364:169–185. doi: 10.1016/s0005-2728(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 13.Schapira AH. Human complex I defects in neurodegenerative diseases. Biochim. Biophys. Acta. 1998;1364:261–270. doi: 10.1016/s0005-2728(98)00032-2. [DOI] [PubMed] [Google Scholar]
- 14.Murphy MP. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 16.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Carroll J, et al. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 18.Balsa E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Yip CY, Harbour ME, Jayawardena K, Fearnley IM, Sazanov LA. Evolution of respiratory complex I: "supernumerary" subunits are present in the alpha-proteobacterial enzyme. J. Biol. Chem. 2011;286:5023–5033. doi: 10.1074/jbc.M110.194993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efremov RG, Sazanov LA. The coupling mechanism of respiratory complex I - A structural and evolutionary perspective. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbabio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Efremov RG, Baradaran R, Sazanov LA. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 22.Efremov RG, Sazanov LA. Respiratory complex I: ‘steam engine’ of the cell? Curr. Opin. Struct. Biol. 2011;21:532–540. doi: 10.1016/j.sbi.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Angerer H, et al. A scaffold of accessory subunits links the peripheral arm and the distal proton-pumping module of mitochondrial complex I. Biochem J. 2011;437:279–288. doi: 10.1042/BJ20110359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta. 2003;1604:135–150. doi: 10.1016/s0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 25.Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 27.Berrisford JM, Sazanov LA. Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 2009;284:29773–29783. doi: 10.1074/jbc.M109.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329:448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 29.Efremov RG, Sazanov LA. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 30.Fearnley IM, Walker JE. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim. Biophys. Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- 31.Mathiesen C, Hagerhall C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi K, Murai M, Miyoshi H. Exploring the binding site of acetogenin in the ND1 subunit of bovine mitochondrial complex I. Biochim. Biophys. Acta. 2009;1787:1106–1111. doi: 10.1016/j.bbabio.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Nouws J, Nijtmans LG, Smeitink JA, Vogel RO. Assembly factors as a new class of disease genes for mitochondrial complex I deficiency: cause, pathology and treatment options. Brain. 2012;135:12–22. doi: 10.1093/brain/awr261. [DOI] [PubMed] [Google Scholar]
- 34.Angerer H, et al. Tracing the tail of ubiquinone in mitochondrial complex I. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbabio.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 35.Wikstrom M, Hummer G. Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc. Natl. Acad. Sci. USA. 2012;109:4431–4436. doi: 10.1073/pnas.1120949109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verkhovsky M, Bloch DA, Verkhovskaya M. Tightly-bound ubiquinone in the Escherichia coli respiratory Complex I. Biochim. Biophys. Acta. 2012;1817:1550–1556. doi: 10.1016/j.bbabio.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 38.Kashani-Poor N, Zwicker K, Kerscher S, Brandt U. A central functional role for the 49-kDa subunit within the catalytic core of mitochondrial complex I. J. Biol. Chem. 2001;276:24082–24087. doi: 10.1074/jbc.M102296200. [DOI] [PubMed] [Google Scholar]
- 39.Ohnishi T, Nakamaru-Ogiso E, Ohnishi ST. A new hypothesis on the simultaneous direct and indirect proton pump mechanisms in NADH-quinone oxidoreductase (complex I) FEBS Lett. 2010;584:4131–4137. doi: 10.1016/j.febslet.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berrisford JM, Thompson CJ, Sazanov LA. Chemical and NADH-induced, ROS-dependent, cross-linking between subunits of complex I from Escherichia coli and Thermus thermophilus. Biochemistry. 2008;47:10262–10270. doi: 10.1021/bi801160u. [DOI] [PubMed] [Google Scholar]
- 41.Bai F, et al. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science. 2010;327:685–689. doi: 10.1126/science.1182105. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 45.Huoponen K, Vilkki J, Aula P, Nikoskelainen EK, Savontaus ML. A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy. Am. J. Hum. Genet. 1991;48:1147–1153. [PMC free article] [PubMed] [Google Scholar]
- 46.Kabsch W. Xds. Acta crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Evans P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta crystallogr. D. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krivov GG, Shapovalov MV, Dunbrack RL., Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 54.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallograph. D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 60.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.