Abstract
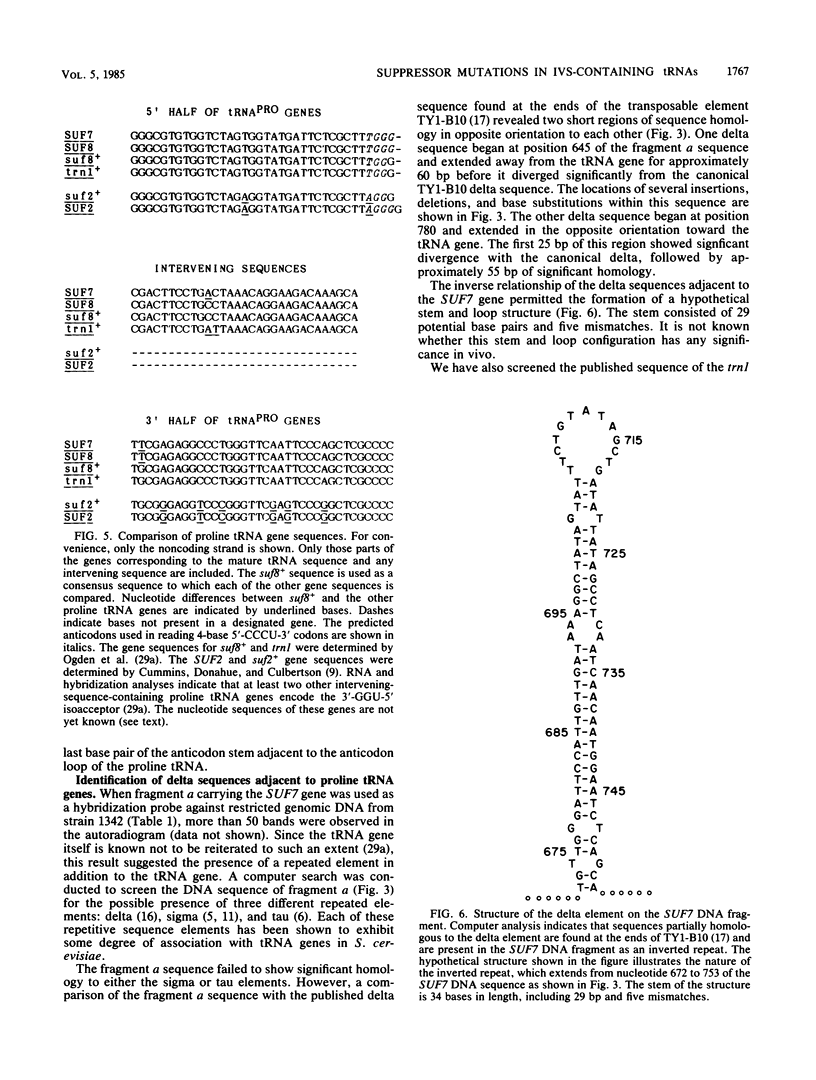
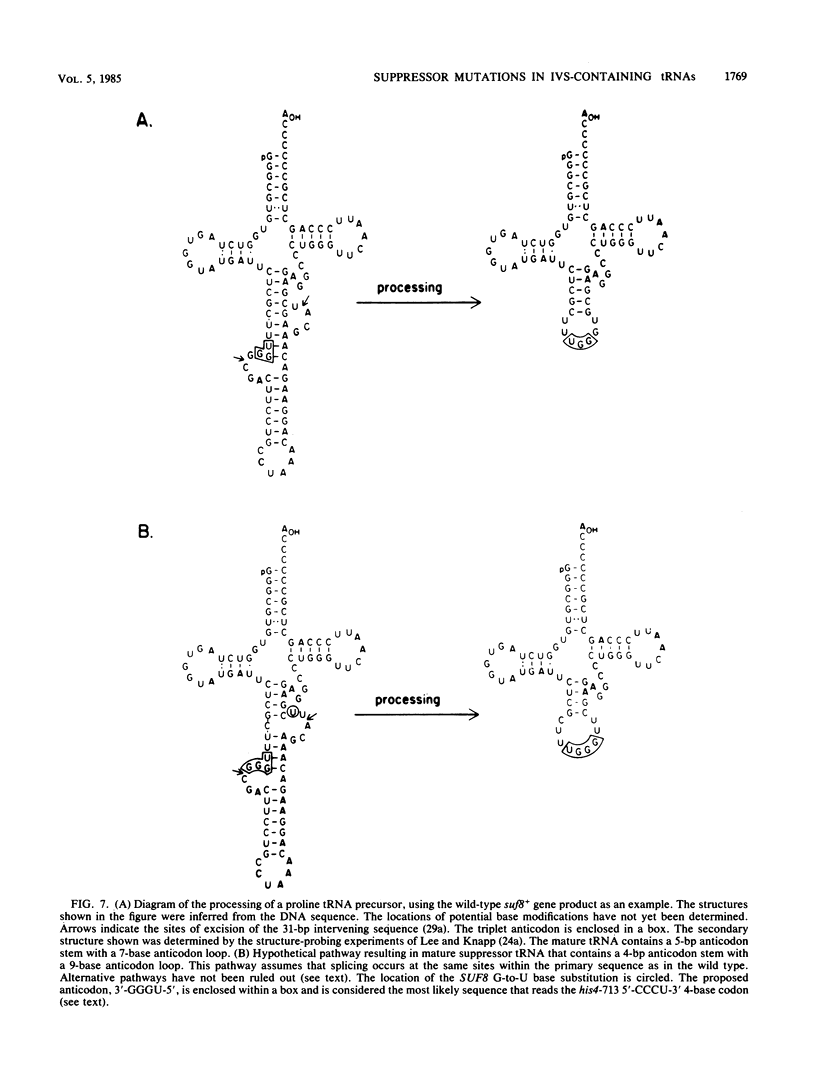
Extragenic suppressors of +1 frameshift mutations in proline codons map in genes encoding two major proline tRNA isoacceptors. We have shown previously that one isoacceptor encoded by the SUF2 gene (chromosome 3) contains no intervening sequence. SUF2 suppressor mutations result from the base insertion of a G within a 3'-GGA-5' anticodon, allowing the tRNA to read a 4-base code word. In this communication we describe suppressor mutations in genes encoding a second proline tRNA isoacceptor (wild-type anticodon 3'-GGU-5') that result in a novel mechanism for translation of a 4-base genetic code word. The genes that encode this isoacceptor include SUF7 (chromosome 13), SUF8 (chromosome 8), trn1 (chromosome 1), and at least two additional unmapped genes, all of which contain an intervening sequence. We show that suppressor mutations in the SUF7 and SUF8 genes result in G-to-U base substitutions at position 39 that disrupted the normal G . C base pairing in the last base pair of the anticodon stem adjacent to the anticodon loop. These anticodon stem mutations might alter the size of the anticodon loop and permit the use of a 3'-GGGU-5' sequence within the loop to read 4-base proline codons. Uncertainty regarding the exact structure of the mature suppressor tRNAs results from the possibility that anticodon stem mutations might affect sites of intervening sequence removal. The possible role of the intervening sequence in the generation of mature suppressor tRNA is discussed. Besides an analysis of suppressor tRNA genes, we have extended previous observations of the apparent relationship between tRNA genes and repetitive delta sequences found as solo elements or in association with the transposable element TY1. Hybridization studies and a computer analysis of the DNA sequence surrounding the SUF7 gene revealed two incomplete, inverted delta sequences that form a stem and loop structure located 165 base pairs from the 5' end of the tRNA gene. In addition, sequences beginning 164 base pairs from the 5' end of the trn1 gene also exhibit partial homology to delta. These observations provide further evidence for a nonrandom association between tRNA genes and delta sequences.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach M. L., Lacroute F., Botstein D. Evidence for transcriptional regulation of orotidine-5'-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Ruth J. R. The influence of codon context on genetic code translation. Nature. 1980 Jul 10;286(5769):123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Brodeur G. M., Sandmeyer S. B., Olson M. V. Consistent association between sigma elements and tRNA genes in yeast. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3292–3296. doi: 10.1073/pnas.80.11.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm G. E., Genbauffe F. S., Cooper T. G. tau, a repeated DNA sequence in yeast. Proc Natl Acad Sci U S A. 1984 May;81(10):2965–2969. doi: 10.1073/pnas.81.10.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Charnas L., Johnson M. T., Fink G. R. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics. 1977 Aug;86(4):745–764. doi: 10.1093/genetics/86.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. M., Culbertson M. R. Molecular cloning of the SUF2 frameshift suppressor gene from Saccharomyces cerevisiae. Gene. 1981 Sep;14(4):263–278. doi: 10.1016/0378-1119(81)90159-1. [DOI] [PubMed] [Google Scholar]
- Cummins C. M., Donahue T. F., Culbertson M. R. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3565–3569. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. M., Gaber R. F., Culbertson M. R., Mann R., Fink G. R. Frameshift suppression in Saccharomyces cerevisiae. III. Isolation and genetic properties of group III suppressors. Genetics. 1980 Aug;95(4):855–879. doi: 10.1093/genetics/95.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. Suppressible four-base glycine and proline codons in yeast. Science. 1981 Apr 24;212(4493):455–457. doi: 10.1126/science.7010605. [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):2052–2061. doi: 10.1128/mcb.4.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Frameshift suppression in Saccharomyces cerevisiae. IV. New suppressors among spontaneous co-revertants of the Group II his4-206 and leu 2-3 frameshift mutations. Genetics. 1982 Jul-Aug;101(3-4):345–367. doi: 10.1093/genetics/101.3-4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. The yeast frameshift suppressor gene SUF16-1 encodes an altered glycine tRNA containing the four-base anticodon 3'-CCCG-5'. Gene. 1982 Sep;19(2):163–172. doi: 10.1016/0378-1119(82)90002-6. [DOI] [PubMed] [Google Scholar]
- Gafner J., Robertis E. M., Philippsen P. Delta sequences in the 5' non-coding region of yeast tRNA genes. EMBO J. 1983;2(4):583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J., Fink G. R. Identification of chromosomal location of yeast DNA from hybrid plasmid p Yeleu 10. Nature. 1977 Sep 15;269(5625):265–267. doi: 10.1038/269265a0. [DOI] [PubMed] [Google Scholar]
- Ingolia T. D., Craig E. A., McCarthy B. J. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell. 1980 Oct;21(3):669–679. doi: 10.1016/0092-8674(80)90430-4. [DOI] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Krayev A. S., Kramerov D. A., Skryabin K. G., Ryskov A. P., Bayev A. A., Georgiev G. P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1201–1215. doi: 10.1093/nar/8.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meyers S., Friedland P. Knowledge-based simulation of genetic regulation in bacteriophage lambda. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):1–9. doi: 10.1093/nar/12.1part1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Pagel F. T., Hijazi K. A. Codon context effects in missense suppression. J Mol Biol. 1984 May 5;175(1):19–27. doi: 10.1016/0022-2836(84)90442-x. [DOI] [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. V., Loughney K., Hall B. D. Identification of the yeast DNA sequences that correspond to specific tyrosine-inserting nonsense suppressor loci. J Mol Biol. 1979 Aug 15;132(3):387–410. doi: 10.1016/0022-2836(79)90267-5. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Broach J. R., Wensink P. C., Hereford L. M., Fink G. R., Botstein D. Isolation and analysis of recombinant DNA molecules containing yeast DNA. Gene. 1978 Sep;4(1):37–49. doi: 10.1016/0378-1119(78)90013-6. [DOI] [PubMed] [Google Scholar]
- Riddle D. L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol. 1973 Apr 25;242(121):230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Thomas M., Kelly J., Selker E., Davis R. W. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rey F. J., Donahue T. F., Fink G. R. sigma, a repetitive element found adjacent to tRNA genes of yeast. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4138–4142. doi: 10.1073/pnas.79.13.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]