Abstract
Bromodomain inhibition comprises a promising therapeutic strategy in cancer, particularly for hematologic malignancies. To date, however, genomic biomarkers to direct clinical translation have been lacking. We conducted a cell-based screen of genetically-defined cancer cell lines using a prototypical inhibitor of BET bromodomains. Integration of genetic features with chemosensitivity data revealed a robust correlation between MYCN amplification and sensitivity to bromodomain inhibition. We characterized the mechanistic and translational significance of this finding in neuroblastoma, a childhood cancer with frequent amplification of MYCN. Genome-wide expression analysis demonstrated downregulation of the MYCN transcriptional program accompanied by suppression of MYCN transcription. Functionally, bromodomain-mediated inhibition of MYCN impaired growth and induced apoptosis in neuroblastoma. BRD4 knock-down phenocopied these effects, establishing BET bromodomains as transcriptional regulators of MYCN. BET inhibition conferred a significant survival advantage in three in vivo neuroblastoma models, providing a compelling rationale for developing BET bromodomain inhibitors in patients with neuroblastoma.
Keywords: BET bromodomain inhibitor, JQ1, MYCN, neuroblastoma, BRD4
Introduction
The interplay between master regulatory transcription factors and specific chromatin-associated co-activators is an emerging hallmark of cancer. Cancer genomic discovery efforts continue to reveal mutations in epigenetic modifiers, and laboratory efforts to validate functional dependencies are ongoing (1). Moreover, there are notable examples of successful commercial development of drugs targeting epigenetic modifiers, including the development of inhibitors of enzymatic “writers,” such as DNA methyltransferases for myelodysplastic syndrome, and inhibitors of enzymatic “erasers,” such as histone deacetylases for cutaneous T-cell lymphoma.
Modulation of the epigenetic regulators known as “readers” has recently emerged as a therapeutic strategy in cancer treatment. These epigenetic “readers” are structurally diverse proteins, which recognize and bind to covalent modifications of chromatin (2). One important modification associated with open chromatin and transcriptional activation is the side-chain acetylation of lysine residues on histone tails (3). The dominant mode of recognition of acetylated lysine residues is by bromodomains, present in 47 human proteins (4–6). We and others have described the therapeutic potential of targeting one bromodomain-containing family important in regulating transcription, epigenetic memory, and cell growth: the BET (bromodomain and extra-terminal domain) family, composed of BRD2, BRD3, BRD4, and BRDT. Numerous hematologic malignancies and the highly malignant solid tumor NUT midline carcinoma are responsive to BET inhibition in vitro and in mouse models (7–12).
While disease-specific indications for drugs modifying epigenetic regulators have been uncovered, precise genomic biomarkers predictive of treatment response remain elusive. To date, the best validated genetic predictor of response to BET inhibitors is in a rare genetically-defined subset of poorly differentiated squamous cell carcinomas (NUT midline carcinoma), where the presence of recurrent t(15;19) chromosomal translocation results in the expression of the twin N-terminal bromodomains of BRD4 as an in-frame fusion with the NUT protein (13). High-throughput pharmacogenomic profiling offers the opportunity to reveal new insights into selective responses to drugs in defined cancer genotypes. Initial efforts to connect drug response with genotype in the NCI60 cell line panel have since been expanded to screening campaigns in large panels of genetically characterized cancer cell lines (14–17). These efforts have revealed both expected and unexpected connections. For example, the anticipated response to ALK inhibitors in tumors with aberrant ALK activation, such as lymphoma, non-small cell lung cancer, and neuroblastoma, was demonstrated in a screen of over 600 tumor cell lines (15). More recently, the unexpected connections between response to poly (ADP-ribose) polymerase (PARP) inhibitors and expression of the EWS/FLI fusion protein in Ewing sarcoma was elucidated in a screen of 130 drugs in over 600 cancer cell lines (16). In an independent study of 24 anti-cancer drugs in 479 human cancer cell lines, new connections were also observed between small-molecule sensitivities and cell lineage, gene expression, and genotype (17).
We performed a high-throughput pharmacogenomic screen to identify biomarkers of response to BET bromodomain inhibitors. The prototype ligand JQ1, a novel thieno-triazolo-1,4-diazepine, which displaces BET bromodomains from chromatin by competitively binding to the acetyl lysine recognition pocket, has been validated in numerous models, nominating it as an excellent chemical probe for high-throughput screening (7–10). In this study, we therefore queried a large compendium of genetically characterized tumor cell lines to identify predictors of sensitivity to JQ1. We identified MYCN amplification as a top predictive marker of response to JQ1 treatment and characterized the mechanistic and translational significance of this finding in neuroblastoma, the most common extra-cranial solid tumor diagnosed in children, and a cancer notable for frequent MYCN amplification in patients with high-risk disease.
Results
High-throughput Pharmacogenomic Profiling Reveals MYCN Amplification as a Predictor of Response to Bromodomain Inhibitors
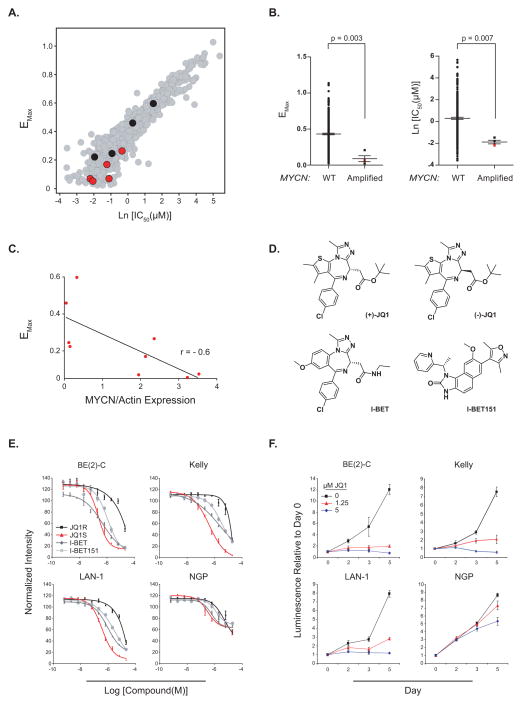
We first conducted an unbiased screen of a collection of 673 genetically characterized tumor derived cell lines (16) to understand response and resistance to BET bromodomain inhibition, so as to discover new opportunities for therapeutic development. Cell lines with response to JQ1 yielding IC50 ≤ 1 μM and Emax ≥ 70 % were designated as sensitive and all other were designated as resistant in a stringent classification schema. Cell lines arising from the pediatric solid tumor of neural crest origin, neuroblastoma, were identified as among the most JQ1-sensitive and MYCN amplification as the most predictive marker of sensitivity; four cell lines out of the 99 sensitive cell lines are MYCN amplified and zero lines out of the 237 resistant cell lines are MYCN amplified. The two-tailed Fisher exact test returns a P value of 0.007 (Fig. 1A–B and Supplementary Table S1). We next determined expression level of MYCN in the neuroblastoma cell lines from the primary screen (Supplementary Fig. S1A) and evaluated the correlation of MYCN protein levels with JQ1 response. MYCN protein level is also substantially correlated with response to JQ1 treatment (Fig. 1C).
Figure 1.
MYCN-amplified neuroblastoma is sensitive to the effects of BET bromodomain inhibition. (A) Anti-proliferative activity of JQ1 was profiled in over 650 cancer cell lines, revealing a broad range of sensitivity and resistance. Red dots are neuroblastoma cell lines with MYCN amplification based on SNP 6.0 arrays and/or high levels of protein expression. Black dots indicate neuroblastoma cell lines wildtype for MYCN and poor MYCN expression. Drug response is presented as the natural log of the half-maximal effective concentration [Ln(IC50)], plotted against the maximum effect corresponding to the minimum measured viability (Emax). (B) Distribution of Emax and Ln(IC50) for MYCN wildtype versus MYCN amplified cancer cell lines based on SNP 6.0 copy number analysis. P value calculated using non parametric Mann-Whitney test. Red squares indicate MYCN-amplified neuroblastoma cell lines. (C) Spearman correlation between minimum measured viability (Emax) and the expression level of MYCN normalized by Actin level (ratio MYCN/Actin). (D) Structures of BET bromodomain inhibitors. (+)-JQ1, I-BET, and I-BET151 are all active, structurally distinct, BET bromodomain inhibitors. JQ1R is the inactive (−)-JQ1 enantiomer. (E) Dose response of neuroblastoma cell line viability with BET bromodomain inhibitor treatment measured by a luminescent ATP detection assay. Data represent mean ± SEM of four biological replicates. (F) Four MYCN-amplified neuroblastoma cell lines were used to determine the effects of JQ1 on growth. Values over time are shown relative to the day zero values, with error bars representing the mean +/− SD of eight replicates per condition.
We retested one of the most highly sensitive MYCN-amplified cell lines from the screen, BE(2)-C, and multiple additional MYCN-amplified neuroblastoma cell lines confirmed to have elevated levels of MYCN protein (Supplementary Fig. S1B). Nearly all of the MYCN-amplified cell lines tested responded to JQ1 and a panel of structurally distinct BET bromodomain inhibitors, each of which conferred a dose-responsive, inhibitory effect on cell viability as well as growth over time (Fig. 1D–F and Supplementary Table S2). The only exception in this cell line panel was NGP, which was comparatively insensitive to all of the BET bromodomain inhibitors tested. Importantly, cell growth was not affected in any cell lines by the (−)-JQ1 enantiomer, which lacks activity against BET bromodomains in biochemical and biological assays, further supporting an on-target mechanism of action. Similarly, we retested one of the least sensitive cell lines from the primary screen, the MYCN-wildtype neuroblastoma cell line SK-N-AS and a second MYCN-wildtype cell line SH-SY5Y not in the primary screen. As predicted, SK-N-AS was insensitive to the effects of JQ1, and SH-SY5Y was less sensitive than the MYCN-amplified cell lines based upon the maximal response (Emax) (Supplementary Fig. S2A–B and Supplementary Table S2).
JQ1 Induces Cell Cycle Arrest and Apoptosis
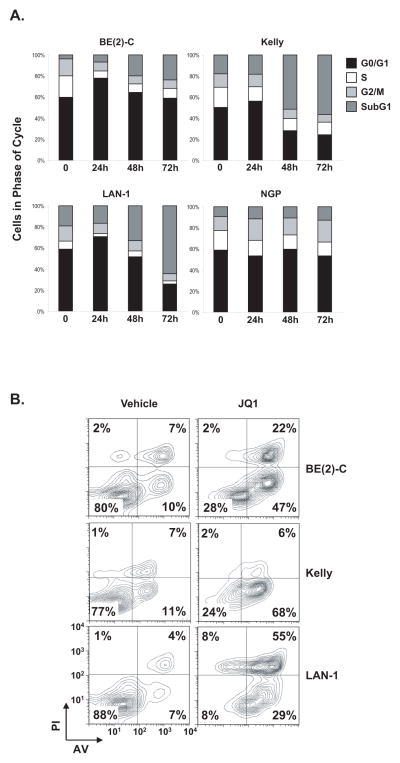
To further characterize the phenotypic consequences of JQ1 treatment on responsive MYCN-amplified neuroblastoma cells, we determined its effects on cell cycle arrest and apoptosis. JQ1 treatment induced a G0/G1 arrest and a decrease in S phase at 24 hours by flow cytometric evaluation of propidium iodide staining (Fig. 2A and Supplementary Fig. S3). There was a progressive increase in cell death, based on the increase in the subG1 fraction, with marked induction of apoptosis confirmed by annexin V staining at 72 hours (Fig. 2B). In contrast, the effect of JQ1 on cell cycle and apoptosis in MYCN-wildtype neuroblastoma was less pronounced (Supplementary Fig. S4A and B).
Figure 2.
JQ1 treatment induces a G0/G1 arrest and apoptosis in neuroblastoma cell lines. Indicated neuroblastoma cell lines were treated with 1 μM JQ1 for (A) 24, 48 and 72 hours before cell cycle analysis or (B) 72 hours before measuring apoptosis by annexin V staining detected by flow cytometry.
Bromodomain Inhibition Downregulates MYCN and c-MYC Transcriptional Programs in Neuroblastoma
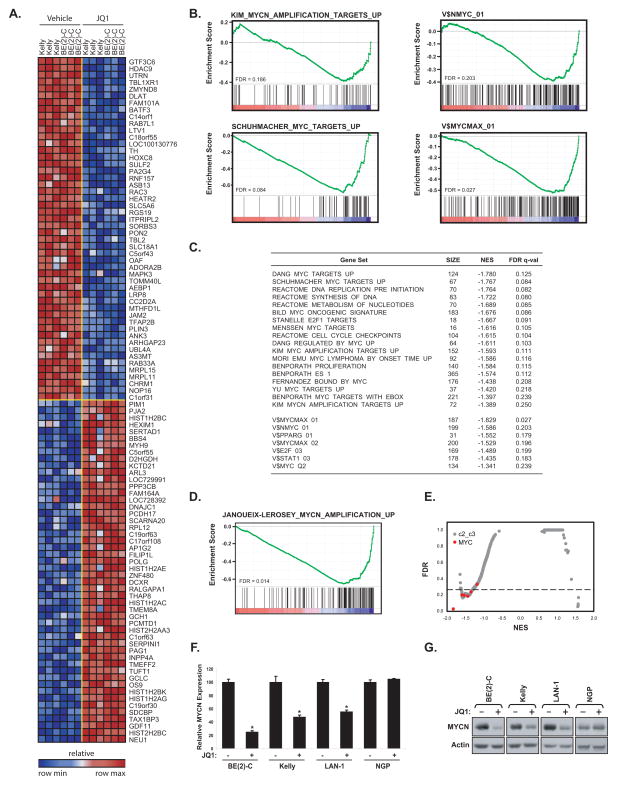
Prior research from our laboratory has demonstrated a selective effect of BET bromodomain inhibition on the expression and function of c-MYC in hematologic malignancies (8–10). We thus hypothesized that the selective sensitivity of neuroblastoma lines to BET inhibition may relate to impaired function of the MYCN and c-MYC transcriptional activators. We profiled two of the highly sensitive neuroblastoma cell lines, BE(2)-C and Kelly, treated with 1 μM JQ1 for 24 hours using genome-wide transcriptional profiling coupled with gene set enrichment analysis (GSEA) (18). The top 50 differentially upregulated and downregulated genes are depicted in the heatmap in Fig. 3A and the full list is reported in Supplementary Table S3. Consistent with the established biological role of BET co-activators in chromatin-dependent transcriptional activation, there was a preponderance of downregulated genes in comparison to upregulated genes. Acute JQ1 treatment prompted a dynamic change in transcription, with 122 genes significantly upregulated and 193 genes significantly downregulated based on permutation P < 0.05 and FDR < 0.05 for signal-to-noise in the comparison of all vehicle-treated versus all JQ1-treated samples. To assess the effects of JQ1 more specifically on transcriptional programs regulated by either MYCN or c-MYC, we interrogated the data with published, validated gene signatures for statistically significant enrichment by GSEA. GSEA seeks to estimate the significance of over-representation of an independently defined set of genes (e.g., c-MYC or MYCN pathways) in the highly correlated or anti-correlated genes in the gene expression data set. The majority of the MYCN and c-MYC-related gene sets were statistically enriched among genes downregulated by BET bromodomain inhibition in neuroblastoma (Fig. 3B and C). Because the MYCN signature in the Molecular Signature Database (MSigDB) was derived in small cell lung cancer cells (19), we sought to develop a custom MYCN upregulated signature in neuroblastoma cells utilizing a publicly available data set of primary neuroblastoma tumors characterized for MYCN amplification status (20). This MYCN signature was also highly downregulated by JQ1 treatment of neuroblastoma cell lines based on GSEA (Fig. 3D and Supplementary Table S4). An open-ended enrichment analysis was next performed, as a measure of specificity, on the entire set of transcription factor target gene signatures available from MSigDB (21). In almost all cases, gene sets defined by adjacency to MYC-binding motifs (both MYCN and c-MYC) were highly enriched in genes whose expression is suppressed by JQ1 in neuroblastoma (Fig. 3E).
Figure 3.
Inhibition of MYCN and c-MYC-dependent transcription by JQ1 treatment of neuroblastoma cells. (A) Heatmap of the top 50 down- and up-regulated genes following 24 hours of 1 μM JQ1 treatment of neuroblastoma cell lines based on an SNR Score and P < 0.05. Data are presented row normalized. (B) GSEA demonstrating downregulation of MYCN and c-MYC-dependent gene sets and representative sets of genes with proximal promoter regions containing MYCN or MYC-MAX binding sites in the transcriptional profiles of neuroblastoma cell lines treated with JQ1. Depicted is the plot of the running sum for the molecular signature data base (MSigDB) gene set within the JQ1 neuroblastoma dataset including the maximum enrichment score and the leading edge subset of enriched genes. (C) Table of selective gene sets enriched among genes downregulated by JQ1 in neuroblastoma cell lines based on GSEA (size = number of genes in each set, NES = normalized enrichment score, and FDR = false discovery rate). (D) GSEA showing downregulation of a custom MYCN gene set derived from the comparison of MYCN-amplified versus MYCN-non-amplified primary neuroblastoma tumors. (E) Quantitative comparison of all transcription factor target gene sets available from the MSigDB by GSEA for downregulation in JQ1-treated neuroblastoma cells. Data are presented as a scatterplot of FDR versus NES for each evaluated gene set. Red indicates sets for either MYCN or c-MYC and gray for other transcription factors. (F) Neuroblastoma cell lines were treated with 1 μM JQ1 or DMSO for 8 hours. After RNA extraction, level of MYCN transcript was quantified. Expression values are shown relative to the DMSO condition for each cell line. Error bars represent mean +/− SD of four technical replicates. *P < 0.001 calculated using a one-way ANOVA with Bonferroni correction comparing JQ1 treatment to DMSO within a cell line. (G) Western-blots for MYCN on whole cell extracts from four neuroblastoma cell lines treated with 1 μM JQ1 or DMSO for 24 hours.
Recently, we reported the unexpected downregulation of MYC expression in multiple myeloma and acute leukemia models by BET bromodomain inhibitors (8–10). Localization of BRD4 to strong enhancer elements proximal to MYC prompted the consideration that expression of amplified MYCN may, indeed, be dependent on BET bromodomains. We therefore evaluated the effects of JQ1 treatment on MYCN expression. While not in the top 100 differentially expressed genes, MYCN expression was significantly downregulated in the genome-wide expression profiling of both neuroblastoma cell lines for three of the four PrimeView representative probes with an average reduction of 50% (Supplementary Table S5). These results were confirmed in the three neuroblastoma cell lines responsive to JQ1 (BE(2)-C, Kelly, and LAN-1) by RT-PCR and immunoblot (Fig. 3F and G). In contrast, MYCN expression was not downregulated in the cell line most insensitive to JQ1, NGP. Finally, c-MYC expression level was reduced in response to JQ1 treatment across all the MYCN amplified or non amplified neuroblastoma cell lines (Supplementary Fig. S5). Taken together, these data provide strong evidence that the MYCN program is downregulated in neuroblastoma cell lines by JQ1 treatment and, as observed for MYC in several hematologic disease models, BET bromodomain inhibition targets the expression of MYCN itself.
Context-Independent and Dependent Transcriptional Effects of BET Bromodomain Inhibition in Cancer Cells
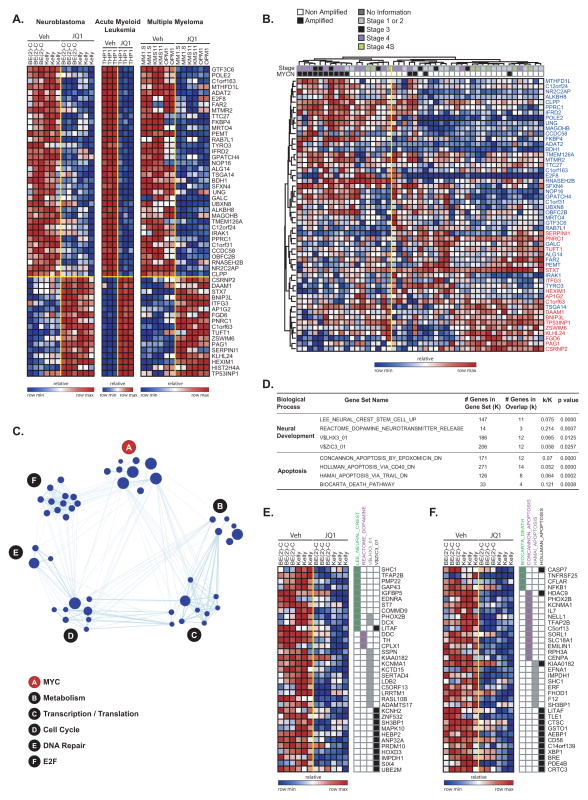
While the transcriptional programs altered by BET bromodomain inhibition in individual diseases have been established, an integrated analysis has not yet been performed. To establish neuroblastoma-specific and canonical BET bromodomain-dependent transcriptional pathways, we analyzed three data sets profiling the transcriptional changes associated with JQ1 treatment in cancer cell lines (multiple myeloma (8), AML (9), and neuroblastoma) to determine whether there is a robust, cell context independent JQ1 signature. First, we generated signatures of genes whose expression is regulated (up and down) by JQ1 treatment in neuroblastoma and used this as a gene set to query both the multiple myeloma and the AML transcriptional profiling data sets. There was significant enrichment of these neuroblastoma JQ1 signatures in the hematopoietic malignancies with an FDR < 0.25 (Supplementary Fig. S6A).
Next, we developed hematopoietic malignancy signatures for both upregulated and downregulated genes with JQ1 treatment using the multiple myeloma and the AML data sets. We applied these signatures as gene sets to query our neuroblastoma transcriptional profiling data by GSEA. Both of these signatures were consistently enriched in the expected directions in neuroblastoma cell lines treated with JQ1 (Supplementary Fig. S6B). Moreover, all of the JQ1 signatures were enriched in the treatment of AML cells with a structurally distinct BET bromodomain inhibitor, I-BET151 (11), consistent with an on-target effect of JQ1 (Supplementary Fig. S7). Finally, we developed a consensus signature across all three data sets (Fig. 4A) and determined the relevance to primary human neuroblastoma. The consensus JQ1 signature was significantly associated with MYCN-amplified neuroblastoma when projected into a primary human neuroblastoma gene expression profiling data set with annotation for MYCN status and stage, suggesting that the signature has relevance beyond cancer cell lines grown in culture to bona fide human disease (Fig. 4B). We next determined the biological and functional connections among those genes. Notably, while many individual gene sets were significant, there was marked enrichment for multiple gene sets associated with MYCN and c-MYC, E2F, cell cycle, transcription/translation, metabolism and DNA damage/repair (Fig. 4C and Supplementary Table S6). Taken together, this analysis suggests that there is indeed a consensus transcriptional signature altered by JQ1 treatment of cancer cells with c-MYC and MYCN gene sets, and their associated biological functions, serving as hubs.
Figure 4.
Transcriptional changes associated with BET bromodomain inhibition by JQ1. (A) Heatmap of the 36 down- and 17 up-regulated genes after JQ1 treatment with a consistent direction of regulation in neuroblastoma, multiple myeloma, and AML and fold-change greater than two and P ≤ 0.05. Data are presented row normalized. (B) Heatmap of the JQ1 consensus signature developed in Fig 4A evaluated in a data set profiling genome-wide expression of primary neuroblastoma tumors. JQ1 consensus signature genes denoted in blue are downregulated and those in red are upregulated with JQ1 treatment. Data are presented row normalized. The neuroblastoma samples cluster into two groups which are associated with the MYCN amplification status of the tumors (P < 0.002 by Fisher exact test) and with high stage (stage 3 and 4) versus low stage (all other) (P < 0.002 by Fisher exact test). (C) A relaxed consensus JQ1 downregulation signature was identified based on the absolute FC ≥ 1.5 and P-value and FDR ≤ 0.05 and was interrogated in a Functional Enrichment Analysis across the MSigDB. The results were visualized with the Enrichment Map software which organizes the significant gene sets into a network called an “enrichment map.” In the enrichment map the nodes correspond to gene sets and the edges reflect significant overlap between the nodes according to a Fisher test. The hubs correspond to collections of genes sets with a unifying class label according to GO biological processes. The size of the nodes is correlated with the number of genes in the gene set. (D) Table describing the results of Fisher tests for the MSigDB signatures enriched with genes selectively downregulated by JQ1 in neuroblastoma cells. Heatmap of the genes uniquely regulated by JQ1 in neuroblastoma in the neural development (E) and apoptosis-related (F) gene sets. Data are presented row normalized.
While BET bromodomain inhibition alters a common transcriptional program across structurally distinct compounds and cancers of different lineages, we were also interested in the transcriptional changes unique to MYCN-amplified neuroblastoma. In this case, we identified a set of genes whose expression is uniquely altered in neuroblastoma but not in the hematopoietic cancer cell lines (Supplementary Fig. S8). Here, there was evidence of loss of lineage related genes with JQ1 treatment, reflected in the downregulation of gene sets associated with neural crest stem cells and neurotransmitter release, as well as the downregulation of sets of genes with proximal promoter regions containing binding sites for LHX3 (LIM homeobox 3, a gene involved in neural development) and of ZIC3 (zinc finger protein of the cerebellum 3) (Fig. 4D and E) (22). In addition, consistent with the observed phenotypic consequences of the compound in neuroblastoma cells, there was enrichment for gene sets associated with apoptosis (Fig. 4F). A full list of enriched gene sets is presented in Supplementary Table S7.
A BET Bromodomain Inhibitor Displaces BRD4 from the MYCN Promoter Region and MYCN Targets Are Downregulated
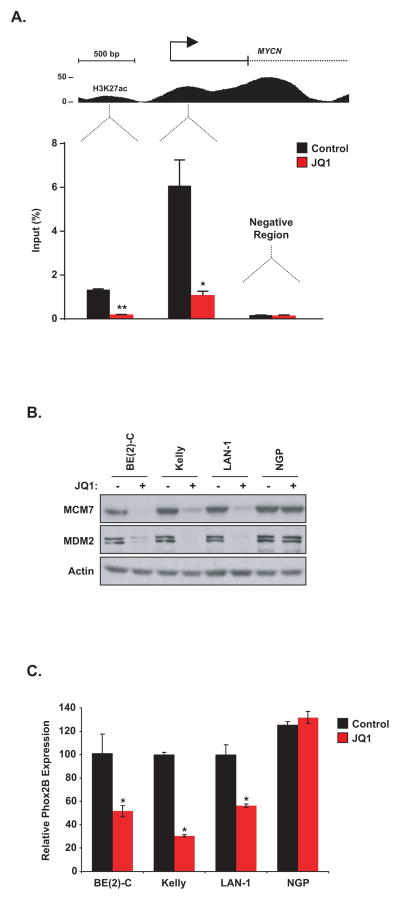
It was previously shown that BET bromodomain Inhibitors displace BRD4 from the c-MYC promoter region suggesting the hypothesis that BET inhibitors would similarly displace BRD4 from the MYCN promoter region (8). Using chromatin immunoprecipitation (ChIP) PCR, we observed BRD4 localization to the MYCN promoter, as well as a putative enhancer region. JQ1 treatment resulted in displacement of the BRD4 co-activator protein from both elements, providing a mechanistic explanation for the observed JQ1-dependent decrease in MYCN transcription in sensitive neuroblastoma cells (Fig. 5A). Moreover, with JQ1 treatment two well-reported targets of MYCN, MCM7 and MDM2, are confirmed to be downregulated by western blotting, and expression of the neuroblastoma specific target identified in our expression profiling analysis, PHOX2B, is also downregulated (Fig 5B and C). As expected, there is no alteration of these targets in NGP, where JQ1 treatment does not lead to downregulation of MYCN expression.
Figure 5.
(A) ChIP with a BRD4 antibody at two sites within the MYCN promoter region in BE(2)-C cells treated with 1 μM JQ1 for 24 hours. Enrichment is shown as the percentage of total input DNA. The negative control region primers amplify within a gene desert region ~1Mb away from MYCN. Error bars represent ± SEM of triplicate data. *P < 0.05, **P < 0.01 (paired t-test). (B) Western blot demonstrating effects of 1 μM JQ1 treatment for 24 hours on the expression of MCM7 and MDM2 in MYCN-amplified neuroblastoma cells. (C) Neuroblastoma cell lines were treated with 1 μM JQ1 or DMSO for 24 hours. After RNA extraction, level of PHOX2B transcript was quantified. Expression values are shown relative to the DMSO condition for each cell line. Error bars represent mean +/minus; SD of four technical replicates. * P < 0.001 calculated using a one-way ANOVA with Bonferroni correction comparing JQ1 treatment to DMSO within a cell line.
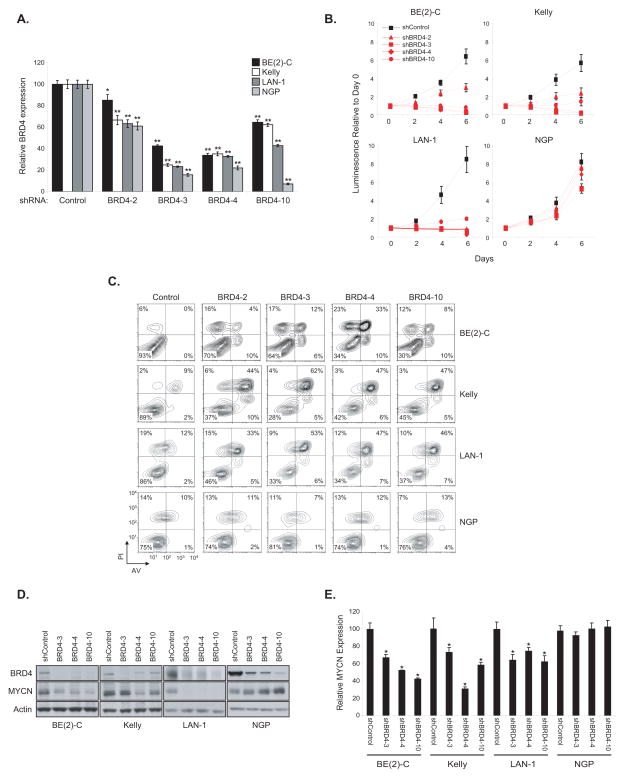
BRD4 Suppression by shRNA Recapitulates the Effects of JQ1 Treatment in Neuroblastoma Cell Lines
Previous studies have focused on the role of a specific BET family member, BRD4, which is potently inhibited by JQ1 and implicated in several malignancies, including NUT midline carcinoma, AML, B-cell acute lymphoblastic leukemia (B-ALL) and multiple myeloma (7–9). Moreover, a recent siRNA screen to identify synthetic lethal interactions with MYC overexpression revealed BRD4 as a candidate gene (23). Therefore, we studied selective suppression of BRD4 by shRNA in the three JQ1-sensitive neuroblastoma cell lines. Four shRNAs directed against BRD4 were evaluated and demonstrated to alter relative cell viability and growth over a six day time course, with a dose response in all three lines sensitive to JQ1 (Fig. 6A and B). As seen with JQ1 treatment, there was a striking induction of apoptosis (Fig. 6C) and downregulation of MYCN expression by RT-PCR and immunoblot with the top three shRNAs in the three JQ1 sensitive cell lines (Fig. 6D and E). Moreover, as expected, direct suppression of MYCN by shRNA resulted in growth arrest and apoptotic cell death (Supplementary Fig. S9A–C). However, in the JQ1 resistant neuroblastoma cell line NGP, BRD4-directed shRNAs did not alter growth/viability, induce apoptosis or lead to suppression of MYCN expression (Fig. 6A–E). Accordingly to these results, BRD4 knock-down did not significantly diminish growth of two MYCN non amplified cell lines, SH-SY5Y and SK-N-AS (Supplementary Fig. S10A–B).
Figure 6.
Effect of BRD4 downregulation on neuroblastoma cell lines. (A–E) The three most JQ1-sensitive neuroblastoma cell lines were transduced with shRNAs targeting BRD4 or a control shRNA. (A) RNA was extracted at day three post-transduction and BRD4 transcript level was quantified. Shown are the expression values relative to the shRNA control-transduced cells. Error bars represent mean +/− SD of four technical replicates. *P < 0.05 and **P < 0.01 calculated using a one-way ANOVA, with Bonferroni correction. (B) Viability values over days post-transduction are shown relative to the day zero (time of seeding) values, with errors bars representing the mean +/− SD of eight replicates per condition. (C) Apoptosis analysis was performed on day 6 post-transduction. (D) Western blots showing BRD4 and MYCN protein levels on day four post-transduction with three BRD4-directed shRNAs. (E) MYCN transcript levels were quantified four days after transduction. The expression values are relative to cells infected with a control shRNA. Error bars represents mean +/− SD of four technical replicates. *P < 0.001 calculated using a one-way ANOVA, with Bonferroni correction, comparing each BRD4-directed shRNA to the control shRNA within a cell line.
Efficacy of JQ1 Treatment in Independent Murine Models of Neuroblastoma
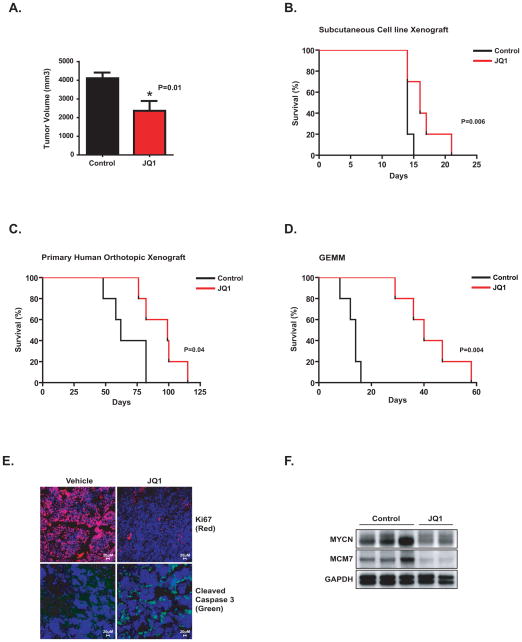
Because there is no consensus regarding the most relevant in vivo mouse model for preclinical testing of a new therapy, we performed in vivo testing of JQ1 in three unique models of MYCN-amplified neuroblastoma: a subcutaneous xenograft, an orthotopic primary human xenograft, and an experimentally-derived mouse model of MYCN-amplified neuroblastoma (24). First, JQ1 was evaluated in a BE(2)-C cell line xenograft model established by subcutaneous injection into NOD-SCID-IL2Rγnul (NSG) mice. Tumor-bearing mice were treated with 50 mg/kg of JQ1 or vehicle delivered by daily intraperitoneal injection (n = 10 per group) until sacrifice. JQ1 treatment significantly diminished tumor volume and prolonged overall survival compared to control-treated mice without an effect on body weight (Fig. 7A and B and Supplementary Fig. S11A) and no overt toxicities were observed. In contrast, treatment of the MYCN-wildtype SH-SY5Y subcutaneous xenograft with JQ1 did not significantly prolong survival or reduce tumor volume (Supplementary Fig. S11B and C).
Figure 7.
Testing of JQ1 in multiple in vivo models of MYCN-amplified neuroblastoma. (A) Mice were injected with BE(2)-C subcutaneously and treated with JQ1 or vehicle once tumors reached 100 mm3. After 15 days, tumor volume was measured. Error bars indicate mean +/− SD of five mice. *P = 0.01 calculated using non parametric Mann-Whitney test. (B) Effects of JQ1 treatment on survival in the BE(2)-C xenograft model. Day 0 indicates the first day of treatment and mice were treated until time of sacrifice. (C) A human MYCN-amplified primary neuroblastoma tumor was implanted into kidney capsule of nude mice and mice were treated once daily with JQ1 for 28 days, starting 7 days after orthotopic transplantation. (D) TH-MYCN mice with palpable tumors were treated with JQ1 or vehicle once daily for 28 days. Statistical significance (A–C) was determined by log-rank (Mantel-Cox) test for the survival curves as shown. Day 0 indicates the day of treatment initiation. (E) Staining for Ki-67 (red), cleaved caspase 3 (green) and DAPI (blue) in GEMM tumors treated with either vehicle or JQ1 as indicated. (F) Western blot indicating the expression of MYCN and MCM7 in GEMM tumors treated with either vehicle or JQ1 as indicated.
In the primary human orthotopic xenograft model, a human MYCN-amplified primary neuroblastoma tumor was obtained from a heavily-pretreated child with relapsed metastatic and drug-resistant disease. Tumor pieces were implanted into the kidney capsule of nude mice, and the mice were treated once daily with 50 mg/kg of JQ1 intraperitoneally for 28 days, starting seven days after orthotopic transplantation. As shown in Figure 7C, JQ1 treatment significantly prolonged survival compared to vehicle treatment.
Finally, we tested the effects of JQ1 therapy in a genetically engineered mouse model (GEMM) of MYCN-amplified neuroblastoma (TH-MYCN) (24). TH-MYCN mice with palpable tumors were treated with JQ1 or vehicle, as above, once daily for 28 days. A marked improvement in survival for JQ1-treated mice was observed (Fig. 7D). Tumor biopsies were obtained on therapy with JQ1 or vehicle and assessed for pharmacodynamic effects. As shown in Figure 7E, JQ1 conferred an inhibitory effect on cellular proliferation evidenced by Ki67 staining, an increase in apoptosis by cleaved caspase 3 staining, and an inhibitory effect on the expression of MYCN itself and its downstream target MCM7 (Fig. 7F).
Discussion
Neuroblastoma, a tumor arising from primitive neural crest cells in the sympathetic nervous system, is the most common extra-cranial solid tumor in children (25). While progress has been substantial for treating patients with low-risk disease, the majority of children with neuroblastoma present with high-risk disease, and many of these children will succumb to neuroblastoma despite intensive chemotherapy with autologous stem cell transplantation, surgery and radiation (26). New treatment approaches are clearly needed for these patients as the delivery of dose-intensified, cytotoxic chemotherapy is already maximized. The pathologic activation of MYCN plays a central role in high-risk neuroblastoma, with MYCN amplification identified in 25% of primary neuroblastoma tumors and nearly half of high-risk cases (26–29). Genomic studies of neuroblastoma tumors, including massively parallel sequencing, have failed to reveal other recurrent and imminently targetable molecular lesions, with the exception of mutations in anaplastic lymphoma receptor kinase (ALK) in neuroblastomas which often co-harbor MYCN amplification (20, 30–34). The central role of the MYCN oncogene in neuroblastoma tumorigenesis, and its limited expression in mature, normal tissues, make it a compelling target for drug development. While transcription factor oncoproteins have largely been refractory to conventional drug discovery approaches, the selective targeting of JQ1 in neuroblastoma raises the possibility of targeting MYCN by bromodomain inhibitors in this disease. Indeed, our data also supports the use of other BET bromodomain inhibitors, expanding the possibilities for future clinical testing.
While numerous examples now exist of genotypic predictors of chemical phenotypes, particularly for kinase inhibitors (35–37), we are still lacking genetic/genomic predictors of response to most drugs in current clinical use and in early clinical testing. In this study, we reveal the power of large-scale screening of highly annotated collections of cancer cell lines to uncover a strong genetic predictor of response to BET bromodomain inhibitors. Many of the cancer cell lines in our high-throughput screening study, particularly among the solid tumors, were insensitive to JQ1, suggesting that JQ1 is not a general cytotoxic agent. While nearly all of the prior literature supports the use of BET bromodomain inhibitors in hematologic malignancies, our work demonstrates a strong indication for the use of BET bromodomain inhibitors in a genetically defined solid tumor, MYCN-amplified neuroblastoma. Similar to the observation that MYC is transcriptionally regulated by bromodomain inhibitors, we demonstrated that MYCN expression is repressed both by JQ1 treatment and by shRNA directed against BRD4. Importantly, in the one MYCN amplified cell line (NGP) with no response to JQ1 or other structurally distinct BET bromodomain inhibitors, no downregulation of MYCN expression at either a transcriptional or protein level occurred with either JQ1 treatment or BRD4-directed shRNA. While the precise mechanism for the failure to suppress MYCN expression in NGP by BET bromodomain inhibition is still under investigation, these data support the assessment of MYCN downregulation as a putative approach to response stratification in clinical trials investigating this compound class. Furthermore, while in primary human neuroblastoma tumors, MYCN amplification is strongly correlated with the JQ1 expression signature derived from cell lines, there are some primary human tumors which cluster with the JQ1 signature but lack MYCN amplification, suggesting that additional predictors of response to JQ1 remain to be discovered.
In addition to identifying a consensus JQ1 gene expression signature shared by JQ1-responsive cell lines, we also identified a gene signature uniquely regulated in neuroblastoma, including genes associated with neural cellular identity. For example, one of the genes whose expression is uniquely modulated by BET bromodomain inhibition in neuroblastoma is PHOX2B, a gene encoding a transcription factor involved in the differentiation of neural crest cells. PHOX2B is mutated in neuroblastoma, particularly in association with familial neuroblastoma and with congenital central hypoventilation syndrome and Hirschprung’s disease (38–40). Similarly, tyrosine hydroxylase expression is uniquely modulated by BET bromodomain inhibition with JQ1 in neuroblastoma, a likely explanation for the downregulation of MYCN expression in our GEMM model of neuroblastoma in which MYCN is driven from a tyrosine hydroxylase promoter. The even more dramatic effects of JQ1 on the downstream target of MYCN, MCM7, could be explained by the contribution of a direct inhibitory effect of JQ1 on the interaction of BRD4 and MYCN at MYCN promoter sites.
While it will be important in future studies to extend testing of BET bromodomain inhibition to other diseases with MYCN amplification, such as medulloblastoma (41), the open-source nature of publicly available screening data infinitely expands the utility of any individual laboratory’s screening efforts by enabling re-mining of the data. For example, if we relax the criteria for identifying candidate biomarkers of sensitivity versus resistance to JQ1 to significant (P ≤ 0.05) values for Fisher’s exact test at the twenty-fifth percentile (sensitivity) or the seventy fifth percentile (resistance) of the Ln(IC50) distribution, NOTCH1 activation is also correlated with response to JQ1, nominating diseases with aberrant NOTCH1 activation, such as T-cell acute lymphoblastic leukemia (T-ALL) (42) and chronic lymphocytic leukemia (CLL) (43), for future study (Supplementary Table S8). Similarly, expression of GNAS, MDM2, and NF2 should be explored as predictors of resistance and the combination of MDM2 inhibitors with bromodomain inhibitors tested. Moreover, the recent report that ALK(F1174L)/MYCN tumors exhibit increased MYCN dosage due to ALK(F1174L)-induced activation of the PI3K/AKT/mTOR and MAPK pathways suggests the possibility of combining ALK inhibitors with BET bromodomain inhibitors in neuroblastoma (44).
In summary, we have leveraged high-throughput, cell-based screening of genetically characterized cancer cell lines to identify MYCN amplification as a strong predictor of sensitivity to BET bromodomain inhibition. We have confirmed this finding in MYCN-amplified neuroblastoma and executed on the preclinical validation of this hypothesis. With a survival advantage observed in three independent in vivo models of neuroblastoma treated with JQ1, including a highly chemotherapy-resistant genetically engineered MYCN mouse model, the clinical development of bromodomain inhibitors for testing in children with relapsed/refractory neuroblastoma is already underway.
Methods
Genomic Characterization of Cancer Cell Lines
A total of 84 of the most frequently mutated, amplified, deleted or rearranged cancer genes were examined in the cell line collection. 65 genes were sequenced to base-pair resolution across all coding exons for each gene by capillary sequencing. The presence of seven of the most commonly rearranged cancer genes (e.g. BCR-ABL and EWS-FLI1) was determined across the drug screen cell line panel by the design of breakpoint-specific sequence primers that enabled the detection of the rearrangement following capillary sequencing. In addition, analysis of microsatellite instability (MSI) was carried out according to the guidelines set down by “The International Workshop on Microsatellite Instability and RER Phenotypes in Cancer Detection and Familial Predisposition” workshop (45). Samples were screened using the markers BAT25, BAT26, D5S346, D2S123 and D17S250 and were characterized as MSI if two or more markers showed instability. Total integral copy number values across the footprints of the cancer genes were determined from Affymetrix SNP6.0 microarray data using the ‘PICNIC’ algorithm to predict copy number segments in each of the cell lines (46). For a gene to be classified as amplified, the entire coding sequence must be contained in one contiguous segment defined by PICNIC, and have a total copy number of eight or more. Deletions must occur within a single contiguous segment with copy number zero. A complete description of the characterization of the cancer cell lines collection used in this study is available from the Cancer Genome Project webpages (47).
Cell Line Screening
Cell lines in an exponential phase of growth were seeded at optimal density previously determined to provide maximum signal in control (untreated) wells, while maintaining growth through the experimental exposure. Cells were seeded on Day 1, with drug delivered the following day. Nine concentrations of (+)-JQ1 were applied to cells at a step-wise two-fold dilution series. After 72 hours of drug exposure, cells were fixed and stained with Syto60 (Invitrogen) for fluorescence measurement as described previously (48). Replicate experiments were variably performed for quality assessment during the screening phase. Stringent quality control criteria were applied and estimates of IC50 were derived from the 9-dose response curves as described previously (16). Drug response is presented as the LN[IC50] plotted against the Emax (the maximum effect corresponding to the minimum measured viability). Enrichment for a given organ of origin (673 cell lines) or genotype (473 cell lines with genotype information) in the sensitive cell lines was tested using a Fisher exact test based approach. Cell lines with response to JQ1 yielding IC50 ≤ 1 μM and Emax ≥ 70% were designated as sensitive and all other were designated as resistant in the most stringent classification schema. Using less stringent criteria, cell lines were assigned to the sensitive or resistant group thresholds corresponding to the twenty-fifth to the seventy-fifth percentile of the distribution of LN(IC50) values. A Fisher exact test was used to determine the statistical enrichment of a given genotype (or organ) in the sensitive or resistant group at each threshold. In this analysis mutation is defined as either the presence of a specific gene fusion, a sequence change or a copy number change across 84 cancer genes.
Cell Culture
Neuroblastoma cell lines were maintained in DMEM (Cellgro, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin-streptomycin with glutamine (Cellgro). BE(2)-C and SH-SY5Y were obtained from the American Type Culture Collection, and Kelly, LAN-1, SK-N-AS and NGP were kindly provided by Dr. Rani George. The identity of all lines was verified by small tandem repeat profiling performed by the Shannon McCormack Advanced Molecular Diagnostics Laboratory at the Dana-Farber Cancer Institute (Boston, MA, USA).
Compounds, Cellular Viability, and Growth
JQ1R, JQ1S, I-BET and I-BET151 were synthesized by Dr. Jun Qi. For the testing of multiple BET bromodomain inhibitors in four neuroblastoma cell lines, cells were seeded onto 384-well tissue culture-treated plates at a density of 2000 cells/well in a volume of 50 μL/well. Addition of inhibitor was performed with a JANUS Workstation (Perkin Elmer, Waltham, MA) using a 384-well pinhead tool that is calibrated to deliver 100 nL drug/well. After 72 hours of incubation with inhibitor, cells were analyzed for cell viability using the ATPlite (Perkin Elmer, Waltham MA) luminescent assay kit per the manufacturer’s instructions using ATP content as a surrogate for viable cell number. Luminescence was read on an EnVision 2104 Multilabel Plate Reader (Perkin Elmer, Waltham, MA). Nonlinear dose response curves were fitted to the data using Graphpad Prism software (La Jolla, CA). To assess growth, neuroblastoma cell lines were incubated with JQ1 in 384-well plates. ATP content was measured on days zero, two, three, and five using CellTiter Glo (Promega, Madison, WI, USA) per the manufacturer’s instructions.
Flow Cytometry
Cell cycle was determined by measuring DNA content using propidium iodide staining. Apoptosis was measured using the Annexin V: FITC Apoptosis Detection Kit as per the manufacturer’s protocol (BD Pharmingen, San Jose, CA, USA).
Protein Extraction and Immunoblotting
Protein was extracted from cells by lysis with Cell Signaling Lysis Buffer (Cell Signaling Technology, Danvers, MA, USA) containing Complete, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany) and PhosSTOP Phosphatase Inhibitor (Roche Diagnostics). Immunoblots were run as previously described (49). Primary antibodies included anti-MYCN (Abcam 16898), anti-BRD4 (Abcam 75898), anti-Vinculin (Abcam 18058), anti-MCM7 (Cell Signaling Technology #4018S), anti-MDM2 (Santa Cruz Biotechnology sc-813) and anti-Actin (Neomarkers MS1295P).
RNA Extraction and Real-time RT-PCR
RNA was extracted from cells with the RNeasy Kit and on-column DNA digestion (Qiagen, Valencia, CA, USA). Primers and probes for MYCN (Hs00232074_m1), BRD4 (Hs04188087_m1), PHOX2B (Hs00243679_m1) and the control gene RPL13A (Hs01926559_g1) were obtained from Applied Biosystems (Carlsbad, CA, USA). Data were collected in technical quadruplicate, analyzed using the ΔΔCT method, and plotted as percentage of transcript compared to the negative control condition.
shRNA Studies
Virus was created by transfecting 293T cells with packaging plasmid (pCMV-d8.9), envelope plasmid (VSV-G), and a pLK0.1 hairpin plasmid (see Supplementary Table S9 for hairpin target sequences) using Fugene 6 per the manufacturer’s instructions (Promega). It was filtered with a 0.4 μM filter before use. Neuroblastoma cell lines were transduced in 10 cm plates with 2 mL virus and 8 μg/ml polybrene (Sigma-Aldrich) for 2 hours at 37°C, and then 8 mL medium was added to each plate. On day two post-transduction, 2 μg/mL (BE(2)-C) or 1 μg/ml (all other cell lines) puromycin (Sigma-Aldrich) was added to select for infected cells and four days post-transduction, experiments to evaluate cell growth, apoptosis, and protein and transcript levels were begun.
Genome-wide Expression Analysis
BE(2)-C and Kelly cells were treated in triplicate with 1 μM JQ1 or DMSO for 24 hours. RNA was extracted and a decrease in MYCN transcript was confirmed by real time RT-PCR as described above. The samples were profiled using the Affymetrix PrimeView Human Gene Expression Array (Affymetrix) at Beth Israel Deaconess Medical Center (Boston, MA, USA). The computational analysis pipeline was performed on the Genome Space bioinformatics platform (50).
Data Processing
The CEL data files were subjected to quality control and passed tests based on distance between arrays, array intensity distribution and variance mean dependence which are implemented in the ArrayQuality R package (Bioconductor). The raw expression data was processed and log2 transformed by applying the RMA algorithm in the Affymetrix package (Bioconductor). All probe sets with an average log2 expression below four were considered under-expressed and were designated as “filtered”. Of the 49,293 probe sets on the Affymetrix PrimeView Human Gene Expression Array, 45,925 probe sets remained after filtering. The data from the 45,925 probe sets was further collapsed to 19,108 non-redundant genes with distinct HUGO symbols, by assigning to each gene the probe set with the maximum average expression intensity. Gene expression data is available from Gene Expression Omnibus Expression (GEO) (accession no. GSE43392).
Comparative Marker Analysis
The 12 samples available in the data were separated into two groups: 1 μM JQ1-treated samples versus DMSO-treated samples. The Comparative Marker Selection module from GenePattern 3.5.0 (50) was employed to identify individual genes differentially expressed between the JQ1- and the DMSO-treated groups (51). A 2-sided signal-to-noise ratio (SNR) test followed by 1000 permutations of phenotype labels was performed for this analysis. The settings for the SNR parameters were log-transformed-data:yes, complete:no, balanced:no, smooth p-values:yes. A permutation P ≤ 0.05, a Benjamini-Hochberg false discovery rate (FDR) ≤ 0.05, accounting for multiple hypothesis testing, and an absolute fold change (FC) ≥ 2 served as a cutoff for significant genes. Heatmaps were created with GENE-E software (50).
Gene Set Enrichment Analysis
The GSEA software (50) was utilized to identify pathways, or groups of functionally related genes, deregulated by the JQ1 treatment (18). The goal of GSEA is to identify groups of genes sharing common biological function (genesets), which are distributed at the top or at the bottom of the ranked list of differentially expressed genes. GSEA assigns to each gene set an enrichment score (ES) calculated as a running sum statistic by walking down the ranked list of differentially expressed genes, increasing the sum when encountering genes in the gene set and decreasing it when encountering genes not in the gene set. The significance of the ES is estimated based on a permutation p-value and adjusted for multiple hypotheses testing through FDR. The set of differentially expressed genes accounting for the enrichment signal is called the leading edge.
GSEA was run on the collections of 3272 curated gene sets (c2) and 615 transcription factor targets (c3) from version 3.1 of the Molecular Signatures Database (50). Gene sets with less than 15 genes or more than 500 genes were excluded from the analysis. Gene sets with an FDR ≤ 0.25 and a nominal P ≤ 0.05 were considered significant. The gene ranking metric in the weighted ES was the 2-sided SNR, and the p-values were calculated using 1,000 permutations of the phenotype.
MYCN Amplification Signature
A gene expression upregulation signature to distinguish primary neuroblastoma tumors with MYCN amplification from those without MYCN amplification was created from the expression profiling of primary neuroblastoma tumors (GSE12460, available at the GEO Database) (20). The signature was generated based on the Comparative Marker Selection algorithm of GenePattern with the cutoff threshold 2 for the absolute fold change ratio and the cutoff 0.05 for the p-value and the FDR.
A Consensus JQ1 Signature
A consensus JQ1 signature was created by intersecting the JQ1 differential expression signature in this study with those from the studies by Delmore et al. in multiple myeloma and Zuber et al. in AML (8, 9). This consensus JQ1 signature consisted of 36 genes significantly downregulated and 17 genes significantly upregulated based on the cutoffs of an absolute FC ≥ 2, and p-value and FDR ≤ 0.05. The consensus JQ1 signature was evaluated on the primary neuroblastoma tumor data of Janoueix-Lerosey et al. (20) describing stage and MYCN amplification status. The primary tumor data were projected onto the consensus JQ1 signature and clustered based on the average linkage hierarchical method. The JQ1 consensus signature separated the tumors into two clusters. The association of the tumor clusters with the MYCN status and stage was computed based on the two tailed exact Fisher test.
A relaxed consensus JQ1 signature for downregulated genes identified based on the absolute FC ≥1.5 and a p-value and FDR ≤ 0.05 was used in a Functional Enrichment Analysis of the consensus JQ1 signature by GSEA. Functional enrichment analysis results were visualized with Enrichment Map software and functional clusters are listed in Supplementary Table S7.
Neuroblastoma JQ1-specific Signature
A collection of 116 genes significantly downregulated (absolute FC ≥ 2, and p-value and FDR ≤ 0.05) in the JQ1-treated neuroblastoma cell lines vs. control, and unchanged in any of the other two JQ1-treated studies, was identified. The small size of the neuroblastoma JQ1 specific signature, even in a relaxed format, necessitated the use of a Fisher test rather than GSEA to identify the functional relatedness of these genes.
Chromatin Immunoprecipitation (ChIP)
ChIP studies were carried out as described previously using specific primers for the MYCN locus (8). Primers were designed to amplify two sites within the promoter region (5′-3′) and a negative control region outside the promoter region: MYCN promoter site 1 (forward) TTTGCACCTTCGGACTACCC and (reverse) TTTGACTGCGTGTTGTGCAG; MYCN promoter site 2 (forward) TCCTGGGAACTGTGTTGGAG and (reverse) TCCTCGGATGGCTACAGTCT; MYCN negative region (forward) GTATCACCGTCCATTCCCCG and (reverse) TTGGAGGCAGCTCAAAGACC. Enrichment data were analyzed by calculating the immunoprecipitated DNA percentage of input DNA for each sample (triplicate PCR reactions).
In Vivo Studies
Cell line xenograft tumors were established by implanting 2 million BE(2)-C-LucNeo cells or 2 million SH-SY5Y cells in 30% Matrigel (BD Biosciences) subcutaneously in the flanks of 6 week old female NOD-SCID-IL2Rcγnull (NSG) mice (Jackson Labs). Serial bioluminescence imaging was performed after injection with 75 mg/kg of D-luciferin (Promega) using a Xenogen IVIS Spectrum instrument (PerkinElmer). Eight days after tumor inoculation, mice with established tumors defined as increasing bioluminescence and measurable tumor volume were divided into cohorts to be treated with 50 mg/kg JQ1 or vehicle (10% DMSO in 5% dextrose) delivered by daily intraperitoneal injection (n=10 per group). Because of rapid tumor growth and saturation, bioluminescence imaging was abandoned as an endpoint. Tumor volume was determined using the equation volume = 0.5 × length × width2. Mice were sacrificed when tumors reached 2 cm in the longest linear dimension. Statistical significance was determined by log-rank (Mantel-Cox) test for survival curves. This study was conducted using protocols approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
TH-MYCN mice with palpable tumors (mean age at detection was 60 +/−15d) were treated with JQ1 via intraperitoneal injection (50 mg/kg in 1:10 solution of DMSO:10% cyclodextrin) or vehicle(1:10 solution of DMSO:10% cyclodextrin), once daily for 28 days (n=5 per group). In the primary human orthotopic model, a human MYCN-amplified primary neuroblastoma tumor was obtained from a child with relapsed, drug-resistant metastatic disease (SFNB-06). Tumor pieces (4mm3) were implanted into kidney capsule of nude mice and maintained as a continuous xenograft. Human primary SFNB-06 mice were treated once daily for 28 days, starting 7 days after orthotopic transplantation, (dosage as above) (n=5 per group). Statistical significance was determined by log-rank (Mantel-Cox) test for survival curves. All mice were monitored until euthanasia was required, in accordance with the University of California, San Francisco, institutional IACUC guidelines.
Supplementary Material
Significance.
Biomarkers of response to small-molecule inhibitors of BET bromodomains, a new compound class with promising anti-cancer activity, have been lacking. Here, we reveal MYCN amplification as a strong genetic predictor of sensitivity to BET bromodomain inhibitors, demonstrate a mechanistic rationale for this finding, and provide a translational framework for clinical trial development of BET bromodomain inhibitors for pediatric patients with MYCN-amplified neuroblastoma.
Acknowledgments
We thank Dr. Rani George for providing cell lines and Ronald Paranal for technical support.
Grant Support: V Foundation for Cancer Research Translational Grant (KS and JEB), Friends for Life Fellowship (KS and ALK), Howard Hughes Medical Institute Physician-Scientist Early Career Award (KS), Damon-Runyon Cancer Research Foundation and Smith Family Foundation (JEB and JQ), Alex Lemonade Stand (WCG), NIH grants R01CA102321 (WAW) P01CA081403 (WAW), K08NS079485 (WCG), and T32CA151022 (YHC); The Wellcome Trust (086357; UM, MJG, PG, CHB); the Katie Dougherty Foundation (WAW), The Cancer League, Inc (WAW), and the Samuel Waxman Cancer Research Foundation (JEB and WAW), L’Association pour la Recherche sur le Cancer (AP). KS is also supported by a Stand-Up-to-Cancer Innovative Research Grant, a program of the Entertainment Industry Foundation (SU2C-AACR-IRG0509).
Footnotes
Disclosures: JEB and the Dana-Farber Cancer Institute established Tensha Therapeutics to translate drug-like inhibitors of BET bromodomains as cancer therapeutic agents. All other authors have no relevant, potential conflicts-of-interest to report.
References
- 1.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marushige K. Activation of chromatin by acetylation of histone side chains. Proc Natl Acad Sci U S A. 1976;73:3937–41. doi: 10.1073/pnas.73.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS letters. 2002;513:124–8. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 5.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Molecular biology of the cell. 2009;20:4899–909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;471:235–9. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-MYC and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012 doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–7. [PubMed] [Google Scholar]
- 14.Weinstein JN, Myers TG, O’Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–9. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 15.McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–95. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 16.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–8. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 20.Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 21.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- 23.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109:9545–50. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Cancer Institute. Surveillance Epidemiology and End Results (SEER) Database. 2008 Available at hhtp://seercancergov.
- 26.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–8. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 28.Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–6. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 29.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 30.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 31.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–71. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.George RE, Sanda T, Hanna M, Frohling S, Luther W, 2nd, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 35.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 36.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trochet D, Bourdeaut F, Janoueix-Lerosey I, Deville A, de Pontual L, Schleiermacher G, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am J Hum Genet. 2004;74:761–4. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raabe EH, Laudenslager M, Winter C, Wasserman N, Cole K, LaQuaglia M, et al. Prevalence and functional consequence of PHOX2B mutations in neuroblastoma. Oncogene. 2008;27:469–76. doi: 10.1038/sj.onc.1210659. [DOI] [PubMed] [Google Scholar]
- 40.van Limpt V, Schramm A, van Lakeman A, Sluis P, Chan A, van Noesel M, et al. The Phox2B homeobox gene is mutated in sporadic neuroblastomas. Oncogene. 2004;23:9280–8. doi: 10.1038/sj.onc.1208157. [DOI] [PubMed] [Google Scholar]
- 41.Korshunov A, Remke M, Werft W, Benner A, Ryzhova M, Witt H, et al. Adult and pediatric medulloblastomas are genetically distinct and require different algorithms for molecular risk stratification. J Clin Oncol. 2010;28:3054–60. doi: 10.1200/JCO.2009.25.7121. [DOI] [PubMed] [Google Scholar]
- 42.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 43.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berry T, Luther W, Bhatnagar N, Jamin Y, Poon E, Sanda T, et al. The ALK(F1174L) Mutation Potentiates the Oncogenic Activity of MYCN in Neuroblastoma. Cancer Cell. 2012;22:117–30. doi: 10.1016/j.ccr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 46.Greenman CD, Bignell G, Butler A, Edkins S, Hinton J, Beare D, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics. 2010;11:164–75. doi: 10.1093/biostatistics/kxp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Characterization of the cancer cell lines collection. available from the Cancer Genome Project webpages http://www.sanger.ac.uk/genetics/CGP/CellLines/
- 48.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, Hahn CK, et al. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. J Clin Invest. 2012;122:935–47. doi: 10.1172/JCI46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.GENE-E version 1, GenePattern version 3.5.0, GSEA version 13, Cytoscape version 2.8.3 and MSigDB version 3.1. Available from http://www.genomespace.org.
- 51.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.