Abstract
The shift to digital systems for the creation, transmission and storage of information has led to increasing complexity in archiving, requiring active, ongoing maintenance of the digital media. DNA is an attractive target for information storage1 because of its capacity for high density information encoding, longevity under easily-achieved conditions2–4 and proven track record as an information bearer. Previous DNA-based information storage approaches have encoded only trivial amounts of information5–7 or were not amenable to scaling-up8, and used no robust error-correction and lacked examination of their cost-efficiency for large-scale information archival9. Here we describe a scalable method that can reliably store more information than has been handled before. We encoded computer files totalling 739 kB of hard disk storage and with an estimated Shannon information10 of 5.2 × 106 bits into a DNA code, synthesised this DNA, sequenced it and reconstructed the original files with 100% accuracy. Theoretical analysis indicates that our DNA-storage scheme scales far beyond current global information volumes. These results demonstrate DNA-storage to be a realistic technology for large-scale digital archiving that may already be cost-effective for low access, multi-century-long archiving tasks. Within a decade, as costs fall rapidly under realistic scenarios for technological advances, it may be cost-effective for sub-50-year archival.
Although techniques for manipulating, storing and copying large amounts of DNA have been established for many years11–13, these rely on the availability of initial copies of the DNA molecule to be processed, and one of the main challenges for practical information storage in DNA is the difficulty of synthesising long sequences of DNA de novo to an exactly-specified design. Instead, we developed an in vitro approach that represents the information being stored as a hypothetical long DNA molecule and encodes this using shorter DNA fragments. A similar approach was proposed by Church et al.9 in a report submitted and published while this manuscript was in review. Isolated DNA fragments are easily manipulated in vitro11,13, and the routine recovery of intact fragments from samples that are tens of thousands of years old14,15 indicates that a well-prepared synthetic DNA sample should have an exceptionally long lifespan in low-maintenance environments3,4. In contrast, systems based on living vectors6–8 would not be reliable, scalable or cost-efficient, having disadvantages including constraints on the genomic elements and locations that can be manipulated without affecting viability, the fact that mutation will cause the fidelity of stored and decoded information to reduce over time and possibly the requirement for storage conditions to be carefully regulated. Existing schemes in the field of DNA computing in principle permit large-scale memory1,16, but data encoding in DNA computing is inextricably linked to the specific application or algorithm17 and no practical schemes have been realised.
We selected computer files to be encoded as a proof of concept for practical DNA-storage, choosing a range of common formats to emphasise the ability to store arbitrary digital information. The five files comprised all 154 of Shakespeare’s sonnets (ASCII text), a classic scientific paper18 (PDF format), a medium-resolution colour photograph of the European Bioinformatics Institute (JPEG 2000 format), a 26 s excerpt from Martin Luther King’s 1963 “I Have A Dream” speech (MP3 format) and a Huffman code10 used in this study to convert bytes to base-3 digits (ASCII text), giving a total of 757,051 bytes (Shannon information10 5.2 × 106 bits). Full details are given in Supplementary Information and Supplementary Table S1.
The bytes comprising each file were represented as single DNA sequences with no homopolymers (runs of ≥ 2 identical bases, which are associated with higher error rates in existing high-throughput sequencing technologies19 and led to errors in Church et al.’s experiment9). Each DNA sequence was split into overlapping segments, generating fourfold redundancy, and alternate segments were converted to their reverse complement (see Fig. 1 and Supplementary Information). These measures reduce the probability of systematic failure for any particular string, which could lead to uncorrectable errors and data loss. Each segment was then augmented with indexing information that permitted determination of the file from which it originated and its location within that file, and simple parity-check error-detection10. In all, the five files were represented by a total of 153,335 strings of DNA, each comprising 117 nt. An additional advantage of our encoding scheme is that the perfectly uniform fragment lengths and absence of homopolymers make it obvious that the synthesised DNA does not have a natural (biological) origin, and so imply the presence of deliberate design and encoded information2.
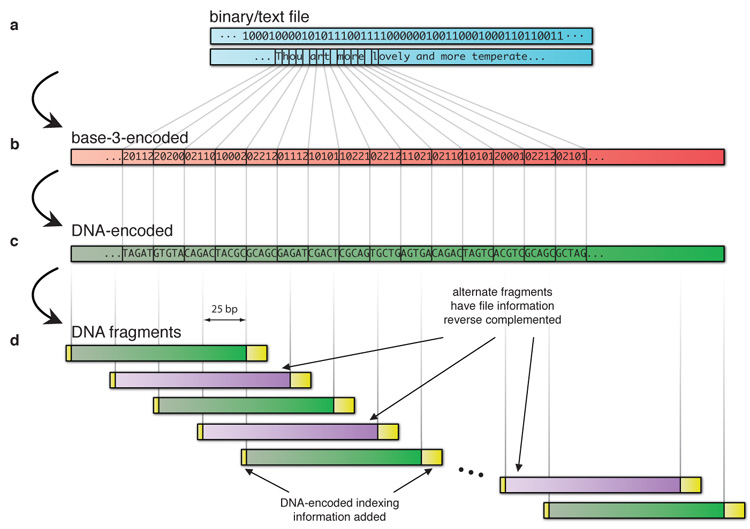
Figure 1. Digital information encoding in DNA.
Digital information (a, in blue), here binary digits holding the ASCII codes for part of Shakespeare’s sonnet 18, was converted to base-3 (b, red) using a Huffman code that replaces each byte with five or six base-3 digits (trits). This in turn was converted in silico to our DNA code (c, green) by replacement of each trit with one of the three nucleotides different from the previous one used, ensuring no homopolymers were generated. This formed the basis for a large number of overlapping segments of length 100 bases with overlap of 75 bases, creating fourfold redundancy (d, green and, with alternate segments reverse complemented for added data security, violet). Indexing DNA codes were added (yellow), also encoded as non-repeating DNA nucleotides. See Supplementary Information for further details.
Oligonucleotides (oligos) corresponding to our designed DNA strings were synthesised using an updated version of Agilent Technologies’ OLS (oligo library synthesis) process20. This created a large number (~2.5 × 106) of copies of each DNA string, with errors occurring only rarely (~1 error per 500 bases) and independently in the different copies of each string, again enhancing our method’s error tolerance. The synthesised DNA was supplied lyophilised, a form expected to have excellent long-term preservation characteristics3,4, and was shipped (at ambient temperature, without specialised packaging) from the USA to Germany via the UK. After resuspension, amplification and purification, a sample of the resulting library products was sequenced in paired-end mode on the Illumina HiSeq 2000. The remainder of the library was transferred to multiple aliquots and re-lyophilised for long-term storage.
Base calling using AYB21 yielded 79.6M read-pairs of 104 bases in length. Full-length (117 nt) DNA strings were reconstructed in silico from the read-pairs, with those containing uncertainties due to synthesis or sequencing errors being discarded. The remaining strings were then decoded using the reverse of the encoding procedure, with the error-detection bases and properties of the coding scheme allowing us to discard further strings containing errors. While many discarded strings will have contained information that could be recovered with more sophisticated decoding, the high level of redundancy and sequencing coverage rendered this unnecessary in our experiment. Full-length DNA sequences representing the original encoded files were then reconstructed in silico. The decoding process used no additional information derived from knowledge of the experimental design. Full details of the encoding, sequencing and decoding processes are given in Supplementary Information.
Four of the five resulting DNA sequences could be fully decoded without intervention. The fifth however contained two gaps: runs of 25 bases each for which no segment was detected corresponding to the original DNA. Each of these gaps was caused by the failure to sequence any oligo representing any of four consecutive overlapping segments. Inspection of the neighbouring regions of the reconstructed sequence permitted us to hypothesise what the missing nucleotides should have been (see Supplementary Information) and we manually inserted those 50 bases accordingly. This sequence could also then be decoded. Inspection confirmed that our original computer files had been reconstructed with 100% accuracy.
To investigate its suitability for long-term digital archiving, we studied how DNA-storage scales to larger applications. The number of bases of synthesised DNA needed to encode information grows linearly with the amount of information to be stored, but we must also consider the indexing information required to reconstruct full-length files from short fragments. As indexing information grows only as the logarithm of the number of fragments to be indexed, the total amount of synthesised DNA required grows sub-linearly. Increasingly-large parts of each fragment are needed for indexing however and, although it is reasonable to expect synthesis of longer strings to be possible in future, we modelled the behaviour of our scheme under the conservative constraint of a constant 114 nt available for both data and indexing information (see Supplementary Information). As the total amount of information increases, the encoding efficiency decreases only slowly (Fig. 2a). In our experiment (megabyte scale) the encoding scheme is 88% efficient; Fig. 2a indicates that efficiency remains > 70% for data storage on petabyte scales and > 65% on exabyte scales, and that DNA-storage remains feasible on scales many orders of magnitude greater than current global data volumes22. Fig. 2a also shows that costs (per unit information stored) rise only slowly as data volumes increase over many orders of magnitude. Efficiency and costs scale even more favourably if we consider the synthesised fragment lengths available using the latest technology (Supplementary Fig. S5).
Figure 2. Scaling properties and robustness of DNA-storage.
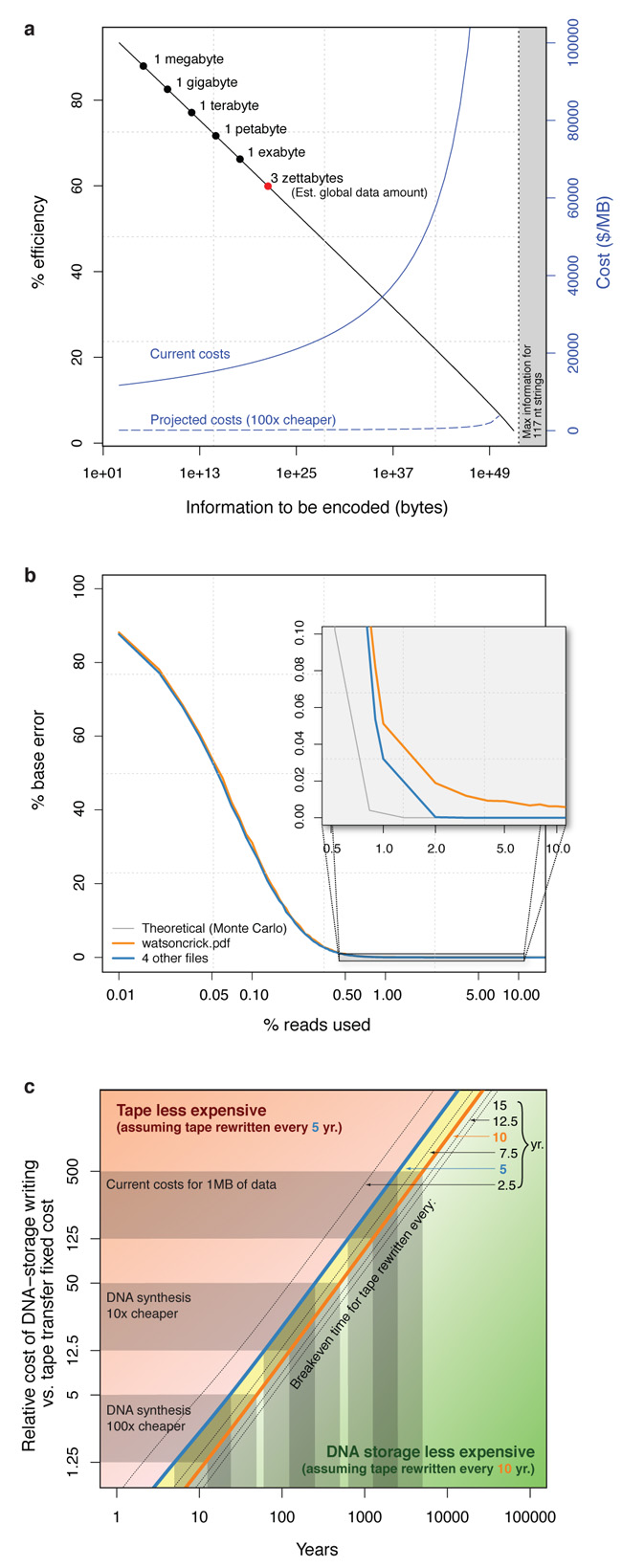
a, Encoding efficiency and costs change as the amount of stored information increases. The x-axis (logarithmic scale) represents the total amount of information to be encoded. Common data scales are indicated, including the 3 ZB (3 × 1021 bytes) global data estimate. The black line (y-axis scale to left) indicates encoding efficiency, measured as the proportion of synthesised bases available for data encoding. The blue curves (y-axis scale to right) indicate the corresponding effect on encoding costs, both at current synthesis cost levels (solid line) and in the case of a two-order of magnitude reduction (dashed line). b, Per-recovered-base error rate (y-axis) as a function of sequencing coverage, represented by the percentage of the original 79.6M read-pairs sampled (x-axis; logarithmic scale). The blue curve represents the four files recovered without human intervention: the error is zero when 2% of the original reads are used. The grey curve is derived from our theoretical error rate model. The orange curve represents the file that required manual correction: the minimum possible error rate is 0.0036%. c, Timescales for which DNA-storage is cost-effective. The blue curve indicates the relationship between break-even time beyond which DNA-storage is less expensive than magnetic tape (x-axis) and relative cost of DNA-storage synthesis and tape transfer fixed costs (y-axis), assuming the tape archive has to be read and re-written every 5 years. The orange curve corresponds to tape transfers every 10 years; broken curves correspond to other transfer periods as indicated. In the green-shaded region, DNA-storage is cost-effective when transfers occur more frequently than every 10 years; in the yellow-shaded region, DNA-storage is cost-effective when transfers occur from 5- to 10-yearly; in the red-shaded region tape is less expensive when transfers occur less frequently than every 5 years. Highlighted ranges of relative costs of DNA synthesis to tape transfer are 125–500 (current costs for 1 MB of data), 12.5–50 (achieved if DNA synthesis costs reduce by one order of magnitude) and 1.25–5 (costs reduced by two orders of magnitude). Note the logarithmic scales on both axes. See Supplementary Information for further details.
As the amount of information stored increases, decoding requires more strings to be sequenced. A fixed decoding expenditure per byte of encoded information would mean that each base is read fewer times and so is more likely to suffer decoding error. We studied this effect by extending our scaling analysis to model the influence of reduced sequencing coverage on the per-decoded-base error rate (see Supplementary Information), and found that error rates increase only very slowly as the amount of information encoded increases to a global data scale and beyond (Supplementary Table S4). This also suggests that our mean sequencing coverage of 1,308× was considerably in excess of that needed for reliable decoding. We confirmed this by subsampling from the 79.6M read-pairs to simulate experiments with lower coverage. Fig. 2b indicates that reducing the coverage by a factor of 10 (or even more) would have led to unaltered decoding characteristics. These results further illustrate the robustness of our DNA-storage method.
DNA-storage might already be economically viable for long-horizon archives with a low expectation of extensive access. Applications include government and historical records23,24 and, in a scientific context, CERN’s CASTOR system25, which stores a total of 80 PB of LHC data and grows at 15 PB/year. Only 10% is maintained on disk, and CASTOR migrates regularly between magnetic tape formats. Archival of older data is needed for potential future verification of events, but access rates decrease considerably 2–3 years after collection. Further examples are found in astronomy, medicine and interplanetary exploration26. With optimised use of the technologies we employed, we estimate current costs to be $12,400/MB for information storage in DNA and $220/MB for information decoding. Computational costs are negligible. Although the latency of DNA-storage writing and reading is high, both processes can be accelerated through parallelisation (Supplementary Information). Modelling relative long-term costs of archiving using DNA-storage or magnetic tape shows that the key parameters are the ratio of the one-time cost of synthesising the DNA to the recurrent fixed cost of transitioning between tape technologies or media, which we estimate to be 125–500 currently, and the frequency of tape transition events (Supplementary Information and Supplementary Fig. S7). We find that with current technology and our encoding scheme, DNA-storage may be cost-effective for archives of several megabytes with a ~600–5,000-year horizon (Fig. 2c). One order of magnitude reduction in synthesis costs reduces this to ~50–500 years; with two orders of magnitude reduction, as can be expected in less than a decade if current trends continue13,27, DNA-storage becomes practical for sub-50-year archives.
The DNA-storage medium has different properties from traditional tape- or disk-based storage. As the basis of life on Earth, methods for manipulation, storage and reading of DNA will remain the subject of continual technological innovation. A large-scale DNA-storage archive would need stable DNA management28 and physical indexing of depositions, but whereas current digital schemes for archiving require active, ongoing maintenance and regular transitioning between storage media, the DNA-storage medium requires no active maintenance other than a cold, dry and dark environment3,4 (such as the Global Crop Diversity Trust’s Svalbard Global Seed Vault, which has no permanent on-site staff29) and remains viable for thousands of years even by conservative estimates. We achieved an information storage density of ~2.2 PB/g (Supplementary Information). Our sequencing protocol consumed just 10% of the library produced from the synthesised DNA (Supplementary Table S2), already leaving enough for multiple equivalent copies. Existing technologies for copying DNA are highly efficient11,13, meaning that DNA-storage is an excellent medium for the creation of copies of any archive for transportation, sharing or security. Overall, DNA-storage has potential as a practical solution to the digital archiving problem and may become a cost-effective solution for rarely accessed archives.
Supplementary Material
Acknowledgements
At the University of Cambridge: David MacKay and Graeme Mitchison for advice on codes for runlength-limited channels. At CERN: Bob Jones for discussions on data archival. At EBI: Ari Löytynoja for custom multiple sequence alignment software, Hazel Marsden for computing base calls and for detecting an error in the original parity-check encoding, Tim Massingham for computing base calls and advice on code theory and Kevin Gori, Daniel Henk, Remco Loos, Sarah Parks and Roland Schwarz for assistance with revisions to the ms. In the Genomics Core Facility at EMBL-Heidelberg: Vladimir Benes for advice on NGS protocols, Dinko Pavlinić for sequencing and Jonathon Blake for data handling. C.D. is supported by a fellowship from the Swiss National Science Foundation (grant 136461). B.S. is supported by a European Molecular Biology Laboratory Interdisciplinary Postdoc under Marie Curie Actions (COFUND).
Footnotes
Supplementary Information is provided as a number of separate files accompanying this document.
Author Contributions N.G. and E.B. conceived and planned the project and devised the information encoding methods. P.B. advised on NGS protocols, prepared the DNA library and managed the sequencing process. S.C. and E.M.L. provided custom oligonucleotides. N.G. wrote the software for encoding and decoding information into/from DNA and analysed the data. N.G., E.B., C.D. and B.S. modelled the scaling properties of DNA-storage. N.G. wrote the paper with discussions and contributions from all other authors. N.G. and C.D. produced the figures.
Author Information Data are available online at http://www.ebi.ac.uk/goldman-srv/DNA-storage and in the Sequence Read Archive (SRA) with accession number ERP002040 (to be confirmed). Correspondence and requests for materials should be addressed to N.G. (goldman@ebi.ac.uk).
Competing Financial Interests The authors declare competing financial interests: details have been uploaded via Nature’s online manuscript tracking system.
References
- 1.Baum EB. Building an associative memory vastly larger than the brain. Science. 1995;268:583–585. doi: 10.1126/science.7725109. [DOI] [PubMed] [Google Scholar]
- 2.Cox JPL. Long-term data storage in DNA. TRENDS Biotech. 2001;19:247–250. doi: 10.1016/s0167-7799(01)01671-7. [DOI] [PubMed] [Google Scholar]
- 3.Anchordoquy TJ, Molina MC. Preservation of DNA. Cell Preservation Tech. 2007;5:180–188. [Google Scholar]
- 4.Bonnet J, et al. Chain and conformation stability of solid-state DNA: implications for room temperature storage. Nucl. Acids Res. 2010;38:1531–1546. doi: 10.1093/nar/gkp1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clelland CT, Risca V, Bancroft C. Hiding messages in DNA microdots. Nature. 1999;399:533–534. doi: 10.1038/21092. [DOI] [PubMed] [Google Scholar]
- 6.Kac E. [accessed online, 10 May 2012];Genesis. 1999 http://www.ekac.org/geninfo.html.
- 7.Ailenberg M, Rotstein OD. An improved Huffman coding method for archiving text, images, and music characters in DNA. Biotechniques. 2009;47:747–754. doi: 10.2144/000113218. [DOI] [PubMed] [Google Scholar]
- 8.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 9.Church GM, Gao Y, Kosuri S. Next-generation digital information storage in DNA. Science. 2012;337:1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 10.MacKay DJC. Information Theory, Inference, and Learning Algorithms. Cambridge University Press; 2003. [Google Scholar]
- 11.Erlich HA, Gelfand D, Sninsky JJ. Recent advances in the polymerase chain reaction. Science. 1991;21:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- 12.Monaco AP, Larin Z. YACs, BACs, PACs and MACs: artificial chromosomes as research tools. Trends Biotech. 1994;12:280–286. doi: 10.1016/0167-7799(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 13.Carr PA, Church GM. Genome engineering. Nature Biotech. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 14.Willerslev E, et al. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science. 2007;317:111–114. doi: 10.1126/science.1141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kari L, Mahalingam K. DNA computing: a research snapshot. In: Atallah MJ, Blanton M, editors. Algorithms and Theory of Computation Handbook. 2nd ed vol. 2. Chapman & Hall; 2009. pp. 31-1–31-24. [Google Scholar]
- 17.Pǎun G, Rozenberg G, Salomaa A. DNA Computing: New Computing Paradigms. Springer-Verlag; 1998. [Google Scholar]
- 18.Watson JD, Crick FHC. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 19.Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, Barron AE. Landscape of next-generation sequencing technologies. Analytical Chemistry. 2011;83:4327–4341. doi: 10.1021/ac2010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeProust EM, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucl. Acids Res. 2010;38:2522–2540. doi: 10.1093/nar/gkq163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massingham T, Goldman N. All your base: a fast and accurate probabilistic approach to base calling. Genome Biol. 2012;13:R13. doi: 10.1186/gb-2012-13-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantz J, Reinsel D. Extracting value from chaos. IDC; 2011. [Google Scholar]
- 23.Brand S. The Clock of the Long Now. Basic Books; 1999. [Google Scholar]
- 24.The Economist. Digital archiving. History flushed. The Economist. 2011;403(8782):56–57. [Google Scholar]
- 25.Bessone N, Cancio G, Murray S, Taurelli G. Increasing the efficiency of tape-based storage backends. J. Phys.: Conf. Ser. 2010;219:062038. [Google Scholar]
- 26.Baker M, et al. Proceedings of the 1st ACM SIGOPS/EuroSys European Conference on Computer Systems. ACM; 2006. A fresh look at the reliability of long-term digital storage; pp. 221–234. [Google Scholar]
- 27.Carlson R. [accessed online, 7 September 2012];New cost curves. 2011 http://www.synthesis.cc/2011/06/new-cost-curves.html.
- 28.Yuille M, et al. The UK DNA banking network: a “fair access” biobank. Cell Tissue Bank. 2010;11:241–251. doi: 10.1007/s10561-009-9150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Global Crop Diversity Trust [accessed online, 10 May 2012];Svalbard Global Seed Vault. 2012 http://www.croptrust.org/main/content/svalbard-global-seed-vault.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.