Abstract
Erythropoetin-producing hepatoma (Eph) receptors are cell surface protein tyrosine kinases mediating cell-cell communication. Upon activation they form signalling clusters. We report crystal structures of the full ectodomain of human EphA2 (eEphA2), alone and in complex with the receptor-binding domain of the ligand ephrinA5 (ephrinA5RBD). Unliganded eEphA2 forms linear arrays of staggered parallel receptors involving two patches of residues conserved across A-class Ephs. eEphA2-ephrinA5RBD forms a more elaborate assembly, whose interfaces include the same conserved regions on eEphA2, but re-arranged to accommodate ephrinA5RBD. Cell surface expression of mutant EphA2s demonstrated that these interfaces are critical for localization at cell-cell contacts and activation-dependent degradation. Our results suggest a ‘nucleation’ mechanism whereby a limited number of ligand-receptor interactions seed an arrangement of receptors which can propagate into extended signalling arrays.
Ephs constitute a quarter of all known human receptor tyrosine kinases. They direct key processes during development and repair of the nervous system, blood vessel formation, insulin secretion, immune system function, intestinal homeostasis and bone tissue integrity1. They are grouped into two classes, A and B2, but domain composition is conserved across the family (Fig. 1a). The extracellular region comprises an N-terminal ligand-binding domain (LBD), a cysteine-rich region and two fibronectin type III domains (FN1 and FN2). FN2 connects to the transmembrane helix (TM), followed by an intracellular region comprising a juxtamembrane region (JM), a tyrosine kinase domain and a sterile-alpha motif (SAM) domain often linked to a C-terminal PDZ binding motif. Structures have been determined for examples of all Eph domains in isolation, with the exception of the cysteine-rich region3,4.
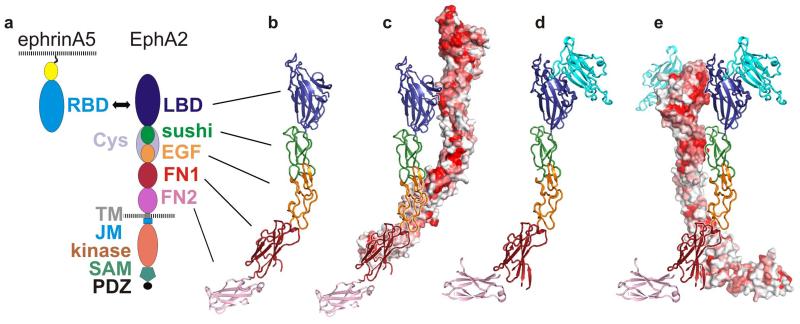
Figure 1. Crystal structures of the complete EphA2 extracellular region (eEphA2) alone and in complex with ephrinA5RBD.
a, Domain composition of ephrinAs and Eph receptors. b, Crystal structure of eEphA2 comprising EphA2 LBD, Cys, FN1 and FN2 domains. Colours are as in a. The previously uncharacterized Cys domain is composed of a sushi domain (CCP) and an EGF domain. c, Arrangement of two eEphA2 molecules within the crystal. Their contacts bury 2661 Å2 of molecular surface. The front molecule is displayed and oriented as in a. The back molecule is shown in surface view and coloured according to human Eph class A receptor sequence conservation (red = strongest conservation, white = weakest conservation). d, eEphA2 (colours as in a) in complex with ephrinA5RBD (cyan). The orientation of the sushi domain is the same as in b and c. e, Arrangement of neighbouring complexes within the crystal. The eEphA2 molecule on the right is oriented and coloured as in d. Its neighbour on the left is in surface view and coloured according to EphA2 sequence conservation. Their contacts bury 1240 Å2.
Activation of Eph receptors depends on the presence of their ligands (ephrins) and involves the packing of Ephs into signalling clusters1,4. Ephrins consist of an N-terminal extracellular receptor-binding domain (ephrinRBD) and a C-terminal extension linked to the plasma-membrane by a lipid anchor (class A ephrins) or transmembrane helix (class B ephrins). Binding affinities vary for different Eph-ephrin pairs, but in general binding within classes is favoured5. Crystal structures for 1:1 complexes of Eph LBDs bound to ephrinRBDs show a hydrophobic ephrin-binding groove on the receptor and provide insight into ligand-receptor specificity, but do not define a mechanism for ligand-dependent signalling6. Members of the Eph family of receptor tyrosine kinases appear to require a more complex form of oligomerisation to trigger signalling than the ligand-induced dimerisation model developed from studies of systems such as the human growth hormone receptor7. Insights into Eph receptor clustering upon ephrin binding are as yet limited4. The crystal structure of the EphB2 LBD - ephrinB2RBD complex revealed a second, putative tetramerization, interface6. However, five subsequent Eph LBD - ephrinRBD complex crystal structures have not shown evidence for a consistent mode of oligomerisaton8-11. Biophysical and functional analyses have also implicated additional regions of Eph ectodomains in ephrin mediated signalling12. We therefore sought to provide structural insight into a full Eph receptor ectodomain architecture and its interactions. We focus on EphA2 which is expressed during embryonic development and in adult epithelia13 where it regulates cell adhesion14,15. EphA2 also acts as a powerful oncogene in many tumours by promoting vascularisation and metastasis16-19.
RESULTS
Structure of EphA2 ectodomain (eEphA2)
To improve crystallization, we expressed eEphA2 (residues K27-N534) in glycosylation-impaired mammalian cells20,21, trimmed the resultant glycans down to single N-acetylglucosamine moieties and di-methylated the amino groups22. The structure (one molecule per asymmetric unit) was refined to a final Rwork = 24% (Rfree = 30%) using data to 3.0 Å resolution. Detailed data and refinement statistics are given in Table 1. A view of the final electron density map is shown in Supplementary Fig. 1. All five domains are well-ordered and the overall linear arrangement results in an elongated molecule (length ~172 Å, width ~22-48 Å, Fig. 1b). The previously uncharacterized cysteine-rich region shows the topology of a sushi (CCP) domain followed by an EGF-like domain. Sequence alignment of class A Ephs mapped onto the eEphA2 structure reveals patches of surface conservation on the LBD and sushi domains (Fig. 1c). Conserved sushi domain residues contribute to a crystal contact with a parallel neighbouring molecule staggered by one domain (Fig. 1c). In total, interfaces between these molecules bury 2661 Å2 of surface area and stabilise linear arrays of staggered, parallel Ephs. These crystal contacts for full length EphA2 ectodomain do not re-iterate the “head-to-head” contacts recently observed for crystals of EphA2 LBD10. Two putative N-linked glycosylation sites, located in the FN1 domain and in the linker between domains FN1 and FN2, are not involved in crystal contacts.
Table 1. Data collection and refinement statistics.
| eEphA2 (refinement) | eEphA2 (phasing) | eEphA2+ephrinA5RBD | |
|---|---|---|---|
| Data collection | |||
| RCSB PDB code | XXX | XXX | XXX |
| Space group | P1 | P1 | P2221 |
| Cell dimensions | |||
| a, b, c (Å) | 41.92, 44.65, 93.42 |
41.47, 44.35, 92.08 |
173.63, 59.63,112.16 |
| α, β, γ (°) | 93.02, 98.22, 112.12 |
93.41, 97.22, 112.44 |
90, 90, 90 |
| Resolution (Å)* | 46-3.0 (3.1-3.0) | 45-3.0 (3.1-3.0) | 173.6-4.8 (4.9-4.8) |
| Rmeas * | 0.143 (0.443) | 0.097 (0.645) | 0.112 (0.743) |
| I/σI* | 9.87 (2.60) | 8.97 (1.74) | 12.95 (2.83) |
| Completeness (%)* | 97.6 (94.5) | 97.8 (97.7) | 98.2 (75.5) |
| Redundancy* | 3.3 (2.3) | 2.2 (2.0) | 6.2 (6.3) |
| Refinement | |||
| Resolution (Å)* | 45.9-3.0 (3.3-3.0) | - | 40.5-4.8 (6.1-4.8) |
| No. reflections | 12037 | - | 5643 |
| Rwork/Rfree | 0.236/0.299 | - | 0.312/0.314 |
| No. atoms | 3743 | - | 4889 |
| Protein | 3728 | - | 4889 |
| Ligand/ion | 15 | - | - |
| Water | - | - | - |
| average B-factors | |||
| Protein | 100.4 | - | 247.2 |
| Ligand/ion | 107.6 | - | - |
| Water | - | - | - |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | - | 0.003 |
| Bond angles (°) | 0.697 | - | 0.675 |
Values in parentheses are for highest-resolution shell.
Structure of eEphA2 in complex with ephrinA5RBD
To compare unliganded eEphA2 with a ligand-bound form, we attempted to crystallize it with a panel of ephrinA ligands and succeeded in producing diffracting crystals of eEphA2 in complex with human ephrinA5 RBD (ephrinA5RBD). Di-methylation of the amine groups and extensive optimization yielded a crystal (containing one eEphA2-ephrinA5RBD per asymmetric unit) from which diffraction data were collected to 4.8 Å resolution. The resultant complex structure was refined (by rigid body and TLS) to Rwork= 31%, Rfree= 31% (Fig. 1d). Crystallographic details are summarized in Table 1 and an example of the final electron density map is shown in Supplementary Fig. 1. The structure of ephrinA5RBD in the complex shows no evidence of major structure changes when compared to the previously reported structure23 of unliganded ephrinA5RBD. Superposition of ligand-bound and unbound eEphA2s reveals domain re-orientations (Supplementary Fig. 2). The largest re-orientation, a relative rotation of 71° by FN2, demonstrates the hinge-like character of the FN1-FN2 linker. Other points of flexion are located between the LBD and sushi domain and within the EGF-like domain.
The crystal consists of parallel and anti-parallel arrays of eEphA2-ephrinA5RBD complexes (Supplementary Fig. 3) formed by interfaces A-F (Supplementary Fig. 4 and Supplementary Table 1). Interfaces A-E mediate parallel arrays (Fig. 1e and Fig. 2a), interface F stabilizes anti-parallel packing (Fig. 2b). The previously reported Eph-LBD - ephrinRBD interaction3 forms interface A and broadly matches that previously reported for the complex of EphA2 LBD with ephrinA1RBD10 (Fig. 2a, c). Interface C is a small contact between eEphA2 and a second ephrinA5RBD (Fig. 2a, c). Interfaces B, D and E connect neighbouring eEphA2 molecules via their LBDs, sushi domains and FN1 domains, respectively (Fig. 2a, c-e). Interfaces B and D use the conserved surface patches which form the staggered LBD-sushi contact in the unliganded eEphA2 crystals, but in the presence of ephrinA5RBD these patches mediate in-register LBD-LBD (interface B) and sushi-sushi (interface D) interactions (Fig. 1e). Finally, the FN1-FN2 domains from eEphA2 form interface F with ephrinA5RBD bound (via interface A) to an anti-parallel eEphA2 neighbour (Fig. 2b, f).
Figure 2. The eEphA2-ephrinA5RBD complex: structural changes within eEphA2 and formation of array-like clusters.
a, b, Crystal packing of eEphA2-ephrinA5RBD. The ribbon diagrams are coloured as in Fig. 1. The cartoon above each panel illustrates the crystal contacts designated interfaces A-F (dotted lines) for eEphA2 (purple) and ephrinA5RBD (cyan). Panel a shows six symmetry-related eEphA2-ephrinA5RBD complexes forming a parallel array; view is approximately along unit cell axis c. The protein arrangement reflects EphA2 and ephrinA5 interaction in trans, i.e. where the proteins are located on opposing cells. Panel b shows six symmetry-related eEphA2-ephrinA5RBD complexes forming an anti-parallel array, view as in f, but tilted by ~45° around unit cell axis a. The arrangement shows EphA2 and ephrinA5 interacting both in trans and in cis, consistent with both proteins presented on both cells. c-f, Interfaces A-F are shown as black lines. Colours are as in a. Interfaces A-E form the crystal contact interactions shown in a, interface F is a major interface but contributes only to the contacts shown in b.
EphA2 cell-cell contact localization is not dependent on the intracellular domains
Recent work has shown that EphA2 localizes at the cell-cell contacts in the mouse eye lens where it binds to ephrinA5 and maintains tight cellular packing24. EphA2 is also known to localize at cell-cell contacts when transfected into a range of epithelial cell lines25-27 and we therefore chose to exploit this system to dissect the interaction characteristics of EphA2 in vivo using a human embryonic kidney epithelial cell line (HEK293T) and fluorescent protein (mVenus)-tagged constructs (Fig. 3). A series of deletion constructs all showed cell surface expression (Supplementary Fig. 5). As anticipated, full length EphA2 accumulated at the contact region between transfected cells. We did not observe EphA2 accumulation in regions of the cell surface not involved in contacts, or in contact regions between EphA2-expressing and non-transfected native cells (Fig. 3a-c). These observations imply that trans presentation of EphA2 (i.e. from opposing cells) is required for clustering. We found that deleting individual cytoplasmic domains, or the entire intracellular region, did not prevent cell surface expression and did not affect the pattern of localization at cell-cell contacts (Supplementary Fig. 6). In contrast, although cell surface localization is still maintained, deletion of the N-terminal four extracellular domains (LBD-FN1) abolishes the specific localization at the cell-cell interface, suggesting that the ectodomain is essential (Supplementary Fig. 6).
Figure 3. EphA2 localization at cell-cell contacts depends on ectodomain clustering.
Representative results are presented of functional studies using HEK293T cells. A full set of results is provided in Supplementary Fig. 6. Filled white arrows indicate cell-cell contacts between transfected cells. Open arrows point to cell-cell contacts between a transfected cell and a non-transfected cell. a-c, mVenus-tagged EphA2 accumulates at cell-cell contacts of transfected cells. d-e, Mutants in interfaces A, B, D and, to some extent, interface F do not cluster at cell-cell contacts of transfected cells but are distributed evenly on the cell surface (the interface B mutant G131Y is shown as an example). a, d Fluorescence images are shown. b, e, Phase contrast images of the corresponding cells. c, f, Overlay images of a, b, d, e.
Mutations in interfaces A, B, D, F affect cell-cell contact localization
To investigate which of the interfaces in our crystal structures have biological relevance, we produced a panel of EphA2 point mutants designed to disrupt interfaces A-F (Supplementary Table 1). Interface B and D mutations result in homogeneous EphA2 cell surface distribution, completely abolishing the wild type localization pattern (Fig. 3d-f and Supplementary Fig. 6). These are the major interfaces for the in-register arrays of liganded eEphA2, the same surfaces form the staggered arrays for unliganded eEphA2, and in vivo could mediate in cis interaction, i.e. of Eph receptors on the same cell. Previous mutagenesis studies have indicated interface B residues can contribute to EphA3 – ephrinA5 binding affinity12, although it is not the main ephrin binding site. Mutations in the minor interfaces C and E had no effect on EphA2 localization (Supplementary Fig. 6).
Introduction of an N-linked glycosylation site (EphA2 A190N + L192S) to disrupt interface A, the well-established ephrin binding site for binding in trans, abolished cell-cell contact localization (Supplementary Fig. 6). As HEK293 cells express class A ephrins endogenously28, these results suggest that binding to endogenous ephrins in trans is necessary to drive localization of expressed EphA2 to cell-cell contacts, a conclusion further supported by studies using insect (SF9) cells (Supplementary Fig. 6c). Interface F (the second largest interface in the complex crystals) would in vivo be consistent with a cis-interaction between ephrinA5 and EphA2 FN1-FN2; mutations at this interface caused an intermediate phenotype where protein is partially de-localized over the entire cell surface (Supplementary Fig. 6). This is consistent with previous functional studies suggesting the FN2 domain of EphA3 binds ephrinA5 in cis29.
Specific proteolytic cleavage of EphA2 depends on clustering
Previous studies have demonstrated that activation of Eph receptors leads to their internalization and degradation30 and specific activation-dependent proteolytic cleavage has been reported for EphB231,32. We performed immunoblot assays which show that EphA2 is proteolytically cleaved in HEK293 cells. Upon wild type EphA2 expression at least three C-terminal tyrosine-phosphorylated EphA2 proteolytic fragments are observed, corresponding to ~70, ~75 and ~95 kDa (including the ~28 kDa mVenus tag, Supplementary Fig. 7). For mutants in interfaces A, B and D the fragments of ~75 kDa and ~95 kDa are absent. For mutants in interface F only the ~95 kDa fragment is absent. Notably, the cleavage patterns observed for EphA2 wild type and mutants correlate with their cell surface localization patterns; interface F mutants having less severe impairment of cell-cell contact localization than interface A, B, and D mutants (Supplementary Fig. 6 and 7). The correlation of specific cleavage with localization to cell-cell contacts suggests that this process depends on the ability of the receptor to cluster via interfaces A, B, D and F.
DISCUSSION
Previously reported data for Eph function point to a requirement for receptor clustering into extended cell surface assemblies4,33-35. However, full length Eph ectodomains (eEphA2, eEphB2 and eEphB3) show no propensity to form oligomers in solution (as assessed by multiple angle light scattering, MALS, and analytical ultracentrifugation) either alone or in complex with monomeric, soluble ephrins (ephrinA5RBD, ephrinA2RBD and ephrinB2RBD) (Supplementary Fig. 8 and data not shown). These data accord with previous reports that full length Eph receptors only cluster if the ephrin ligand is presented in a membrane-bound or oligomeric form36. Nevertheless, at very high concentration in crystals both unliganded and complexed eEphA2 form array-like networks of interfaces which functional experiments demonstrate are relevant to the cell surface signalling. The eEphA2 arrays involve staggered interactions between the LBD and sushi domains. Ephrin binding snaps the Eph arrays into register with LBD-LBD and sushi-sushi interactions involving the same eEphA2 surface patches. Whilst we were completing our analysis an independently determined crystal structure of unliganded human EphA2 ectodomain was deposited in the Protein Data Base37. The 3FL7 lattice includes crystal contacts similar to the in-register form we observe indicating that this mode of Eph-Eph interaction can also occur without ephrin-binding. This implies that in the absence of ligand transient cis associations between cell surface Ephs may occur as a dynamic equilibrium between LBD-LBD/sushi-sushi and LDB-sushi interactions. For such a system ephrin-binding at “nucleation” points could trigger more widespread recruitment of EphA2 into in-register arrays containing unliganded receptor and facilitating, in a cooperative manner, additional ephrin-binding (Fig. 4).
Fig.4. A mechanism for the nucleation and propagation of EphA2 signalling assemblies.
a, b, Eph and ephrin are shown schematically with ectodomains colour coded as in Fig. 1a and the Eph intracellular region in grey. Lightening flashes indicate the strength of activation. In panel a transient arrays of staggered Ephs snap into in-register signalling arrays on nucleation by ephrins. The intermediate phenotype of interface F mutations in functional assays may reflect weaker interactions between Eph receptors and their ligands, resulting in a weakened signalling activity. In panel b additional cis (interface F) interactions inter-connect anti-parallel arrays further stabilising the signalling assemblies.
Indeed, it has been reported that once an Eph receptor signalling cluster is nucleated by ephrin-binding, further Eph are recruited to the cluster in an ephrin-independent manner33. The above steric ‘seeding’ mechanism provides a model for how relatively low levels of ephrins can trigger the formation of extended assemblies of EphA2. These assemblies may be further stabilized by transmembrane38 and cytoplasmic39 region interactions. Our results illustrate a nucleation mechanism for ectodomain-driven array-based signalling which could be broadly applicable to other cell surface receptor systems.
METHODS
Vectors and Cloning
Secreted polyhistidine-tagged ectodomains of the human EphA2 (eEphA2, residues 27-534, Uniprot: P29317) and ephrin A5 (ephrinA5RBD, residues 27-166, Uniprot: P52803) were cloned into the AgeI-KpnI sites of pHLsec21 using standard PCR methods (EphA2 residues 1-23 and ephrinA5 residues 1-20 belong to the secretion signals in EphA2 and ephrin A5 as predicted by SignalP40). For cell surface expression transmembrane constructs of human EphA2, fused to a C-terminal monomeric fluorescent protein mVenus (green)41, were cloned into pHLsec (coding additionally for a C-terminal poly-histidine tag). The following constructs were produced as mVenus fusions: EphA2 (residues 27-976), EphA2ΔSAM (residues 27-889), EphA2ΔKSAM (residues 27-564), EphA2Δe (residues 436-976). EphrinA5 (residues 27-166) was fused at its C-terminus to the monomeric transmembrane helix of receptor protein tyrosine phosphatase RPTPμ42 (residues 742-769, Uniprot: P28827) followed by the monomeric fluorescent protein mCherry, and cloned into pHLsec.
Protein Crystallization
Details of the protein expression and purification are given in the Supplementary Material Online. eEphA2 was treated with endoglycosidase F143,44 and lysine-methylated 22. Crystals diffracting to 3 Å resolution grew at 20 °C from drops45 where protein solution was mixed 1:1:1 with reservoir solution (20 % polyethylene glycol 6000, 1 M LiCl, 0.1 M Tris pH 8) and 0.4 M non-detergent sulfobetaine 256 (Hampton). Selenomethionine labeled protein21 was purified and crystallized as the native protein. Crystals of eEphA2 and ephrinA5RBD complex grew in conditions similar to the that specified below, but diffracted only to 6-8 Å resolution. Only one crystal diffracted well to 4.8 Å resolution. This crystal grew at 20 °C in a drop where two parts of protein solution were mixed with one part water, one part reservoir solution (8 % polyethylene glycol 6000, 0.8 M LiCl, 0.08 M citrate pH 5) and one part 1 % polyvinylpyrrolidone K15 (Hampton).
Data collection and processing
Data statistics are summarized in Table 1. Selenomethionine derivative crystals of eEphA2 were flash-frozen after briefly dipping the crystal containing loop into perfluoropolyether oil PFO-X125/03 (Lancaster). A data set was collected at 100 K at the selenium edge (wavelength = 0.98 Å) at the ESRF beamline BM14 using the inverse beam option (rotation every 20 images, corresponding to 10 degrees). This crystal diffracted to 3 Å resolution. The data were processed using XDS46 and MOSFLM47. Molecular replacement using models for the ligand binding domain of EphA2 (PBD entry 3C8X), the fibronectin type III domain 2 of EphB4 (PDB entry 2E7H), and fibronectin type III domain 2 of EphA8 (PDB entry 1X5L) yielded initial phases that were used to locate selenium sites in PHASER48. Electron density modification was done with DM49 and manual improvement of the protein model in COOT50. Cycles of phase calculation plus model building and refinement iteratively lead to a satisfactory model. A second crystal yielded better data at high resolution at the ESRF beamline ID23-EH2 (100 K, wavelength = 0.87 Å) and was used for the final rounds of refinement. eEphA2 carries two predicted N-glycosylation sites, the first is on the FN1 domain where N-acetyl-glucosamine (NAG) was placed into the electron density. This NAG molecule is in close proximity to, but not in contact with the FN2 domain of a neighbouring EphA2 chain. The second putative N-glycosylation site located in the poorly defined linker region between FN1 and FN2 and lacked electron density indicating the presence of sugar, possibly due to the relative disorder of this region. This region is also not involved in crystal contacts. COOT and MOLPROBITY51 were used for model validation. 3 residues (0.6 %) are located in outlier regions of the Ramachandran plot.
Crystals of eEphA2 in complex with ephrinA5RBD were frozen in a cryoprotectant solution containing one part reservoir solution and one part reservoir solution supersaturated with LiCl. After brief equilibration in cryoprotectant, the crystals were mounted on a loop which was then dipped into perfluoropolyether oil PFO-X125/03 and flash-frozen. The best dataset was collected at 100 K on ESRF beamline ID14-EH2 (wavelength = 0.93 Å) and processed using XDS. A model of human EphA2 ligand binding domain in complex with ephrin A2 receptor binding domain (PDB code 3CZU) was used for molecular replacement in PHASER (RFZ=11.1, TFZ=9.6, LLG=162). We placed the models of individual eEphA2 domains sequentially into the improving density and refined each using rigid body refinement in PHASER. Molecular replacement was initially done using the 3.0 Å resolution eEphA2 model presented here. After deposition of the 2.5 Å resolution eEphA2 model in the Protein Data Bank (PBD code 3FL7) by the Structural Genomics Consortium, Toronto, we used this higher resolution model. The model was refined using PHENIX52 (Supplementary Material Online). 6 residues are in outlier regions of the Ramachandran plot as found in the input molecular replacement model downloaded from the Protein Data Bank. The two putative N-glycosylation sites lie outside crystal contact areas.
To calculate the angle between the orientations eEphA2 FN2 adopts in the unliganded and ephrinA5RBD-bound structures, we first generated two models of liganded eEphA2 superposed onto the FN1 and FN2 domains of unliganded eEphA2. We then used LSQKAB within the CCP4 suite53 to calculate the rotation applied to superpose the two resulting structures via their FN2 domain.
Multiangle Light Scattering (MALS)
Proteins were purified by size-exclusion chromatography and concentrated to approximately 1.6 ml/ml (eEphA2) and 1.8 mg/ml (eEphA2/ephrinA5RBD complex). Separation for the MALS was achieved using an analytical Superdex S75 10/30 column (GE Heathcare) and the eluate was passed through online static light scattering (DAWN HELEOS II, Wyatt Technology, Santa Barbara, CA), differential refractive index (Optilab rEX, Wyatt Technology) and Agilent 1200 UV (Agilent Techologies) detectors. Data were analysed using the ASTRA software package (Wyatt Technology).
Live cell assays
HEK293T cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM high glucose, Sigma) supplemented with L-glutamine, non-essential amino-acids (Gibco) and 10% foetal calf serum (FCS, Sigma). Plasmid DNA was transfected with lipofectamine 2000 following the recommended protocol (Invitrogen). Final concentrations were 2 μg plasmid DNA per ml, 5 μl lipofectamine per ml and 1.5% FCS. After 4-6 hours, cells were returned to growth medium, which contains 10% FCS, and plated on poly-L-lysine-coated glass bottom culture plates (MatTek). To observe trans interactions of different constructs, cells were transfected separately and plated out together at this stage. 15-20 hours later cells were imaged using a Zeiss LSM510 confocal microscope (with variable emission detectors [Meta-Head]). For this time-scale, we do not observe cell-rounding and death upon EphA2 expression for either HEK293 or COS7 cells (data not shown). All images were taken with the same detector and laser settings and the images showing fluorescent signal were not subsequently modified, thus allowing direct comparison.
Immunoblot analysis
To assess cell surface expression of truncated EphA2 constructs, cells were incubated for 10 minutes at 4 °C to slow down endocytosis and then incubated 30 min with 1 mg/ml No-Weigh Sulfo-NHS-LC-Biotin (Pierce) at 4 °C. The biotinylation reaction was stopped by adding 100 mM Tris pH 7.5. Cells were lysed in phosphate buffer saline supplemented with 100 mM Tris pH 7.5 and 1% Triton X-100 (Sigma). Cell lysates were incubated for 30 minutes with streptavidin-coated agarose beads (Pierce) at 4 °C, the beads washed three times with lysis buffer and then re-suspended in standard protein denaturing gel loading buffer for immunoblot analysis using anti-pentahistidine (penta-His, Qiagen, dilution 1:1000). Cells intended for anti-phosphotyrosine or anti-green fluorescent protein immunoblot analyses were re-suspended in chilled phosphate buffer saline 15-20 hours post transfection, immediately mixed with standard protein gel loading buffer and analyzed using anti-phosphotyrosine (4G10® Platinum, Millipore, dilution 1:2000), anti-GFP (Cat. No. A-11122, Invitrogen, dilution 1:1000), anti-actin (ACTN05 (C4), AbCam, dilution 1:400), anti-mouse IgG peroxidase-conjugated secondary antibody (Sigma, dilution 1:10000) and anti-rabbit IgG peroxidase-conjugated secondary antibody (AbCam, dilution 1:5000). To rule out that FCS or trace amounts of trypsin used for cell propagation are responsible for EphA2 cleavage, we performed control experiments in cells cultured with 0% or 1.5% FCS, in the presence of 0%, 0.5% or 5% of the trypsin-EDTA solution used for tissue culture maintenance (Invitrogen, Cat. No. 25300-054) (data not shown).
Supplementary Material
Acknowledgements
We thank M. Walsh for assistance during data collection, B. Jannsen for aiding crystallographic data analysis, T.S. Walter for advice on protein crystallization, M. Jones and W. Lu for technical assistance, R. Gilbert for analytical ultracentrifugation measurements, K.J. Morris and N. Alsamhouri for their help with confocal microscopy, D.I. Stuart for critical reading of the manuscript. This research was funded by Cancer Research UK (CR-UK). A.R.A is funded by the UK Medical Research Council and E.Y.J. by CR-UK.
Footnotes
Author information
Atomic coordinates and structure factors will be deposited within the Protein Data Bank.
References
- 1.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Eph Nomenclature Committee Unified nomenclature for Eph family receptors and their ligands, the ephrins. Cell. 1997;90:403–4. doi: 10.1016/s0092-8674(00)80500-0. [DOI] [PubMed] [Google Scholar]
- 3.Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 4.Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–42. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gale NW, et al. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- 6.Himanen JP, et al. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–8. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham BC, et al. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science. 1991;254:821–5. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 8.Himanen JP, et al. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci. 2004;7:501–9. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- 9.Chrencik JE, et al. Structural and biophysical characterization of the EphB4*ephrinB2 protein-protein interaction and receptor specificity. J Biol Chem. 2006;281:28185–92. doi: 10.1074/jbc.M605766200. [DOI] [PubMed] [Google Scholar]
- 10.Himanen JP, et al. Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 2009;10:722–8. doi: 10.1038/embor.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowden TA, et al. Structural Plasticity of Eph Receptor A4 Facilitates Cross-Class Ephrin Signaling. Structure. 2009;17:1386–1397. doi: 10.1016/j.str.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith FM, et al. Dissecting the EphA3/Ephrin-A5 interactions using a novel functional mutagenesis screen. J Biol Chem. 2004;279:9522–31. doi: 10.1074/jbc.M309326200. [DOI] [PubMed] [Google Scholar]
- 13.Lindberg RA, Hunter T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the eph/elk family of protein kinases. Mol Cell Biol. 1990;10:6316–24. doi: 10.1128/mcb.10.12.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao H, Burnett E, Kinch M, Simon E, Wang B. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat Cell Biol. 2000;2:62–9. doi: 10.1038/35000008. [DOI] [PubMed] [Google Scholar]
- 15.Carter N, Nakamoto T, Hirai H, Hunter T. EphrinA1-induced cytoskeletal re-organization requires FAK and p130(cas) Nat Cell Biol. 2002;4:565–73. doi: 10.1038/ncb823. [DOI] [PubMed] [Google Scholar]
- 16.Lin YG, et al. EphA2 overexpression is associated with angiogenesis in ovarian cancer. Cancer. 2007;109:332–40. doi: 10.1002/cncr.22415. [DOI] [PubMed] [Google Scholar]
- 17.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–6. [PubMed] [Google Scholar]
- 18.Kinch MS, Carles-Kinch K. Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin Exp Metastasis. 2003;20:59–68. doi: 10.1023/a:1022546620495. [DOI] [PubMed] [Google Scholar]
- 19.Wykosky J, Debinski W. The EphA2 receptor and ephrinA1 ligand in solid tumors: function and therapeutic targeting. Mol Cancer Res. 2008;6:1795–806. doi: 10.1158/1541-7786.MCR-08-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–24. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aricescu AR, Lu W&, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–50. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 22.Walter TS, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–22. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolov D, Li C, Lackmann M, Jeffrey P, Himanen J. Crystal structure of the human ephrin-A5 ectodomain. Protein Sci. 2007;16:996–1000. doi: 10.1110/ps.062665807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper MA, et al. Loss of ephrin-A5 function disrupts lens fiber cell packing and leads to cataract. Proc Natl Acad Sci U S A. 2008;105:16620–5. doi: 10.1073/pnas.0808987105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zantek ND, et al. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 1999;10:629–38. [PubMed] [Google Scholar]
- 26.Miura K, Nam JM, Kojima C, Mochizuki N, Sabe H. EphA2 engages Git1 to suppress Arf6 activity modulating epithelial cell-cell contacts. Mol Biol Cell. 2009;20:1949–59. doi: 10.1091/mbc.E08-06-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orsulic S, Kemler R. Expression of Eph receptors and ephrins is differentially regulated by E-cadherin. J Cell Sci. 2000;113:1793–802. doi: 10.1242/jcs.113.10.1793. [DOI] [PubMed] [Google Scholar]
- 28.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–71. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho RF, et al. Silencing of EphA3 through a cis interaction with ephrinA5. Nat Neurosci. 2006;9:322–30. doi: 10.1038/nn1655. [DOI] [PubMed] [Google Scholar]
- 30.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–75. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 31.Lin KT, Sloniowski S, Ethell DW, Ethell IM. Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. J Biol Chem. 2008;283:28969–79. doi: 10.1074/jbc.M804401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litterst C, et al. Ligand binding and calcium influx induce distinct ectodomain/gamma-secretase-processing pathways of EphB2 receptor. J Biol Chem. 2007;282:16155–63. doi: 10.1074/jbc.M611449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimmer-Kleikamp SH, Janes PW, Squire A, Bastiaens PI, Lackmann M. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J Cell Biol. 2004;164:661–6. doi: 10.1083/jcb.200312001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vearing CJ, Lackmann M. Eph receptor signalling; dimerisation just isn’t enough. Growth Factors. 2005;23:67–76. doi: 10.1080/08977190500055869. [DOI] [PubMed] [Google Scholar]
- 35.Egea J, et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron. 2005;47:515–28. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 36.Davis S, et al. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science. 1994;266:816–9. doi: 10.1126/science.7973638. [DOI] [PubMed] [Google Scholar]
- 37.Walker JR, et al. Deposited in the RCSB Protein data Bank on 2008-12-18 by the Structural Genomics Consortium (SGC) 2009. [Google Scholar]
- 38.Bocharov EV, et al. Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J Biol Chem. 2008;283:29385–95. doi: 10.1074/jbc.M803089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. 1999;283:833–6. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- 40.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 42.Chin CN, Sachs JN, Engelman DM. Transmembrane homodimerization of receptor-like protein tyrosine phosphatases. FEBS Lett. 2005;579:3855–8. doi: 10.1016/j.febslet.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 43.Grueninger-Leitch F, D’Arcy A, D’Arcy B, Chene C. Deglycosylation of proteins for crystallization using recombinant fusion protein glycosidases. Protein Sci. 1996;5:2617–22. doi: 10.1002/pro.5560051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang VT, et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–73. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61:651–7. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 47.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cowtan K. dm: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994:34–38. [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 53.Collaborative Computational Project 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.