Abstract
Antibodies can be carried into the cell during pathogen infection where they are detected by the ubiquitously expressed cytosolic antibody receptor TRIM21. Here we show that TRIM21 recognition of intracellular antibodies activates immune signaling. TRIM21 catalyses K63-ubiquitin chain formation, stimulating transcription factor pathways NF-κB, AP-1 and IRF3, IRF5, IRF7. Activation results in proinflammatory cytokine production, modulation of natural killer (NK) stress ligands and the induction of an antiviral state. Intracellular antibody signaling is abrogated by genetic deletion of TRIM21 and is recovered by ectopic TRIM21 expression. Antibody sensing by TRIM21 can be stimulated upon infection by DNA or RNA non-enveloped viruses or intracellular bacteria. The antibody-TRIM21 detection system provides potent, comprehensive innate immune activation, independent of known pattern recognition receptors.
The sensing of intracellular pathogens is critical to the immune response. Known methods of detection rely on the recognition of pathogen-associated molecular patterns (PAMPs) by germline-encoded pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs)1 and cytoplasmic nucleic acid receptors RIG-I and MDA52,3. Alternatively the host may sense physiological changes that accompany pathogen infection or sterile injury through the detection of danger associated molecular patterns (DAMPs)4. DAMPs are host-derived molecules which, when detected in a specific context, can induce an inflammatory response5. In the non-inflammatory resting state, the location of DAMPs must be tightly regulated. For instance, antibodies patrol the extracellular spaces and mediate extracellular immune responses.
Antibodies can be carried into cells when attached to infecting virus particles6. Once inside the cell, antibody-coated viruses are bound by the cytosolic antibody receptor TRIM21 via its C-terminal PRYSPRY domain. The binding affinity of TRIM21 to antibody is subnanomolar, making TRIM21 the highest affinity human Fc receptor7. After binding incoming virus-antibody complexes in the cytoplasm, TRIM21 targets virions for proteasome and VCP-dependent degradation in a process known as antibody-dependent intracellular neutralization (ADIN)6,8,9. Depletion of TRIM21 prevents efficient neutralization of adenovirus by pooled human serum IgG6. Conversely, high expression of TRIM21 permits neutralization by fewer than two antibody molecules per virus particle10. ADIN is dependent on the ability of TRIM21 to synthesize K48-linked ubiquitin chains via its RING domain6. TRIM21 is a close homologue of TRIM5α, which restricts infection of retroviruses in a species-specific manner11. Human TRIM5α responds to infection by restricted viruses by synthesizing unanchored K63-linked ubiquitin chains12. This activity stimulates the downstream kinase TAK1, resulting in a signaling cascade activating NF-κB and AP-1 transcription factors.
In this study we asked whether antibody entering the cytoplasm while bound to a pathogen acts in a context-dependent manner to initiate immune signaling. We found that cytoplasmic antibodies are a potent DAMP and that TRIM21 is necessary and sufficient for detection. TRIM21 synthesizes unanchored K63-linked ubiquitin chains in a RING domain-dependent manner. Incoming virus-antibody complexes activate NF-κB, AP-1 and IRF signaling pathways resulting in proinflammatory cytokine production and the induction of an antiviral state. TRIM21 signaling is not pathogen specific, since non-enveloped viruses, bacteria, as well as antibody-coated latex beads are able to elicit signaling. These findings demonstrate the existence of a potent detection mechanism that allows cells to stimulate broad-spectrum immunity upon penetration of their cytosol by antibody-bound pathogens.
Results
Detection of adenovirus-antibody complexes elicits NF-κB signaling
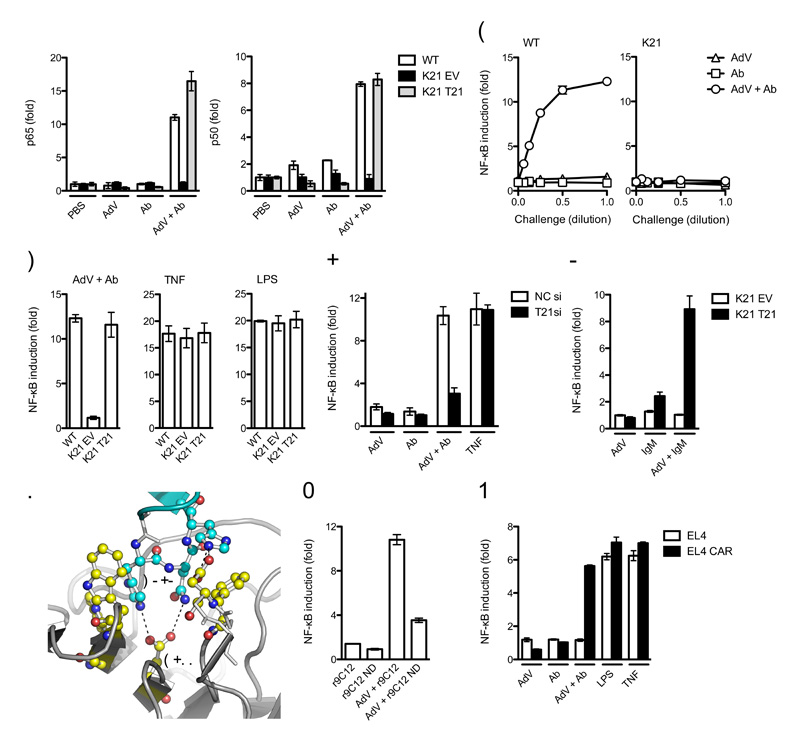
To test whether antibody entering the cytoplasm while bound to a pathogen initiates immune signaling, we assayed activated NF-κB subunits p65, p50 and p52 4 h post-challenge with an adenovirus type 5 vector (AdV) in the presence of antibody (Ab) by analyzing binding of the NF-κB subunits to consensus NF-κB DNA oligonucleotides in an ELISA assay. In wild-type mouse embryonic fibroblast (MEF) cells, a substantial increase in activated NF-κB was observed upon infection with adenovirus-antibody complex (AdV + Ab) but not with either component alone (Fig. 1a and Supplementary Fig. 1). The response was dependent upon TRIM21, as activation was not observed in MEFs derived from Trim21-deficient mice. Furthermore, activation in Trim21-deficient MEFs could be restored by ectopic expression of human TRIM21 (Fig. 1a), confirmed by immunoblot analyses (Supplementary Fig. 1). Titration of AdV, Ab or AdV + Ab onto wild-type and Trim21-deficient MEFs revealed that activation of an NF-κB luciferase reporter was dose-dependent and approached saturation at high multiplicity of infection (moi) but was absent at all multiplicities in Trim21-deficient MEFs (Fig. 1b). TRIM21 was not required for constitutive NF-κB signaling in response to other stimuli, as similar activation was observed in wild-type MEFs and Trim21-deficient MEFs transduced with empty vector or human TRIM21 when challenged with lipopolysaccharide (LPS) or tumor necrosis factor (TNF) (Fig. 1c and Supplementary Fig. 1). These results demonstrate that TRIM21 detects intracellular AdV + Ab and activates NF-κB signaling.
Figure 1. TRIM21 senses intracellular Ab-bound virus.
(a) DNA binding ELISA showing NF-κB subunits p65 and p50 binding to consensus oligonucleotides 4 h post challenge of wild-type (WT) MEFs or Trim21-deficient MEFs transduced with empty vector (K21 EV) or expressing human Trim21 (K21 T21) with PBS, anti-adenovirus monoclonal antibody 9C12 (Ab), adenovirus (AdV) or adenovirus-antibody complex (AdV + Ab). (b) Induction of NF-κB luciferase reporter activity in wild-type or Trim21-deficient (K21) MEFs 7 h after challenge with a serial dilution of goat anti-adenovirus, AdV or adenovirus-antibody complex over PBS-treated controls. (c) Induction of NF-κB luciferase reporter activity in wild-type, Trim21-deficient and Trim21-deficient cells expressing human TRIM21. (d) Induction of NF-κB luciferase reporter activity in HeLa cells or HeLa cells depleted of TRIM21 by siRNA. Ab is pooled human serum IgG. (e) Induction of NF-κB luciferase reporter activity after challenge of Trim21-deficient MEFs transduced with empty vector or human TRIM21 with AdV, human serum IgM or AdV + IgM complex. (f) Hot-spot interactions between IgG Fc (cyan) and TRIM21 PRYSPRY domain (yellow) required for complex formation (based on PDB structure 2IWG). (g) Induction of NF-κB luciferase reporter activity in wild-type MEFs challenged with AdV incubated with recombinant 9C12 (r9C12) or point mutant N434D (r9C12 ND). (h) Induction of NF-κB luciferase reporter activity in AdV non-permissive EL4 cells and AdV permissive EL4 CAR 7 h after challenge. For panels a-e, g, h error bars represent SEM from three replicates.
TRIM21 recognizes antibodies of diverse species origin and class
TRIM21-IgG molecular interaction and ADIN are conserved among mammalian species10,13. HeLa cells infected with AdV incubated with human serum IgG activated NF-κB (Fig. 1d), suggesting TRIM21 signaling is similarly conserved. AdV + Ab induced a similar level of signaling to TNF, whereas poor activation was observed in cells depleted of TRIM21 or challenged with virus or antibody alone. Furthermore, cells of human (HeLa) and mouse (MEF) origin responded to both conspecific and heterospecific antibodies (Supplementary Fig. 2). Early stage humoral immune responses are governed by IgM, which also interacts with TRIM216. We observed NF-κB stimulation from AdV + Ab complex formed using pooled human IgM in a similar manner to IgG, but not from either component alone (Fig. 1e). No stimulation was observed in Trim21-deficient MEFs, confirming that Trim21-IgM recognition is able to initiate signaling. These data demonstrate that TRIM21 detection of IgG is conserved between human and mouse and can respond to antibodies of diverse species origin. Moreover, TRIM21 can detect antibody classes IgG and IgM.
Direct TRIM21-Fc molecular interaction is required for detection
Point mutations at the TRIM21-IgG binding interface prevent TRIM21 binding and disrupt intracellular neutralization10,13. To demonstrate that direct TRIM21-antibody interaction is also required for signaling, we made the point mutation N434D in the anti-hexon antibody 9C12, a residue at the TRIM21 binding site (Fig. 1f). NF-κB luciferase response following challenge by mutant N434D was impaired compared to wild-type recombinant 9C12 (Fig. 1g). To demonstrate that infection, rather than a cell surface interaction, is required for TRIM21-mediated signaling, we compared challenge of the AdV infection of a non-permissive cell line, EL4, to EL4 cells stably transduced with the coxsackie and adenovirus receptor (CAR), which renders the cells permissive for AdV infection (Supplementary Fig. 3). NF-κB luciferase induction was observed in both cell lines in response to LPS or TNF but only cells expressing CAR were responsive to AdV + Ab challenge (Fig. 1h). We therefore conclude that an intracellular molecular interaction between IgG and TRIM21 is required for signaling in response to AdV + Ab.
Signaling is dependent on TRIM21 RING domain
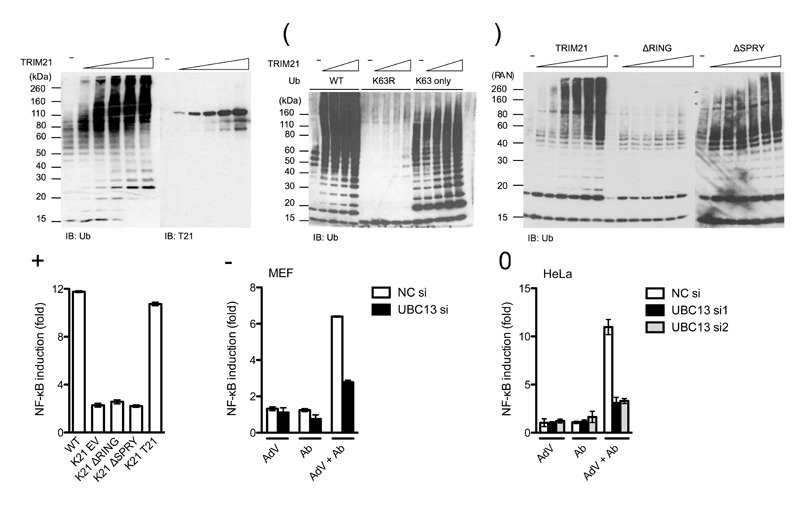
TRIM5α has been shown activate AP-1 and NF-κB signaling pathways, both constitutively within the LPS stimulation pathway and upon infection with retroviruses, by catalyzing the formation of free K63-linked ubiquitin chains12. In the presence of K63-specific E2 enzymes UBC13 and UEV1A, we found that TRIM21 catalyzed K63-linked chain formation in vitro(Fig. 2a). Both TRIM21 and the ubiquitin E2 conjugation enzymes remained unmodified suggesting that the resulting chains are unanchored (Fig. 2a and Supplementary Fig. 4). Ubiquitin chain formation was observed with wild-type or K63-only ubiquitin but not K63R, confirming that linkage is through lysine 63 (Fig. 2b). Ubiquitination was dependent on the RING domain (Fig. 2c) and accordingly, TRIM21 signaling was RING dependent, as Trim21-deficient MEFs expressing human TRIM21 lacking the RING domain (ΔRING) failed to activate NF-κB upon challenge with AdV + Ab (Fig. 2d). However, TRIM21 signaling, but not in vitro ubiquination activity was dependent on the PRYSPRY domain (Fig. 2c, d). This is suggestive of a greater complexity of regulation of TRIM21 activation in cells. Depletion of UBC13 using siRNA substantially reduced signaling in response to AdV + Ab in both MEF and HeLa cells (Fig. 2e, f). This reduction was similar to that observed upon challenge with TNF (Supplementary Fig. 5), a finding which is in accordance with UBC13’s reported involvement in TNF receptor signaling14. Together, these data show that TRIM21 catalyzes K63-linked ubiquitin chain formation in a RING-dependent manner and that deletion of the RING or depletion of the K63 E2 conjugating enzyme, UBC13, prevents efficient signaling in response to infection with antibody-coated adenovirus.
Figure 2. TRIM21 RING domain synthesizes K63-linked ubiquitin chains.
(a-c) Immunoblots of in vitro ubiquitination reactions with K63-specific E2 conjugation enzymes UBC13 and UEV1A with (a) titration of TRIM21, (b) ubiquitin or mutants K63R or K63 only and (c) full-length TRIM21 or RING (ΔRING) and PRYSPRY (ΔSPRY) domain deletions. The target of each immunoblot is indicated below each panel, TRIM21 (T21), Ubiquitin (Ub). (d) Induction of NF-κB luciferase reporter in wild-type (WT) or Trim21-deficient (K21) MEF cells transduced with indicated TRIM21 constructs (EV, empty vector) upon infection with AdV + Ab. (e) Induction of NF-κB luciferase reporter upon challenge with Ab, AdV or AdV + Ab in wild-type MEF treated with negative control siRNA (NC si) or UBC13-directed siRNA. (f) Induction of NF-κB luciferase reporter upon challenge with Ab, AdV or AdV + Ab in HeLa cells treated with control or one of two UBC13-directed siRNA treatments. For panels d-f, error bars represent SEM from three replicates.
TRIM21 signals via NF-κB, AP-1 and IRF pathways
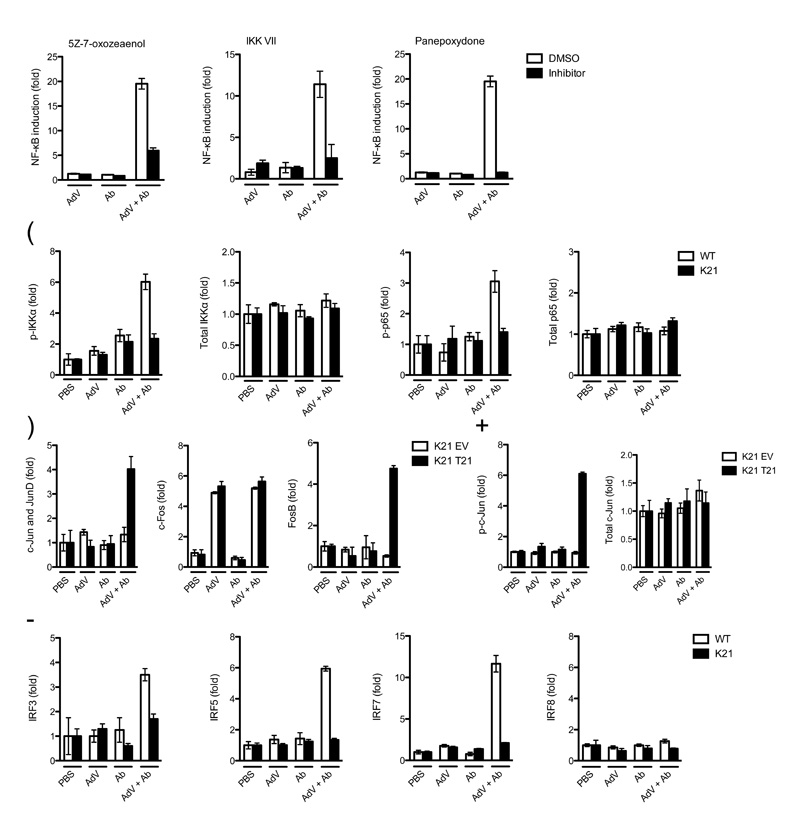
K63-linked ubiquitin chains can promote signaling via the kinase complexes TAK1-TAB1-TAB212,15 and IKKα-IKKβ-NEMO16,17. To test whether these complexes are activated by TRIM21, we used inhibitors of TAK1 (5Z-7-oxozeaenol) and IKKα (IKK VII). We also used an inhibitor of the NF-κB pathway at the downstream component, IκB (panepoxydone). NF-κB signaling in response to AdV + Ab was suppressed after treatment with any of the three inhibitors (Fig. 3a), confirming the involvement of both kinase complexes. Next, we analysed the phosphorylation state of individual pathway components. Upon AdV + Ab challenge, we observed an increase in both phosphorylated IKKα and NF-κB p65 in wild-type, but not in Trim21-deficient MEFs (Fig. 3b). Activation of the TAK1 complex can also stimulate the AP-1 transcriptional complex, composed of dimers of Jun and Fos family members. To test whether the AP-1 pathway is also stimulated by TRIM21, we measured Jun phosphorylation and the ability of Fos and Jun to interact with TPA response element (TRE) by DNA binding ELISA. c-Jun was phosphorylated and FosB and Jun bound TRE oligonucleotides only in wild-type, but not Trim21-deficient MEFs stimulated by AdV + Ab complex (Fig. 3c, d). Of interest, c-Fos activation was not TRIM21-dependent and was stimulated by both AdV alone and AdV + Ab, indicative of the activity of other pattern-recognition receptors. TRIM21 has been reported to modulate the activity of IRF transcription factors both positively18,19 and negatively20-22. We tested whether AdV + Ab recognition by TRIM21 was able to stimulate IRFs using a DNA binding assay. We found that IRF3, IRF5 and IRF7, but not IRF8, were specifically activated by AdV + Ab in a TRIM21-dependent manner (Fig 3e). Taken together, our data suggest that recognition of AdV + Ab by TRIM21 stimulates the three principle signaling pathways NF-κB, AP-1 and IRF.
Figure 3. TRIM21 signaling is dependent upon TAK1 and stimulates κB, AP-1 and IRF pathways.
(a) NF-κB luciferase induction in wild-type MEF cells when treated with DMSO or inhibitors 7 h post challenge with AdV, Ab or AdV + Ab. (b) ELISA showing induction of phosphorylated (p-) and total IKKα (Ser176 and Ser180) and p65 (Ser536) 4 hours post-challenge of wild-type (WT) or Trim21-deficient (K21) MEF cells. (c) DNA binding ELISA showing of induction of AP-1 components binding to consensus oligonucleotides 4 h post challenge of Trim21-deficient MEF cells transduced with empty vector (K21 EV) or human TRIM21 (K21 T21). (d) ELISA showing phosphorylated (Ser73) and total c-Jun induction after treatment with indicated stimuli in Trim21-deficient MEF cells or Trim21-deficient MEF cells transduced with human TRIM21. (e) DNA binding ELISA showing induction of IRF3, IRF5, IRF7 and IRF8 binding to DNA response elements 4 h post-infection in wild-type and Trim21-deficient MEF cells. For all panels, error bars represent SEM from three replicates.
Detection of virus-antibody complex elicits a proinflammatory response
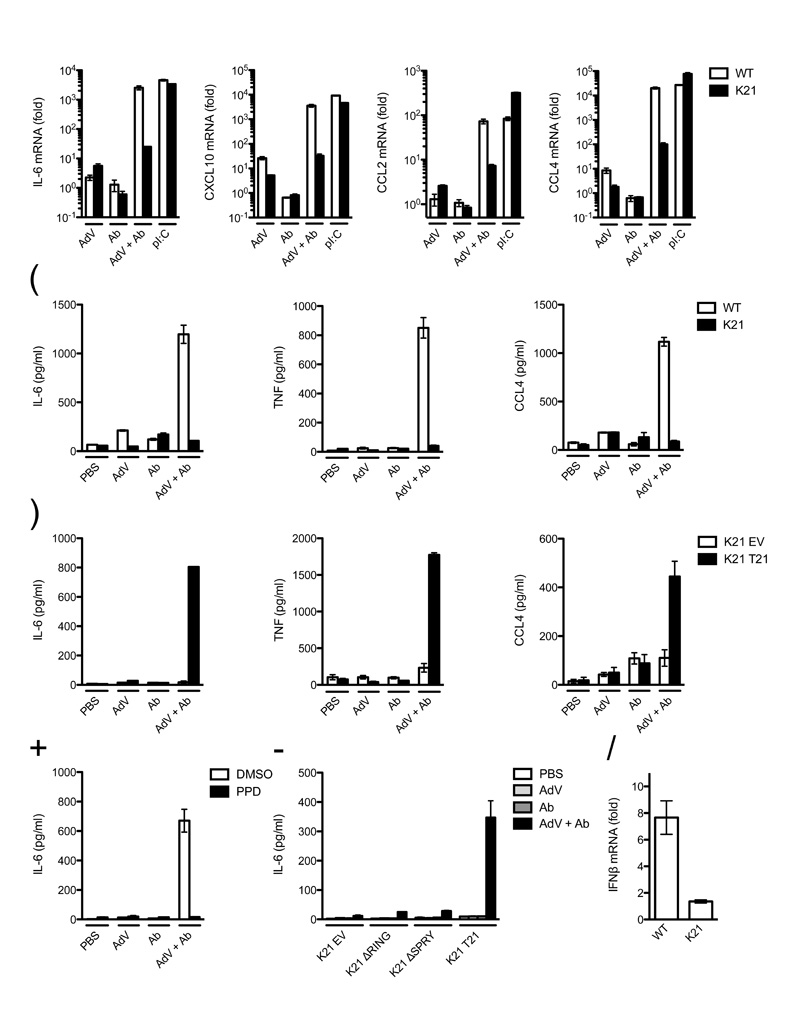
To determine the effect of TRIM21 and intracellular antibody immune activation on the inflammatory immune response, we compared inflammatory cytokine induction in wild-type and Trim21-deficient MEFs after AdV + Ab challenge. A 100 to 10,000-fold induction of interleukin 6 (IL-6), CXCL10, CCL2 and CCL4 transcripts was observed in wild-type cells after AdV + Ab challenge with substantially lower levels of induction by either component alone (Fig. 4a). In Trim21-deficient MEFs, cytokine mRNA levels were between 10 and 1,000-fold lower after challenge with AdV + Ab, whereas treatment with polyinosinic:polycytidylic acid (pI:C) gave similar induction to wild-type. Transcriptional upregulation in AdV + Ab challenged wild-type MEFs was accompanied by protein production of tested cytokines IL-6, CCL4 and TNF (Fig. 4b). Moreover, ectopic expression of TRIM21 in Trim21-deficient MEFs was sufficient to rescue cytokine expression in response to AdV + Ab (Fig. 4c).
Figure 4. TRIM21 signaling initiates production of proinflammatory cytokines.
(a) Quantitative RT-PCR showing induction of cytokine transcripts after challenge with human serum IgG (Ab), AdV, AdV + Ab or poly I:C in wild-type (WT) or Trim21-deficient (K21) MEFs. Data are represented as fold change above PBS-treated control at 7 h post-infection. (b) ELISAs showing concentration of cytokine protein in wild-type and Trim21-deficient MEF supernatant 72 h post-challenge with indicated treatment. (c) ELISAs showing concentration of cytokines produced by Trim21-deficient expressing empty vector (K21 EV) or human TRIM21 (K21 T21) 72 h post-challenge. (d) ELISA showing concentration of IL-6 of wild-type MEF cell supernatant incubated with DMSO or panepoxydone (PPD) 72 h after challenge with indicated treatment. (e) ELISA showing concentration of IL-6 in supernatant of Trim21-deficient MEF expressing the indicated constructs or transduced with empty vector 72 h post-challenge. (f) Quantitative RT-PCR showing induction of IFN-β transcripts in wild-type and Trim21-deficient MEFs by AdV + Ab over AdV only. For all panels error bars represent SEM from three replicates.
We tested if production of cytokines was dependent on the NF-κB signaling pathway by infecting wild-type MEF cells in the presence of the IκB inhibitor panepoxydone. Addition of the inhibitor prevented substantial IL-6 production after challenge with AdV + Ab (Fig. 4d). In addition, we tested whether IL-6 production was TRIM21 domain-dependent. Expression of full-length TRIM21 was sufficient to recover IL-6 production in Trim21-deficient MEFs challenged with AdV + Ab, whereas empty vector or expression of TRIM21 constructs lacking the RING or PRYSPRY domains was not (Fig. 4e). We tested whether activation of signaling by Trim21 was sufficient to induce interferon production. Challenge of wild-type MEFs with AdV + Ab resulted in upregulation of IFNβ (Fig. 4f). In contrast, little upregulation was observed in Trim21-deficient MEFs. These results demonstrate that TRIM21-mediated detection of antibody-bound antigen induced a proinflammatory state in the cells.
Intracellular antibody recognition by TRIM21 triggers an antiviral state
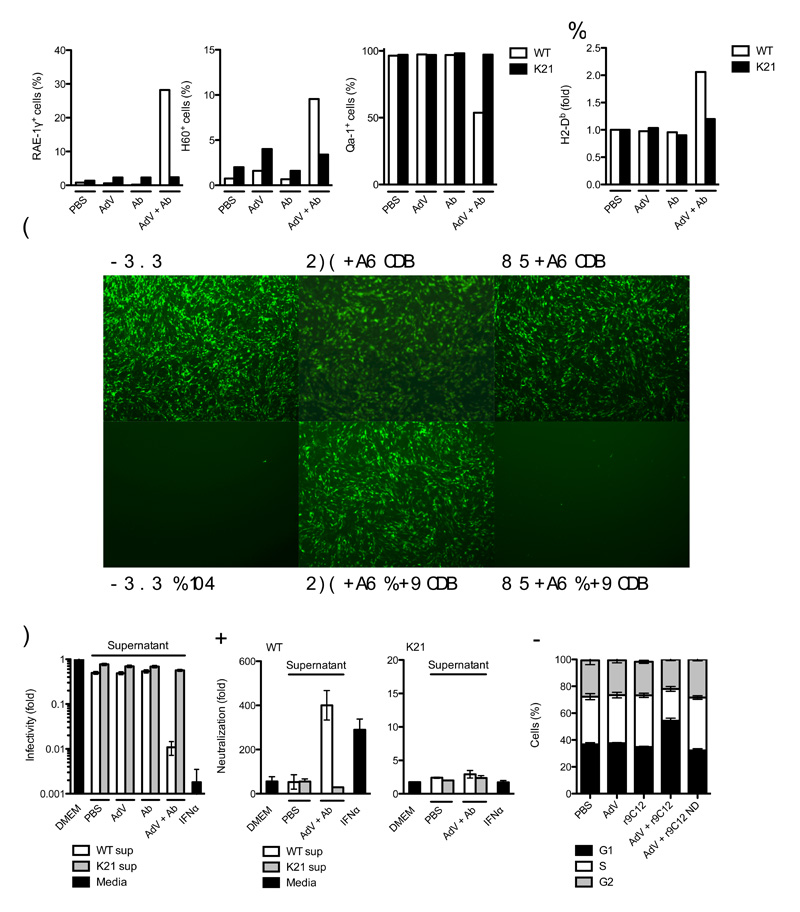
The antiviral state encompasses changes in cell physiology that reduce viral infection and replication and flag infected cells for destruction by leukocytes. For instance, infected cells can stimulate effector T cells and natural killer (NK) cells by upregulating the major histocompatibility complex (MHC) class I and MHC class I-like cell surface molecules such as NKG2 ligands23. We investigated whether TRIM21 activation is sufficient to trigger such an antiviral state. AdV + Ab infection resulted in a TRIM21-dependent upregulation of RAE-1γ and H60 (mouse NKG2D ligands) and downregulation of Qa-1 (mouse HLA-E homolog) (Fig. 5a). Normalized expression of MHC class Ia H2-Db was elevated in wild-type MEFs upon infection with AdV + Ab, whereas there were no substantial changes in Trim21-deficient MEFs or in wild-type MEFs treated with AdV or Ab alone (Fig. 5b).
Figure 5. Detection of intracellular antibody-bound pathogens promotes an antiviral state.
(a) Fluorescence-activated cell sorting (FACS) data showing percent of wild-type (WT) or Trim21-deficient (K21) MEF cells positive for surface expression of NKG2D ligands RAE-1γ and H60 and NKG2A ligand Qa-1. (b) FACS data showing change in fluorescence intensity after staining for surface MHC class Ia allele H2-Db expression over PBS-treated control in wild-type and Trim21-deficient MEFs. (c, d) Sindbis GFP virus infection of wild-type MEF ‘reporter’ cells which were incubated with fresh media (DMEM), media supplemented with mouse IFNα or supernatant (sup) derived from wild-type or Trim21-deficient MEFs challenged with AdV, Ab or AdV + Ab. (c) Fluorescence microscope images showing wild-type MEF reporter cell monolayers 12 h after Sindbis GFP infection. (d) Percent GFP positive wild-type reporter MEF cells relative to DMEM-treated control. (E) Fold neutralization of adenovirus by monoclonal antibody 9C12 relative to PBS-treated adenovirus after treatment of wild-type and Trim21-deficient reporter cells with Fresh media (DMEM), media supplemented with IFNα or supernatant transferred from wild-type or Trim21-deficient MEFs that were either PBS-treated or challenged with AdV + Ab. (f) Propidium iodide staining showing proportion of cells in G1, S or G2 24 h after challenge with AdV or AdV complexed with recombinant 9C12 (r9C12) or 9C12 bearing a point mutation N434D (r9C12 ND). For panels a, b, d-f, error bars represent SEM from three replicates.
To examine whether the cytokines produced upon TRIM21 stimulation are sufficient to reduce viral infection, we transferred supernatant from wild-type and Trim21-deficient MEFs which had been challenged with PBS, antibody, AdV or AdV + Ab onto fresh ‘reporter’ MEF cells. As a measure of host cell permissivity to viral infection, we challenged reporter cells with a Sindbis virus GFP vector that is sensitive to IFNα treatment. Supernatant from AdV + Ab-challenged wild-type cells reduced Sindbis virus infection in reporter MEF cells to a degree similar to IFNα treatment (Fig. 5c, d). In contrast, Trim21-deficient supernatants did not substantially reduce infectivity of reporter MEFs. These data demonstrate that the cytokines produced following stimulation of TRIM21 are sufficient to confer an antiviral state.
TRIM21 is upregulated following IFNα treatment, resulting in the ability to alter experimentally the levels of antibody-dependent intracellular neutralization10. Transfer of PBS or AdV + Ab treated Trim21-deficient MEF supernatant onto fresh reporter MEF cells did not alter AdV neutralization levels, as measured by the infectious titer of PBS-treated virus divided by the titer of Ab-treated virus (Fig. 5e). However transfer of supernatant from wild-type cells that had been challenged with AdV + Ab substantially increased neutralization in reporter wild-type MEF cells. The increase in neutralization was the result of TRIM21’s activity as an effector of ADIN, as no increase in neutralization was observed when Trim21-deficient MEFs were used as reporter. These experiments demonstrate that the TRIM21 signaling following challenge with antibody-bound adenovirus results in an increase in neutralization of the same pathogen.
Cells in S phase provide optimal conditions for virus replication24. It has been demonstrated that activation of AP-1 and IRF pathways or cytokine production can induce cell cycle arrest25-27. We tested whether stimulation of TRIM21 was able to alter the proportion of cells in G1, S and G2 phases, by using propidium iodide staining. Wild-type MEF cells responded to challenge with AdV + Ab by a reduction in the percent of cells in S phase (Fig. 5f). However, antibodies bearing the N434D mutation, which is unable to induce TRIM21 signaling, were unable to provoke this change. Together, these experiments show that TRIM21 stimulation renders cells refractory to virus infection and modulates expression of cell surface ligands recognized by professional immune cells.
TRIM21 recognizes intracellular viruses and bacteria
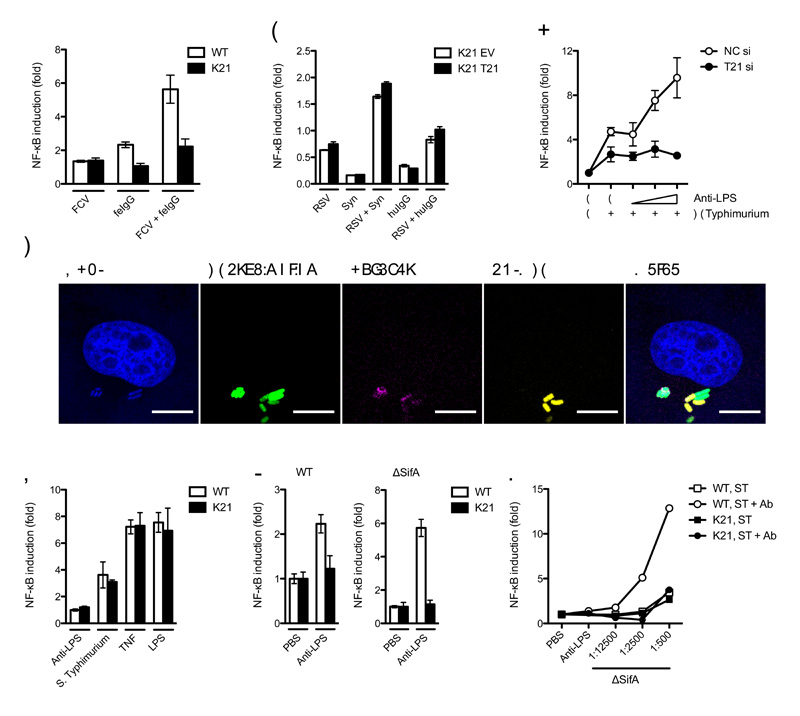
Next we investigated whether TRIM21 signaling is pathogen-dependent. We reasoned that a cytoplasmically accessible Ab-bound pathogen should provoke signaling whereas a pathogen that sheds its antibody before penetrating the cytosol should not. We chose two contrasting viruses to test this hypothesis; the non-enveloped feline calicivirus (FCV), which may carry antibodies with it into the cell upon infection and enveloped respiratory syncytial virus (RSV), which fuses at or near the plasma membrane28 and should lose its attached antibodies during membrane fusion. FCV activated a NF-κB luciferase reporter in the presence of pooled feline serum IgG in wild-type but not Trim21-deicient MEFs. In contrast, infection with RSV pre-incubated with antibody resulted in similar NF-κB luciferase reporter activitation in wild-type and Trim21-deficient MEFs (Fig. 6a, b). Similarly, and in contrast to AdV6,10, incubation of RSV with neutralizing antibody resulted in similar levels of neutralization between untreated, TRIM21 shRNA-treated or IFNα-treated HeLa cells (Supplementary Fig. 6).
Figure 6. TRIM21 promotes NF-κB signaling in response to viral and bacterial pathogens.
(a) NF-κB luciferase reporter induction in response to infection with feline calicivirus (FCV) with feline (fe) serum IgG in wild-type (WT) or Trim21-deficient (K21) MEFs. (b) Respiratory syncytial virus (RSV) with Synagis (Syn) or human (hu) serum IgG in Trim21-deficient MEFs expressing human TRIM21 (K21 T21) or empty vector (K21 EV). (c) Confocal micrographs of HeLa cells 4 h post-infection with Ab-coated GFP-expressing Salmonella enterica serovr Typhimurium (S. Typhimurium). Staining is for anti-LPS Ab and TRIM21. (d) NF-κB luciferase reporter induction in HeLa treated with control (NC) or TRIM21-directed (T21) siRNA 7 h post-challenge with Salmonella in the presence of increasing concentrations of anti-LPS Ab. (e) NF-κB luciferase reporter induction after challenge of wild-type or Trim21-deficient MEFs with Salmonella, LPS or TNF. (f) NF-κB luciferase induction in wild-type or Trim21-deficient MEFs after challenge with Ab-coated wild-type and ΔSifA Salmonella over uncoated bacteria. (g) NF-κB luciferase induction after infection of wild-type MEF or Trim21-deficeint MEFs with a dilution series of ΔSifA Salmonella incubated with PBS or anti-LPS Ab. For panels a, b, d-g, error bars represent SEM from three replicates.
To determine whether antibody-coated bacteria can also be sensed by TRIM21, we studied infection by the facultative intracellular bacterium Salmonella enterica serovar Typhimurium, which can reside within vacuoles or in the cytosol29,30. Confocal microscopy revealed that Salmonella carries Ab into the cell upon infection and that TRIM21 co-localizes with a subset of Ab-bound bacteria (Fig. 6c). Furthermore, Salmonella infection of HeLa cells provokes antibody dependent NF-κB signaling that is inhibited upon depletion of TRIM21 by siRNA (Fig. 6d). In MEF cells, although we observed TRIM21 and antibody-independent NF-κB stimulation in response to infection (Fig. 6e), pre-incubation of bacteria with Ab resulted in a further increase in signaling in wild-type, but not Trim21-deficient MEFs (Fig. 6f). ΔSifA Salmonella, a strain that loses vacuolar integrity resulting in a greater proportion of bacteria in the cytosol31,32, stimulated greater NF-κB activation than wild-type Salmonella when coated with Ab (Fig. 6f). The signaling induced by ΔSifA Salmonella was TRIM21 and Ab-dependent and exceeded the signaling induced by uncomplexed bacteria at all concentrations assayed (Fig. 6g). Together these data suggests that TRIM21 can activate innate immune signaling upon sensing any cytosolic antibody-bound pathogen.
TRIM21 signaling is independent of Fc receptors, PRRs and ADIN
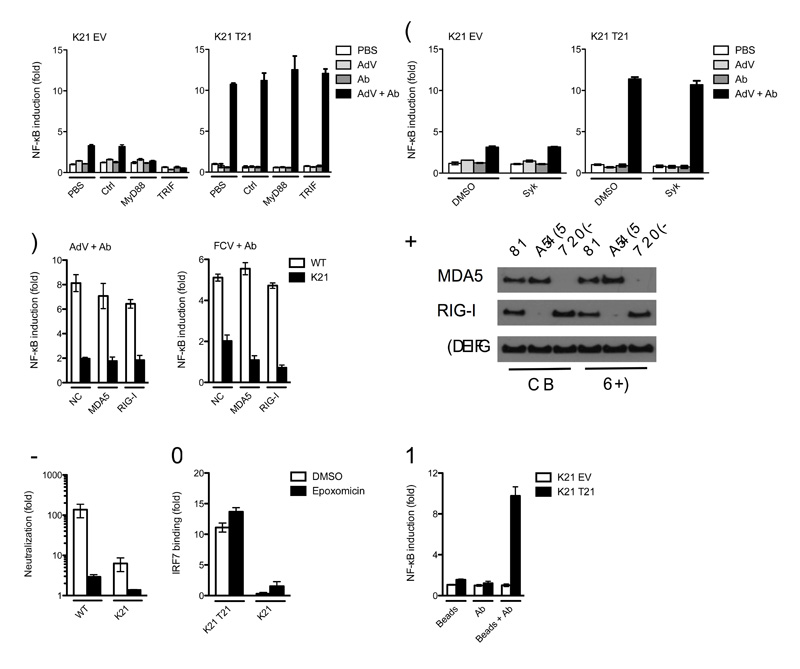
To exclude the possibility that signaling is due to recognition of AdV + Ab immune complexes by FcR or innate sensors other than TRIM21, we performed infection experiments in the presence of specific inhibitors or knockdown of these pathways. Addition of MyD88 and TRIF inhibitors efficiently blocked signaling in response to LPS (Supplementary Fig. 7), but had no effect on AdV + Ab stimulation of NF-κB (Fig. 7a), confirming that signaling is TLR-independent. Inhibition of cytosolic tyrosine kinase Syk prevented signaling in response to cross-linked surface bound antibody (Supplementary Fig. 7) but did not prevent signaling in response to AdV + IgG or AdV + IgM (Fig. 7b), confirming that TRIM21 signaling is FcR-independent. Similarly, knockdown of intracellular nucleic acid receptors, MDA-5 and RIG-I failed to impair signaling in response to antibody-bound AdV or FCV (Fig. 7c,d).
Figure 7. TRIM21 signaling is independent of TLR, FcR, ADIN and PAMPs.
(a) NF-κB luciferase reporter induction in Trim21-deficient MEFs expressing empty vector (K21 EV) or human TRIM21 (K21 T21) with control or inhibitor peptides of TLR signaling pathway components MyD88 and TRIF. (b) NF-κB luciferase reporter induction in MEFs after treatment with DMSO or an inhibitor of FcR kinase Syk. (c) NF-κB luciferase reporter induction in wild-type (WT) or Trim21-deficient MEFs after control (NC), MDA-5-directed or RIG-I-directed siRNA treatment by antibody-coated AdV or FCV. (d) Immunoblot for MDA-5, RIG-I and β–actin from wild-type or Trim21-deficient MEFs after control, MDA-5-directed or RIG-I-directed siRNA treatment. (e) Fold neutralization of adenovirus, measured as relative infectivity of PBS-treated AdV over 9C12-treated AdV on wild-type and Trim21-deficient MEFs following treatment with proteasome inhibitor epoxomicin or DMSO. (f) DNA binding ELISA showing induction of IRF7 activation over PBS-treated controls in DMSO or epoxomicin treated Trim21-deficient MEFs expressing empty vector or human TRIM21 4 h after challenge with AdV + Ab. (g) NF-κB luciferase reporter induction in Trim21-deficient MEFs expressing empty vector or human TRIM21 following transfection of biotin beads, anti-biotin Ab or complexed beads and Ab. Error bars in panels a-c, e, g represent SEM of three replicates.
During ADIN, TRIM21 directs incoming virions to the proteasome, potentially exposing PAMPs that could initiate or promote immune signaling in the cytosol. We investigated whether ADIN is required for immune signaling by inhibiting the proteasome with epoxomicin. We measured neutralization by incubating AdV with PBS or 9C12 and dividing the infectious titer of PBS-treated virus by that of 9C12-treated virus. Treatment with epoxomicin reversed neutralization of AdV + Ab in wild-type MEF cells from approximately 150-fold to 3-fold and in Trim21-deficient MEF cells, residual neutralization of 6-fold is almost completely ablated (Fig. 7e). Proteasomal function is required for NF-κB and AP-1 processing but not IRF7 activation. We therefore examined whether IRF7 DNA binding is preserved during proteasome inhibtion in Trim21-deficient expressing empty vector or human TRIM21. Addition of epoxomicin did not prevent activation of IRF7 (Fig. 7f), demonstrating that TRIM21 signaling is independent of ADIN and can occur under conditions of proteasome inhibition.
The preceding data suggest that the minimum requirement for TRIM21-mediated immune signaling is the presence of antibody in the cytosol. To test this, we transfected cells with biotin-labeled latex beads pre-incubated with anti-biotin Ab. Such Ab-coated beads are detected by TRIM21 and become positive for ubiquitin6. Transfection of antibody-coated beads stimulated strong NF-κB reporter induction in Trim21-deficient MEFs expressing human TRIM21 (Fig. 7g). However, no signaling was observed with either beads or antibody alone, or Trim21-deficient MEFs transduced with empty vector. This result confirms that TRIM21 detection of cytosolic antibody is sufficient to activate immune signaling and is PAMP and ADIN-independent.
TRIM21 activates NF-κB and cytokine production in primary cells
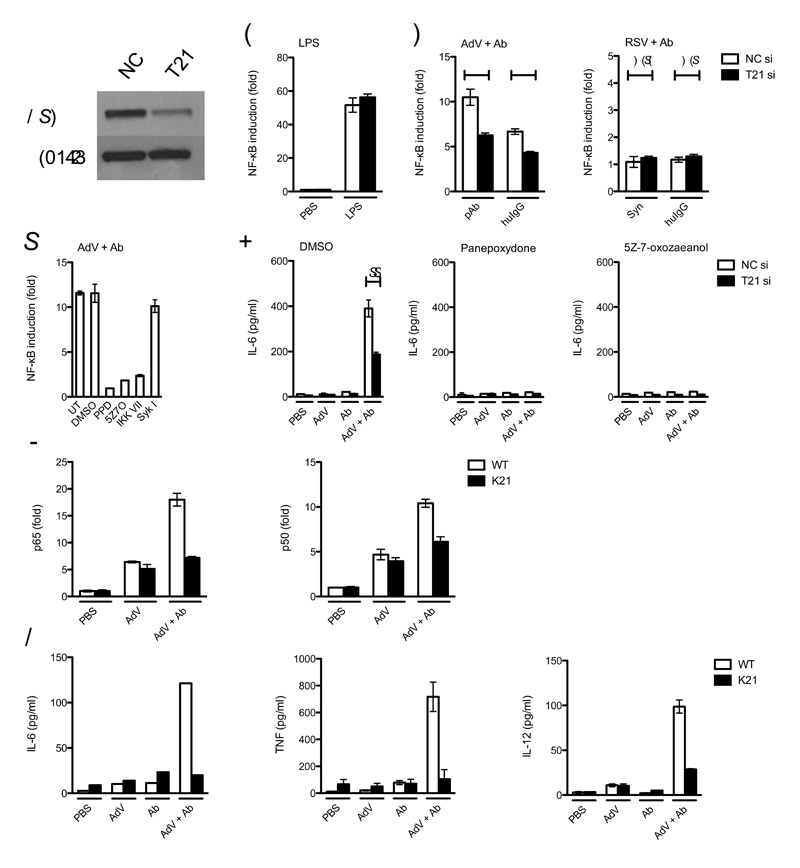
We next investigated whether TRIM21 is able to activate immune signaling in primary cells. Human adenovirus type 5 causes infection of the upper and lower respiratory system33. We therefore examined infection of primary normal human lung fibroblasts (NHLFs). An NF-κB reporter was introduced into NHLFs by lentiviral transduction and TRIM21 was depleted by siRNA treatment alongside siRNA controls (Fig. 8a). Whilst LPS treatment induced NF-κB activation regardless of TRIM21 levels (Fig. 8b), the response to AdV + Ab was significantly diminished by TRIM21 depletion (Fig. 8c). NHLFs challenged with antibody-complexed RSV resulted in no greater NF-κB activation than when treated with RSV alone, confirming that cytoplasmically-accessible antibody is required for activation. As with infection of MEF cells, TAK1 inhibitor 5Z-7-oxozeaenol, IKK inhibitor VII and IκB inhibitor panepoxydone prevented NF-κB activation by AdV + Ab in NHLFs, confirming the involvement of these pathway components. Also, in a similar manner to MEF infection, inhibition of Syk had no effect on NF-κB activation, excluding FcR-mediated activation in NHLFs (Fig. 8d).
Figure 8. TRIM21 mediates an inflammatory response in primary human and mouse cells.
(a) Immunoblot for TRIM21 and β-actin in normal human lung fibroblasts (NHLF) treated with control (NC) or TRIM21-directed (T21) siRNA. NF-κB luciferase reporter induction in control or TRIM21-directed siRNA (si) treated NHLF after treatment with (b) LPS or (c) AdV complexed with goat polyclonal antibody (pAb) or human serum IgG (huIgG) or RSV complexed with Synagis or huIgG. Data are represented as fold increase in NF-κB induction of virus-antibody complex over PBS-treated virus. For both adenovirus antibodies, P < 0.01, n=6, unpaired t-test (*); for both RSV antibodies, the difference was not significant (N.S.). (d) NHLF NF-κB luciferase induction after treatment with the indicated inibitors. Untreated (UT), panepoxydone (PPD), 5Z-7-oxozeaenol (5Z7O), IKK inhibitor (IKK VII), Syk inhibitor (Syk I). For panels b-d error bars represent SEM of six replicates. (e) ELISA showing IL-6 concentration in supernatant of NHLFs treated with DMSO or inhibitors PPD or 5Z7O. The concentration of IL-6 produced was significantly reduced after TRIM21 knockdown P < 0.001, n=3, unpaired t-test (**). (f) DNA binding ELISA showing binding of NF-κB subunits p65 and p50 4 h post-infection in bone marrow derived macrophages (BMDM) collected from C57BL/6 WT or Trim21−/− mice aged 8-10 weeks. (g) ELISA showing cytokine concentration in supernatant from BMDM 72 h post-challenge. Error bars in panels e-g represent SEM of three replicates.
To confirm that TRIM21 dependent signaling is sufficient to activate cytokine production in primary cells, we measured IL-6 secretion by infected NHLFs. Strong IL-6 production was observed in cells challenged with AdV + Ab and this could be diminished by TRIM21 depletion or by inhibition of the NF-κB pathway (Fig. 8e). Of note, adenovirus infection itself was not sufficient to activate IL-6 secretion in NHLFs, indicating the potential importance of TRIM21-antibody signaling in detection of adenovirus in the lung. These experiments demonstrate that innate immune detection of adenovirus in lung fibroblasts is dependent on stimulation of TRIM21, signaling via TAK1, IKKα and IκB and that NF-κB activation is a requirement for the production of proinflammatory cytokine IL-6.
Next we examined signaling in primary immune cells. We infected bone marrow-derived macrophages (BMDM) harvested from wild-type C57BL/6 or Trim21-deficient mice34 and prepared by selection for CD11b using magnetic antibody selection. To study the effects of infection only, we inhibited potential FcR-mediated responses by performing experiments in the presence of Fc fragments. Challenge with AdV + Ab resulted in NF-κB activation in BMDMs derived from wild-type but not Trim21-deficient mice as measured by DNA binding assays for p65 and p50 (Fig. 8f). Unlike other cell types tested in this study, some NF-κB activation was observed in response to AdV in the absence of antibody, indicative of other PRRs in macrophages. However, TRIM21 activation was required for significant levels of cytokine production in BMDM. IL-6, TNF and IL-12 were present in the supernatant of wild-type-derived but not Trim21-deficient cells after challenge (Fig. 8g). These results demonstrate that TRIM21 detects intracellular antibody-bound pathogens in primary cells, including professional cells of the immune system, leading to the production of proinflammatory cytokines.
Discussion
We have shown that antibodies provide a context-dependent danger signal when associated with intracellular pathogens. Cytosolic antibody-bound particles are detected by TRIM21, triggering immune signaling. We have demonstrated that TRIM21 can catalyze the formation of free K63 ubiquitin chains via the E2 conjugating enzyme UBC13 in a RING-dependent manner and promote signaling through three key immune pathways, NF-κB, AP-1 and the IRF family. TRIM21 activation of these pathways results in proinflammatory cytokine production, modulation of cell-surface ligands and adoption of an antiviral state. Antibody sensing by TRIM21 occurs in primary cells of both immune and non-immune origin.
We have defined the minimal requirements for TRIM21 sensing, the mechanism by which it occurs and the characteristics of susceptible pathogens. TRIM21 signaling is independent of its neutralization activity (ADIN), Fc receptors and known PAMPs, such as the TLRs, RIG-1 and MDA5 but dependent upon TAK1, IKKα and IκB. TRIM21 is activated by the presence of antibody in the cytosol and PAMP-free antibody-coated latex beads are sufficient to induce signaling. The full range of entities that can be detected by TRIM21 remains to be determined. However we have shown that both non-enveloped viruses and intracellular bacteria are both capable of carrying antibody into the cytosol and are targets for TRIM21. It is possible that other classes of pathogens such as eukaryotic parasites are similarly recognized. Given the consequences of recognition, evading TRIM21 detection may be a requirement for successful invasion by certain pathogens. We have shown that virus lifecycle features such as cell entry mechanism can determine susceptibility to TRIM21. For instance, the enveloped virus RSV that sheds antibodies upon membrane fusion escapes detection. Some viruses such as the Picornaviridae (eg rhinovirus), although non-enveloped, may avoid the cytoplasmic detection of capsid-bound antibodies by the manner in which they escape their endosome. Entry for rhinovirus involves the fusion of the virus capsid with the endosome resulting in a size-selective pore through which viral genomic RNA is ejected35. Future research should ascertain whether separation of capsid and RNA is sufficient for evasion of TRIM21-mediated neutralization and signaling. Pathogens may also have evolved specific antagonists that prevent activation of, disable or degrade TRIM21.
Identification of intracellular antibody-bound pathogens as ligands for TRIM21 should help elucidate contradictory findings of negative18,19 and positive20-22 regulation of IRF activity by TRIM21. Differences in the relative levels IRF family member activation can greatly influence cytokines production. For instance IRF5 and IRF3 compete for binding at the Il12b locus, with IRF5 activating and IRF3 suppressing activation36,37. In this study we observed greatest activation of IRF5 after challenge with AdV + Ab and, accordingly, observed IL-12 production. IL-12, along with other cytokines strongly upregulated in this study (CXCL10, CCL4 and IFN-γ) are likely to bias T cell activation towards a TH1-type response. However, it is likely that simultaneous recognition of different pathogens by TRIM21 and PRRs elicits different IRF activation profiles and subsequent immune responses appropriate to the nature of the pathogen. Our findings have revealed that TRIM21 concurrently targets viruses for destruction and alerts the body to infection. This dual signaling and effector capability is shared by TRIM5α, which is able to direct retroviral capsid to the proteasome as well as activate innate immunity12,38, but distinct from classical PRRs like the TLRs, which do not themselves exert antimicrobial activity. TRIM21 and TRIM5α may therefore exemplify a new emerging class of innate immune sensors, of which other TRIM proteins and pathogen restriction factors are likely candidates. There may be commonalities between TRIM21 and TRIM5α mechanisms but certain key differences are known to exist. For instance, proteasome inhibition prevents TRIM21 neutralization, whereas it does not relieve TRIM5α restriction. TRIM5α has a high rate of protein turnover but exchange of TRIM5α RING domain with that of TRIM21 increases protein half-life considerably yet preserves anti-retroviral activity39. Low rates of endogenous TRIM21 turnover have also been observed6, marking a potentially important phenotypic difference between the paralogues. Understanding differences such as these will therefore likely be of value in determining the molecular mechanisms of TRIM activity.
The discoveries of TRIM21-mediated signaling and neutralization represent important considerations in the development of viral vectors for vaccination, since pre-existing humoral immunity to the vector may have consequences in the effectiveness of delivery and in the nature of the immunity conferred due to the combined activities of TRIM21. Pre-existing humoral immunity to the adenovirus type 5 vector used in the Step trial, immunizing against HIV-1 components, resulted in qualitatively different HIV-specific immune responses, with a substantial impairment to CD8+ T-cell immunity40,41. Furthermore, the safety of adenovirus vectors has been a cause for concern, given the robust innate immune responses and ‘cytokine storms’ witnessed in certain gene therapy trails42. Understanding the role of TRIM21 in such responses may inform strategies to mitigate their effects. Finally, it may be feasible to advantageously exploit TRIM21-mediated detection of vectors to enhance or direct the nature of acquired immunity.
Materials and Methods
Cells
HeLa and MEF cell lines were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum, penicillin at 100 U/ml and streptomycin at 100 μg/ml. MEF cells were obtained from WT or TRIM21 knockout C57/B6 mice34 and have previously been described10. Bone marrow-derived macrophages (BMDM) were isolated from mouse femurs and selected via CD11b magnetic sorting (Miltenyi Biotec). Cells were maintained as above and 50 ng/ml recombinant mouse GM-CSF (Miltenyi Biotec) and used in assays 7 d post-harvest. Normal Human Lung Fibroblasts (NHLFs) were obtained from Lonza, and were maintained in Fibroblast Growth Medium 2 (Lonza), supplemented with 10% fetal calf serum, 0.1% insulin, 0.1% amphotericin-B and 0.1% gentamicin.
Viruses
E1-deleted adenovirus GFP vector was prepared by CsCl banding as previous6. Sindbis virus GFP vector was produced by in vitro transcription of linearlized plasmids DH-BB and pSin-eGFP using mMessage mMachine SP6 kit (Life Technologies) and transfection of RNA into 293T cells using Lipofectamine 2000. rgRSV was expanded in HeLa cells and cell-free supernatant was harvested and frozen 2 d post-infection. FCV strain F9 was expanded in feline embryonic airway (FEA) cells and harvested 24 h post-infection.
Retroviral transduction
Human TRIM21 and domain deletion mutants ΔRING and ΔPRYSPRY and human CXADR were PCR amplified and cloned into retroviral vector pDON-AI-2-Neo (Takara Biosciences) using NotI and SalI restriction sites. Stably transduced cells were selected using G418 at 500 μg/ml.
Small interfering RNA knockdown
MEF cells stably transfected with NF-κB luciferase reporter were transfected twice with UBC13 siRNA6 (Qiagen) or control oligo (Ambion) using RNAiMAX (Life Technologies). HeLa cells were transiently transfected with pGL4.32 NF-κB luciferase followed after 2 d by transfection with On-Target UBC13 (Thermo Scientific) or control siRNA. After a further 2 d cells were used in downstream assays. MEF cells were transiently transfected with psiRNA-mRIG-I and psiRNA-mMDA-5 plasmids with GFP (Invivogen) along with NF-κB luciferase using Lipofectamine 2000. TRIM21 siRNA was carried out as previous3.
Antibodies and Immunoblotting
Pooled human serum IgG and IgM were obtained from Athens Research and Technology, Georgia, USA and pooled cat serum IgG was obtained from Equitech-Bio, Texas USA. Synagis (Palivizumab) was obtained from MedImmune.
Luciferase reporter assay
Cells stably transfected with pGL4.32 NF-κB luciferase were plated at 1 × 104 per well in 96 well plates. After 24 h, virus and Ab were incubated 1:1 for 1 h before addition to cells. Viruses were added to cells at the following titers: AdV, 7.5 × 105 IU per well; RSV, 2 × 103 IU per well; FCV, 2.8 × 105 TCID50 per well. For controls, LPS was used at 50 pg/ml or TNF at 10 pg/ml. Cells were incubated for 7 h at 37° C before addition of 100 μl Steadylite Plus luciferase reagent (Perkin Elmer) and reading on a BMG Pherastar FS platereader. Data are represented as fold increase over the mean of triplicate PBS-treated control wells unless stated otherwise. NHLFs were infected with Cignal Lenti NF-κB reporter (Qiagen), and treated in the same manner. AdV + IgM complexes were prepared by centrifugation at 28,000 xg at 4° C over 30% sucrose. Biotin-coated latex beads (Sigma) in complex with goat anti-biotin Ab (Sigma) were transfected using Lipofectamine 2000 (Life Technologies). Overnight cultures of wild-type or ΔsifA (strain 12023), Salmonella enterica serovar Typhimurium were diluted 1/33 into 5 ml fresh LB medium and grown at 37 °C, 400 rpm for 3.5 h. Bacteria were incubated with anti-LPS (AbD Serotec) for 20 min before further dilution and addition of 5 μl per well. After 1 h, media was aspirated and cells were washed with PBS before addition of DMEM-FCS supplemented with gentamycin at 50 μg/ml. Luciferase was read as above.
Cytokine Analysis by Quantitative RT-PCR
Cells were infected as above. PolyI:C (Sigma) was transfected at 100 ng per well using Lipofectamine 2000. Cells were incubated at 37° C for 7 h, before washing with PBS and preparation of cDNA using Cells to CT Kit (Ambion). Gene expression was monitored by TaqMan Gene Expression Assays (Applied Biosystems) on a StepOnePlus Real Time PCR System (Life Technologies). Relative expression was quantified using the 2−ΔΔCt method.
ELISAs
MEF cells were plated and infected as above. Phosphorylated transcription factors were measured using Phosphotracer IKKα, IκBα, NF-κB kit (Abcam). Analysis of total protein abundance was performed using IKKα (eBioscience) and NF-κB (Insight Biotechnology) ELISA kits. For analysis of cytokine production, supernatant was harvested 72 h post-infection and analysed by ELISA kits: IL-6 (Life Technologies), IL-12 (Life Technologies), TNF-α (AbFrontier) and CCL4 (Abnova).
DNA binding assay
1.5 × 106 MEF cells or 7.5 × 105 BMDMs were plated in 10cm tissue culture dishes. 3.0 × 107 IU adenovirus was incubated with 9C12 at 100 μg/ml for 1 h. Cells were harvested 4 h post-infection by scraping and subjected to the nuclear fraction protocol of the TransAM NF-κB, IRF3, IRF7 and AP-1 Family ELISA kits (Active Motif). The IRF5 and 8 assays were carried out by binding onto the IRF3 ELISA plate, followed by detection with CHIP grade anti-IRF5 and IRF8 antibodies (Abcam). 2.5 μg nuclear extracts protein was permitted to bind to immobilized oligonucleotides for 1 h.
Inhibitors
Pepinh-MyD (Invivogen) and Pepinh-TRIF (Invivogen) were added 2 h before infection alongside control peptide at 50 μM. 0.5 μM 5Z-7-Oxozeaenol (Sigma), 2 μg/ml panepoxydone (Enzo Life Sciences), 600 nM Syk I (Sigma), 10 μM Syk Inhibitor III (Sigma), 200 nM IKK Inhibitor VII (Millipore) and 2 μM epoxomicin (Sigma) were added 1 h prior to infection.
In vitro ubiquitylation assay
In vitro assays were carried out as described6. UBA1, ubiquitin and E2 enzymes were obtained from Boston Biochem. Full-length, ΔSPRY and ΔRING recombinant TRIM21 were expressed as MBP-fusion proteins in E. coli and purified using amylose resin and size-exclusion chromatography6. Reaction mixtures were incubated at 37 °C for 1 h.
Confocal microscopy
HeLa cells were infected with wild-type Salmonella as above. After 4 h, cells were fixed with 4% paraformaldehyde before permeabilization with 0.1% Triton X-100 and blocking with 5% BSA. Labelling was detected using AlexaFluor-conjugated secondary antibodies (Life Technologies). Coverslips were imaged using a Jena LSM 710 microscope (Carl Zeiss MicroImaging).
Cell Surface Staining
MEF cells were infected with AdV + Ab complexes and harvested 18 h post-infection. Labelling was performed with anti-RAE-1γ-Alexa Fluor 647 (BioLegend), anti-H60-phycoerythrin (PE) (R&D Systems), anti-H2D2-PE (Abnova) and mouse anti-Qa-1 (BD Biosciences) followed by Hamster anti-mouse-PE-Cy7 (BD Biosciences). FACS analysis was performed using a Fortessa (BD Biosciences).
Cell Cycle Progression Analysis
MEFs were infected as above and fixed in 70% ethanol 16 h post-infection. Cells were resuspended in PBS, treated with 10 U/ml RNase One (Promega) and stained with 40 μg/ml propidium iodide (Life Technologies). Data were fitted using FlowJo (Tree Star).
Supplementary Material
Acknowledgements
We thank Mark Peeples Ph.D for providing RSV, Nationwide Children’s Hospital, Ohio, USA and Dr David Brown, University of Cambridge, Cambridge, UK for providing feline calicivirus.
This work was funded by Medical Research Council Grant U105181010 and European Research Council Grant 281627-IAI. J.C.H.T was funded by the Frank Edward Elmore Fund, University of Cambridge School of Clinical Medicine.
Footnotes
Author Contributions W.A.M. & L.C.J. initiated the study. All authors conceived and performed experiments and analysed data. W.A.M., J.C.H.T. and L.C.J. wrote the manuscript. All authors edited the manuscript and prepared figures.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. doi:10.1038/nri1391 nri1391 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. doi:10.1038/ni1087 ni1087 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. doi:nature04734 [pii] 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 4.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. doi:10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. doi:jlb.0306164 [pii] 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 6.Mallery DL, et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci U S A. 2010;107:19985–19990. doi: 10.1073/pnas.1014074107. doi:1014074107 [pii] 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci U S A. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. doi:0609174104 [pii] 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauler F, Mallery DL, McEwan WA, Bidgood SR, James LC. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc Natl Acad Sci U S A. 2012;109:19733–19738. doi: 10.1073/pnas.1210659109. doi:1210659109 [pii] 10.1073/pnas.1210659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEwan WA, Mallery DL, Rhodes DA, Trowsdale J, James LC. Intracellular antibody-mediated immunity and the role of TRIM21. Bioessays. 2011;33:803–809. doi: 10.1002/bies.201100093. doi:10.1002/bies.201100093. [DOI] [PubMed] [Google Scholar]
- 10.McEwan WA, et al. Regulation of Virus Neutralization and the Persistent Fraction by TRIM21. J Virol. 2012;86:8482–8491. doi: 10.1128/JVI.00728-12. doi:JVI.00728-12 [pii] 10.1128/JVI.00728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. doi:10.1038/nature02343 nature02343 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. doi:nature09976 [pii] 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci U S A. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. doi:0800159105 [pii] 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. doi:10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. doi:nature08247 [pii] 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. doi:S1097-2765(06)00192-4 [pii] 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 17.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. doi:ncb1384 [pii] 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 18.Yang K, et al. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol. 2009;182:3782–3792. doi: 10.4049/jimmunol.0803126. doi:182/6/3782 [pii] 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 19.Kong HJ, et al. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. doi:179/1/26 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Young JA, et al. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J Biol Chem. 2011;286:6521–6531. doi: 10.1074/jbc.M110.172288. doi:M110.172288 [pii] 10.1074/jbc.M110.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgs R, et al. Self protection from anti-viral responses--Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-Like receptors. PLoS One. 2010;5:e11776. doi: 10.1371/journal.pone.0011776. doi:10.1371/journal.pone.0011776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgs R, et al. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. doi:181/3/1780 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. doi:IMR893 [pii] 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Israel H, Kleinberger T. Adenovirus and cell cycle control. Front Biosci. 2002;7:d1369–1395. doi: 10.2741/ben. [DOI] [PubMed] [Google Scholar]
- 25.Morse L, Chen D, Franklin D, Xiong Y, Chen-Kiang S. Induction of cell cycle arrest and B cell terminal differentiation by CDK inhibitor p18(INK4c) and IL-6. Immunity. 1997;6:47–56. doi: 10.1016/s1074-7613(00)80241-1. doi:S1074-7613(00)80241-1 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Schreiber M, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liebermann DA, Hoffman B. Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia. 2002;16:527–541. doi: 10.1038/sj.leu.2402477. doi:10.1038/sj.leu.2402477. [DOI] [PubMed] [Google Scholar]
- 28.Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 2009;276:7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. doi:EJB7401 [pii] 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beuzon CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- 30.Birmingham CL, Brumell JH. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. doi: 10.4161/auto.2825. doi:2697 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Beuzon CR, et al. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. doi:10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrovska L, et al. Salmonella enterica serovar Typhimurium interaction with dendritic cells: impact of the sifA gene. Cell Microbiol. 2004;6:1071–1084. doi: 10.1111/j.1462-5822.2004.00419.x. doi:CMI419 [pii] 10.1111/j.1462-5822.2004.00419.x. [DOI] [PubMed] [Google Scholar]
- 33.Schmitz H, Wigand R, Heinrich W. Worldwide epidemiology of human adenovirus infections. Am J Epidemiol. 1983;117:455–466. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimi R, et al. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. doi:182/12/7527 [pii] 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs R, Blaas D. Uncoating of human rhinoviruses. Rev Med Virol. 2010;20:281–297. doi: 10.1002/rmv.654. doi:10.1002/rmv.654. [DOI] [PubMed] [Google Scholar]
- 36.Negishi H, et al. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat Immunol. 2012;13:659–666. doi: 10.1038/ni.2307. doi:ni.2307 [pii] 10.1038/ni.2307. [DOI] [PubMed] [Google Scholar]
- 37.Koshiba R, et al. Regulation of cooperative function of the Il12b enhancer and promoter by the interferon regulatory factors 3 and 5. Biochem Biophys Res Commun. 2012 doi: 10.1016/j.bbrc.2012.11.006. doi:S0006-291X(12)02147-X [pii] 10.1016/j.bbrc.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Chatterji U, et al. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J Biol Chem. 2006;281:37025–37033. doi: 10.1074/jbc.M606066200. doi:M606066200 [pii] 10.1074/jbc.M606066200. [DOI] [PubMed] [Google Scholar]
- 39.Diaz-Griffero F, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. doi:S0042-6822(05)00850-0 [pii] 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 40.Barouch DH. Novel adenovirus vector-based vaccines for HIV-1. Curr Opin HIV AIDS. 2010;5:386–390. doi: 10.1097/COH.0b013e32833cfe4c. doi:10.1097/COH.0b013e32833cfe4c 01222929-201009000-00006 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng C, et al. Decreased pre-existing Ad5 capsid and Ad35 neutralizing antibodies increase HIV-1 infection risk in the Step trial independent of vaccination. PLoS One. 2012;7:e33969. doi: 10.1371/journal.pone.0033969. doi:10.1371/journal.pone.0033969 PONE-D-11-21868 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15:1157–1166. doi: 10.1089/hum.2004.15.1157. doi:10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.