SUMMARY
Inhibitors of the ALK and EGF receptor tyrosine kinases provoke dramatic but short-lived responses in lung cancers harboring EML4-ALK translocations or activating mutations of EGFR, respectively. We used a large-scale RNAi screen to identify MED12, a component of the transcriptional MEDIATOR complex that is mutated in cancers, as a determinant of response to ALK and EGFR inhibitors. MED12 is in part cytoplasmic where it negatively regulates TGF-βR2 through physical interaction. MED12 suppression therefore results in activation of TGF-βR signaling, which is both necessary and sufficient for drug resistance. TGF-β signaling causes MEK/ERK activation, and consequently MED12 suppression also confers resistance to MEK and BRAF inhibitors in other cancers. MED12 loss induces an EMT-like phenotype, which is associated with chemotherapy resistance in colon cancer patients and to gefitinib in lung cancer. Inhibition of TGF-βR signaling restores drug responsiveness in MED12KD cells, suggesting a strategy to treat drug-resistant tumors that have lost MED12.
INTRODUCTION
Cancer therapy is often hampered by the rapid emergence of drug resistance. This is true not only for the conventional chemotherapies but also for the new generation of drugs targeting those components that are mutated or deregulated in tumor cells. For example, treatment of metastatic non-small-cell lung cancers (NSCLCs) harboring activating mutations in the gene encoding the epidermal growth factor receptor (EGFR) leads to significant increases in progression-free survival. However, such responses are often short-lived, resulting in much less impressive patient benefit in terms of overall survival (Maemondo et al., 2010). This lack of long-term benefit is due to the emergence of drug-resistant variants. Development of resistance to targeted therapies is a general phenomenon and is also seen in BCR-ABL-translocated chronic myelogenous leukemias (CML) treated with imatinib (Gorre et al., 2001), BRAF mutant melanomas treated with the BRAF inhibitor vemurafenib (Chapman et al., 2011), and EML4-ALK-translocated NSCLCs treated with the ALK inhibitor crizotinib (Kwak et al., 2010).
About half of the resistance seen in EGFR mutant NSCLCs treated with EGFR inhibitors can be explained by secondary mutations in the EGFR gene itself (Sequist et al., 2011). The T790M “gatekeeper” mutation in EGFR is critical for binding of competitive inhibitors to the ATP-binding pocket (Yun et al., 2008), allowing continued proliferation in the presence of the drug. Similar gatekeeper mutations have been found in BCR-ABL-positive CMLs treated with imatinib (Shah et al., 2002) and in EML4-ALK mutant NSCLCs treated with crizotinib (Choi et al., 2010).
Resistance to targeted therapies that does not involve secondary mutations in the drug target itself is often caused by mutations in the signaling pathway downstream of the target. Thus, primary resistance to EGFR-targeted therapy in colon cancer is associated with mutations in KRAS (Karapetis et al., 2008). Similarly, acquired resistance to BRAF inhibition in melanoma can result from an activating mutation in the MEK1 kinase that was not detectable in the primary tumor (Wagle et al., 2011). Alternatively, resistance can result from activation of a parallel pathway or in genes that feed into the downstream signaling of the drug target. Thus, amplification of the MET oncogene is found in EGFR drug-resistant NSCLC (Sequist et al., 2011), and overexpression of COT, leading to activation of MEK, can be a causal agent in BRAF resistance in melanoma (Johannessen et al., 2010). At present, some 30% of the resistance to EGFR-targeted therapies in NSCLCs cannot be explained by any of the mechanisms described above (Sequist et al., 2011).
Functional genetic screens provide a powerful tool to identify novel components of signaling pathways and can help to identify mechanisms of drug resistance in preclinical models of cancer (Berns et al., 2007; Hölzel et al., 2010). We describe here the use of a large-scale loss-of-function genetic screen to identify genes whose suppression can confer resistance to crizotinib in a NSCLC cell line harboring an EML4-ALK translocation. We identify a key component of the transcriptional MEDIATOR complex, MED12, as a determinant of crizotinib response in NSCLC. Remarkably, we find that suppression of MED12 also confers resistance to a range of cancer drugs, including chemotherapy, in colon cancer, melanoma, and liver cancer. We identify an unexpected activity of MED12 in regulating transforming growth factor β (TGF-β) receptor signaling, as the major mechanism of drug-resistance induction.
RESULTS
MED12 Suppression Confers Resistance to Multiple Tyrosine Kinase Inhibitors in NSCLCs
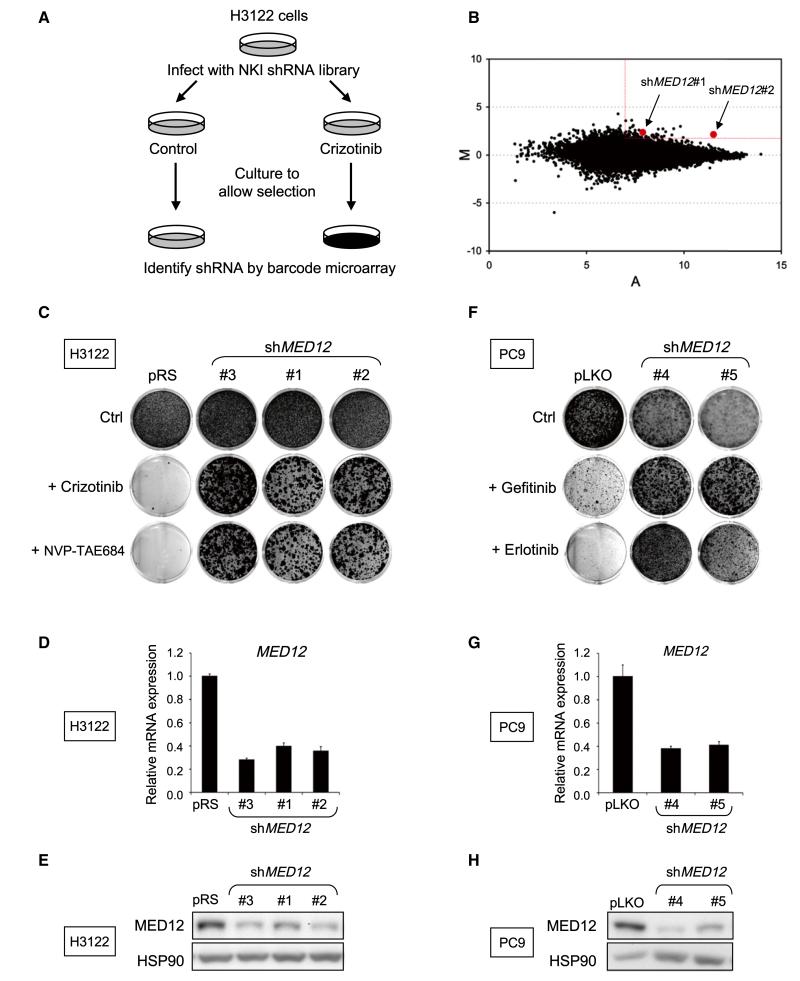
The NSCLC cell line H3122 harbors an EML4-ALK translocation and is exquisitely sensitive to the ALK inhibitors PF-02341066 (crizotinib) and NVP-TAE684 (McDermott et al., 2008). To identify genetic determinants of resistance to ALK inhibitors in EML4-ALK-translocated NSCLC, we performed a large-scale RNA interference (RNAi) genetic screen with a collection of 24,000 short hairpin RNA (shRNA) vectors targeting 8,000 human genes (Berns et al., 2004). As outlined in Figure 1A, we used a barcoding technology to identify genes whose suppression causes resistance to crizotinib in H3122 cells (Brummelkamp et al., 2006;Hölzel et al., 2010). The results are shown in Figure 1B. Each dot in the M/A-plot represents one individual shRNA vector. M and A values reflect relative enrichment and hybridization signal intensity. Low-intensity spots are prone to technical artifacts and are thus unreliable. Therefore we restricted our candidate selection by applying M/A cut-off values as indicated in Figure 1B. To rule out “off-target” effects, we prioritized genes that are present with multiple shRNAs. Only one gene fulfilled these criteria: MED12 encoding a component of the large MEDIATOR transcriptional adaptor complex.
Figure 1. A Genome-wide RNAi Screen Identifies MED12 as a Critical Determinant of Drug Response to Tyrosine Kinase Inhibitors in NSCLCs.
(A) Schematic outline of the crizotinib resistance barcode screen performed in H3122 cells. NKI human shRNA library polyclonal virus was used to infect H3122 cells, which were then left untreated (control) or treated with 300 nM crizotinib for 14 or 28 days, respectively. After selection, shRNA inserts from both populations were recovered, labeled, and hybridized to DNA oligonucleotide barcode arrays.
(B) Analysis of the relative abundance of the recovered shRNA cassettes from crizotinib barcode experiment. Averaged data from three independent experiments were normalized and 2log transformed. Among the 43 top shRNA candidates (M > 2 and A > 7), two independent shMED12 vectors (in red) were identified.
(C-E) Three independent shRNAs targeting MED12 confer resistance to ALK inhibitors. (C) The functional phenotypes of nonoverlapping retroviral shMED12 vectors (#1–3) in H3122 cells are indicated by colony formation assay in 300 nM crizotinib or 2.5 nM NVP-TAE684. The pRS vector was used as a control. The cells were fixed, stained, and photographed after 14 (untreated) or 28 days (treated). (D) The level of MED12KD by each of the shRNAs was measured by examining the MED12 mRNA levels by qRT-PCR. Error bars denote standard deviation (SD). (E) The level of knockdown of MED12 protein was measured by western blotting. (F–H) Suppression of MED12 also confers to EGFR inhibitors. (F) Colony formation assay of PC9 cells that express pLKO control or independent lentiviral shMED12 vectors (#4 and #5) and that were cultured in 50 nM gefitinib or erlotinib. The cells were fixed, stained, and photographed after 10 (untreated) or 28 days (treated). (G) The level of MED12KD by each of the shRNAs was measured by examining the MED12 mRNA levels by qRT-PCR. Error bars denote SD. (H) The level of knockdown of MED12 protein was measured by western blotting.
See also Figures S1 and S2.
To validate MED12 as a gene whose suppression confers resistance to crizotinib, we introduced the two MED12 shRNAs (#1 and #2) from the library and one newly generated shRNA (#3) into H3122 cells by retroviral infection. Empty vector (pRS) or shRNA-targeting GFP (shGFP) served as controls. All three distinct MED12 shRNAs conferred resistance to both crizotinib and NVP-TAE684 (Figure 1C) and also suppressed MED12 mRNA and protein expression (Figures 1D and 1E). Expression of additional independent lentiviral shMED12 vectors (#4 and #5) in H3122 cells also conferred resistance to ALK inhibitors (Figures S1A–S1C available online and data not shown). Furthermore, reconstitution of the RNAi-resistant murine Med12 cDNA in MED12 knockdown (MED12KD) H3122 cells restored the sensitivity of these cells to ALK inhibition (Figure S1). Suppression of MED12 also conferred resistance to the EGFR inhibitors gefitinib or erlotinib in the EGFR mutant NSCLC cell lines PC9 and H3255 (Figures 1F–1H and data not shown). These results establish a potential role for MED12 in resistance to ALK and EGFR inhibitors.
MED12 Loss Leads to MEK/ERK Activation and Drug Resistance in Different Cancer Types
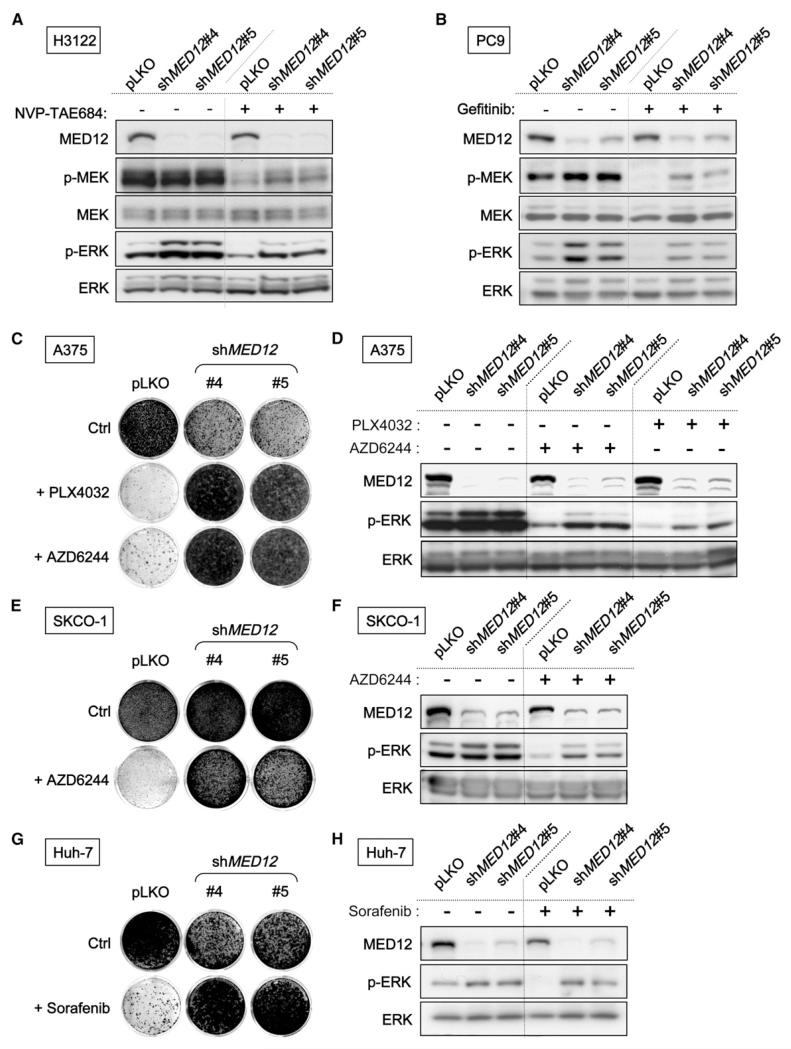
Our finding that MED12 suppression confers resistance to both ALK and EGFR inhibitors suggests that MED12 might act on a core pathway downstream of both ALK and EGFR, such as RAS signaling. Indeed, H3122 cells expressing shMED12 vectors maintained higher levels of phosphorylated MEK (p-MEK) and ERK (p-ERK) in the presence of ALK inhibitor (Figure 2A). Similarly, MED12KD in PC9 and H3255 cells leads to higher levels of p-MEK and p-ERK in both absence and presence of EGFR inhibitors (Figures 2B and 4J). These findings suggest that MED12 loss confers resistance to ALK and EGFR inhibitors in NSCLCs by enhancing MEK/ERK activation.
Figure 2. MED12 Suppression Leads to MEK/ERK Activation and Confers Multidrug Resistance in Different Cancer Types.
(A and B) MED12KD results in an elevated level of phosphorylated MEK (p-MEK) and phosphorylated ERK (p-ERK). (A) MED12KD H3122 cells have higher p-MEK and p-ERK levels. H3122 cells expressing pLKO or shMED12 vectors were grown in the absence or presence of 20 nM NVP-TAE684 for 6 hr, and the cell lysates were harvested for western blotting analysis. (B) Elevated p-MEK and p-ERK levels in MED12KD PC9 cells were documented by western blotting. PC9 cells expressing pLKO or shMED12 vectors were grown in the absence or presence of 25 nM gefitinib for 6 hr.
(C and D) MED12KD confers resistance to BRAF and MEK inhibitors in melanoma cells. (C) BRAFV600E A375 cells expressing pLKO or shMED12 vectors were cultured in the absence or presence of 2.5 μM PLX4032 or 0.5 μM AZD6244. The cells were fixed, stained, and photographed after 10 (untreated) or 28 days (treated). (D) MED12KD results in an elevated level of p-ERK in melanoma cells documented by western blotting. A375 cells expressing pLKO or shMED12 vectors were grown in the absence or presence of 1 μM PLX4032 or 0.5 μM AZD6244 for 6 hr.
(E and F) MED12KD confers resistance to MEK inhibitor in CRC cells. (E) KRASV12 SK-CO-1 cells expressing pLKO or shMED12 vectors were cultured in the absence or presence of 0.5 μM AZD6244. The cells were fixed, stained, and photographed after 14 (untreated) or 28 days (treated). (F) MED12 suppression results in an elevated level of p-ERK in CRC cells documented by western blotting. SK-CO-1 cells expressing pLKO or shMED12 vectors were grown in the absence or presence of 1 μM AZD6244 for 6 hr.
(G and H) MED12KD confers resistance to multikinase inhibitor sorafenib in HCC Huh-7 cells. (G) Colony formation assay of Huh-7 cells that express pLKO or shMED12 vectors (#4 and #5) and that were cultured in 2 μM sorafenib. The cells were fixed, stained, and photographed after 14 (untreated) or 21 days (treated). (H) MED12KD results in an elevated level of p-ERK in HCC cells. Huh-7 cells expressing pLKO or shMED12 vectors were grown in the absence or presence of 4 μM sorafenib for 6 hr.
See also Figure S2.
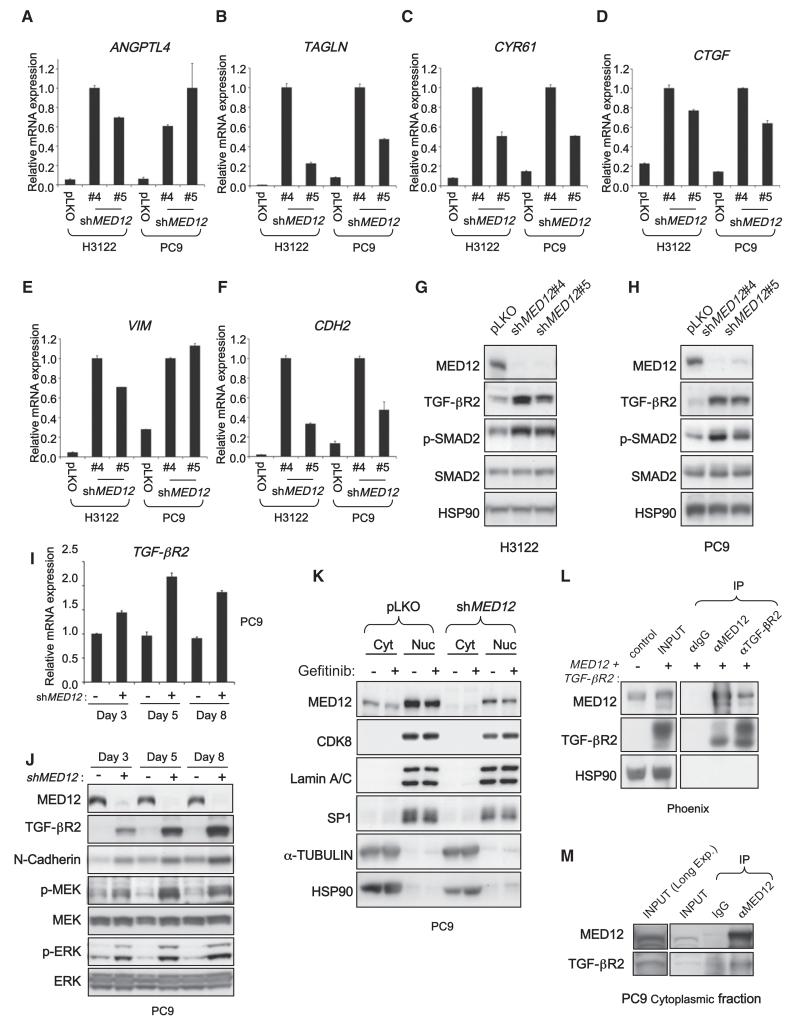
Figure 4. MED12 Suppresses TGF-β Signaling by Negatively Regulating TGF-βR2.
(A–F) MED12KD leads to induction of a panel of TGF-β target genes and EMT marker genes. mRNA expression analysis by qRT-PCR of TGF-β target genes ANGPTL4 (A), TAGLN (B), CYR61 (C), and CTGF (D) and EMT marker genes VIM (E) and CDH2 (F) in H3122 and PC9 cells expressing pLKO controls or shRNAs targeting MED12 is shown. Cells were cultured in normal condition without TGF-β stimulation. Error bars denote SD.
(G and H) MED12KD results in strong induction of TGF-βR2 protein and SMAD2 phosphorylation. Western blot analysis of H3122 (G) and PC9 (H) cells expressing pLKO or shMED12 vectors. HSP90 was used as a loading control.
(I) MED12KD results in a modest induction of TGF-βR2 mRNA in a time course experiment. RNA samples from PC9 cells expressing pLKO or shMED12 were collected at days 3, 5, and 8 post lentiviral infection, and TGF-βR2 mRNA was analyzed by qRT-PCR. Error bars denote SD.
(J) There is a progressive increase in TGF-βR2 protein levels in time after MED12KD, and the increase is associated with increased p-MEK, p-ERK, and N-cadherin. Western blotting analysis of the total lysates from the PC9 cells described in (I). All cells were treated with 25 nM gefinitib for 6 hr before lysate collection.
(K) MED12 localizes to both nucleus and cytoplasm. Western blotting analysis of the nuclear and cytoplasmic fractions prepared from PC9 cells expressing control vector or shMED12 with or without 16 hr of 25 nM gefitinib treatment. Lamin A/C and SP1 were used as marker controls for nuclear fractions, whereas α-TUBULIN and HSP90 were used as controls for cytoplasmic fractions.
(L) MED12 is capable of physically interacting with TGF-βR2. Western blotting analysis of coimmunoprecipitation experiments using Phoenix cells cotransfected with TGF-βR2 and MED12 in a ratio of 5:1 is shown.
(M) Cytoplasmic MED12 interacts with TGF-βR2 in PC9 cells. Western blotting analysis of coimmunoprecipitation experiments with a cytoplasmic fraction of parental PC9 cells is shown.
See also Figures S5 and S6.
Because MED12 suppression leads to ERK activation, one would expect that MED12 loss might also confer resistance to other cancer drugs targeting the kinases upstream of ERK. A375 melanoma cells (BRAFV600E) are highly sensitive to the BRAF inhibitor PLX4032 (vemurafenib) and the MEK inhibitor AZD6244 (selumetinib). We found that MED12KD in A375 cells caused MEK/ERK activation (Figures S2A and 2D) and conferred resistance to both PLX4032 and AZD6244 (Figure 2C). Similar results were obtained in the melanoma cell line SK-MEL-28 (BRAFV600E) (Figures S2B and S2C). MED12KD in SK-CO-1 (KRASV12) colorectal cancer (CRC) cells also resulted in activation of MEK/ERK (Figure 2F and data not shown) and conferred resistance to AZD6244 (Figure 2E). Identical results were observed in the CRC cell line SW1417 (BRAFV600E) (Figures S4D and S4E). Similarly, Huh-7 hepatocellular carcinoma cells became resistant to the multikinase inhibitor sorafenib after MED12KD (Figures 2G and 2H). In addition, MED12KD also conferred resistance to chemotherapy drugs such as cisplatin and 5-Fluorouracil (5-FU) (Figures S2F and S2G). We conclude that the effects of MED12 suppression are mostly context independent as its consequences are readily apparent in several cancer types.
TGF-β Signaling Is Required for Drug Resistance Caused by MED12 Loss
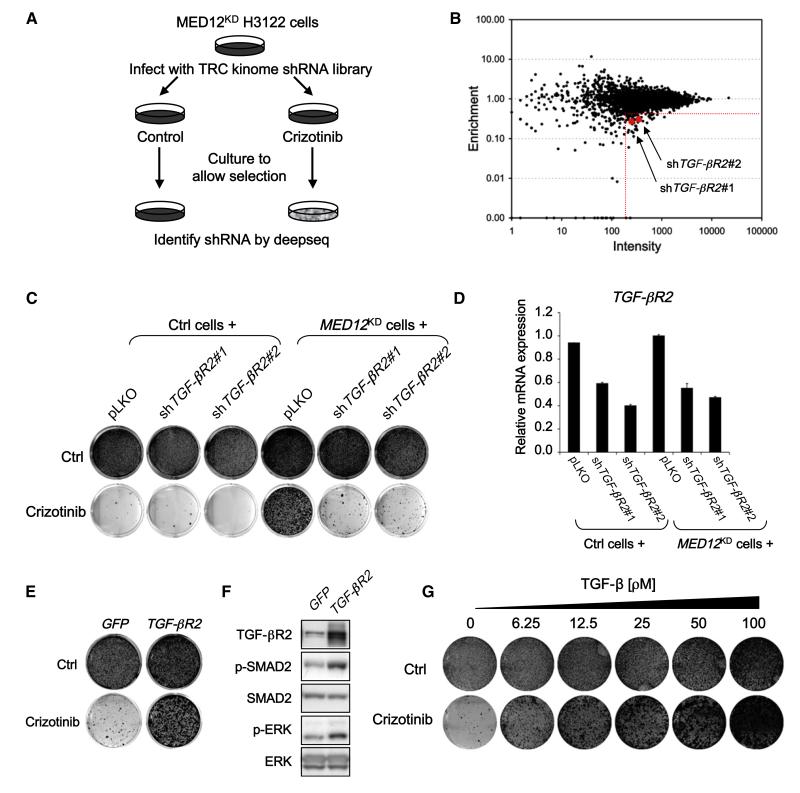
To address through which pathway MED12 acts to mediate these drug-resistance effects, we screened a lentiviral shRNA library representing all 518 human kinases (the “kinome,”Manning et al., 2002) (Table S1) for genes whose inhibition restores sensitivity to ALK inhibitors in MED12KD cells. This “dropout” screen (Figure 3A; see Experimental Procedures) is the inverse of the resistance screen shown in Figures 1A and 1B as here we select for shRNAs that are depleted upon drug treatment rather than enriched. Among the top 51 candidates that met the selection criterion described in Figure 3B, only one gene, transforming growth factor β receptor II (TGF-βR2), was represented by two independent shRNAs. This suggests that suppression of TGF-βR2 synergizes with ALK inhibition in MED12KD cells. To validate this, we infected the same MED12KD H3122 cells with each of these two shTGF-βR2 vectors (both reduced TGF-βR2 levels; Figure 3D) and cultured these cells with or without crizotinib for 2 weeks. Suppression of TGF-βR2 in combination with crizotinib caused a marked inhibition of proliferation in MED12KD cells (Figure 3C). These findings indicate that suppression of TGF-βR2 resensitizes the MED12KD cells to ALK inhibition and suggest that TGF-β signaling is required for the drug resistance caused by MED12 loss.
Figure 3. TGF-β Signaling Is Required for the Drug Resistance Driven by MED12 Suppression.
(A) Schematic outline of the “dropout” RNAi screen for kinases whose inhibition restores sensitivity to crizotinib in MED12KD cells. Human TRC kinome shRNA library polyclonal virus was produced to infect H3122 cells stably expressing shMED12#3, which were then left untreated (control) or treated with 300 nM crizotinib for 10 days. After selection, shRNA inserts from both populations were recovered by PCR and identified by next-generation sequencing.
(B) Representation of the relative abundance of the shRNA barcode sequences from the shRNA screen experiment depicted in (A). The y axis is enrichment (relative abundance of crizotinib treated/untreated), and x axis is the intensity (average sequence reads in untreated sample) of each shRNA. Among the 51 top shRNA candidates (more than 2.5-fold depleted by crizotinib treatment and more than 200 reads in untreated as indicated by the red dash lines), two independent shTGF-βR2 vectors (in red) were identified.
(C) Suppression of TGF-βR2 restores the crizotinib sensitivity in MED12KD cells. Using lentiviral infection, pLKO or two independent shTGF-βR2 vectors were introduced into H3122 control or MED12KD cells. After this, cells were cultured in the absence or presence of 300 nM crizotinib. The cells were fixed, stained, and photographed after 14 (untreated) or 21 days (treated). The level of knockdown of TGF-βR2 by each of the shRNAs was measured by examining the TGF-βR2 mRNA levels by qRT-PCR. Error bars denote SD.
(E and F) Activation of TGF-β signaling by TGF-βR2 overexpression was sufficient to confer resistance to crizotinib in H3122 cells. (E) H3122 cells expressing pQXCIP-GFP control or pQXCIP-TGF-βR2-HA were cultured in the absence or presence of 300 nM crizotinib. The cells were fixed, stained, and photographed after 14 (untreated) or 21 days (treated). (F) Western blotting analysis showing that TGF-βR2 overexpression resulted in elevated levels of p-SAMD2 and p-ERK.
(G) Activation of TGF-β signaling by recombinant TGF-β treatment also leads to resistance to crizotinib in H3122 cells in a TGF-β dosage-dependent manner.
See also Table S1 and Figures S3 and S4.
TGF-β Signaling Is Sufficient to Confer Resistance to a Variety of Cancer Drugs
Overexpression of exogenous TGF-βR2 was sufficient to activate TGF-β signaling (Figures 3F and S3A–S3D) and confer resistance to crizotinib in H3122 cells (Figure 3E). Consistently, recombinant TGF-β treatment also caused resistance to crizotinib in H3122 cells (Figure 3G). Furthermore, TGF-β treatment also caused MEK/ERK activation, consistent with the established activity of TGF-β in non-SMAD pathway signaling (Zhang, 2009) (Figure S4B). These data indicate that TGF-β activation is sufficient to confer resistant to ALK inhibitors in EML4-ALK-positive NSCLCs.
TGF-β treatment also caused MEK/ERK activation and conferred resistance to EGFR inhibitors in PC9 and H3255 NSCLC cells (Figures S3E and S4A and data not shown). Similarly, TGF-β-induced resistance to AZD6244 and PLX4032 was also observed in CRC cells and melanoma cells (Figures S3F, S3G, and S4C). Finally, TGF-β treatment also conferred resistance to cisplatin (Figures S3H and S3I). In some cells such as A375 and Huh-7 (Figure S3G and data not shown), recombinant TGF-β treatment alone resulted in growth inhibition but clearly became beneficial when cells were cultured in the presence of targeted cancer drugs, mimicking the effects of MED12KD in the same cells (Figures 2C and 2G). These results demonstrate that activation of TGF-β signaling is sufficient to confer resistance to multiple cancer drugs in the cancer types in which MED12KD also confers drug resistance.
Downregulation of MED12 Activates TGF-β Signaling by Elevating TGF-βR2 Protein Levels
Our findings suggested that MED12 acts as a suppressor of TGF-β signaling. We explored this by studying gene-expression analysis using transcriptome sequencing (RNA-Seq) in a panel of cells lines (H3122, PC9, SK-CO-1, A375, and Huh-7) and multiple MED12KD derivatives thereof. The genes deregulated by MED12KD (>2-fold) in at least three out of five cell lines used are listed in Table S2A and are referred to as MED12KD signature genes henceforth (237 genes up- and 22 genes downregulated). Strikingly, many of these genes are bona fide TGF-β targets. Upregulation of these TGF-β target genes upon MED12KD was confirmed by quantitative RT-PCR (qRT-PCR) (Figures 4A–4D). We also observed induction of these TGF-β target genes upon MED12KD in other tumor types including melanoma, colon cancer, and hepatocellular carcinoma (HCC) (Figures S5A–S5D). It is well-established that TGF-β induces an epithelial-mesenchymal transition (EMT), leading to the induction of several mesenchymal markers such as Vimentin (VIM) and N-cadherin (CDH2) (Thiery et al., 2009). MED12KD also induced expression of VIM and CDH2, indicating that an EMT-like process is initiated in MED12KD cells (Figures 4E, 4F S5E, and S5F). Accordingly, the protein products of these mesenchymal-specific genes were also detected in MED12KD cells (Figures 4J and S5L and data not shown) at levels similar to those induced by TGF-β treatment in the same cells (Figure S5M). Expression of the epithelial marker E-cadherin (CDH1) was not lost in MED12KD cells (data not shown), suggesting that MED12KD induces a partial EMT. Together these unbiased gene-expression studies support the notion that MED12 is a suppressor of TGF-β signaling in a wide range of cancer types and that its loss activates TGF-β signaling.
To further study the mechanism by which MED12 suppresses TGF-β signaling, we investigated the effect of MED12KD on key components of the TGF-β pathway. We found that MED12KD resulted in a strong induction of TGF-βR2 protein levels (Figures 4G and 4H). As a result of the TGF-βR2 upregulation, SMAD2, the key mediator of TGF-β signaling, was activated as indicated by a strong increase in SMAD2 phosphorylation. Consistently, affinity-labeling assays with 125I-TGF-β1 showed strong increase of the 125I-labeled cell-surface TGF-βR2 upon MED12KD in H3122 cells (Figure S5H). As controls, 125I-BMP9 affinity-labeling experiments showed no significant change in labeled BMP receptors upon MED12KD. Similar results were obtained in A375 melanoma and in SK-CO-1 CRC cells indicating that this interplay between MED12 and TGF-β signaling is conserved across different tumor types (Figures S5I and S5J). Thus the upregulation of TGF-βR2 in MED12KD cells causes the activation of TGF-β signaling, which in turns leads to MEK/ERK activation (Figure 4J). Supporting this notion that downregulation of TGF-βR2 by RNAi suppressed the MEK/ERK activation in MED12KD cells (Figures S4D-S4F and data not shown).
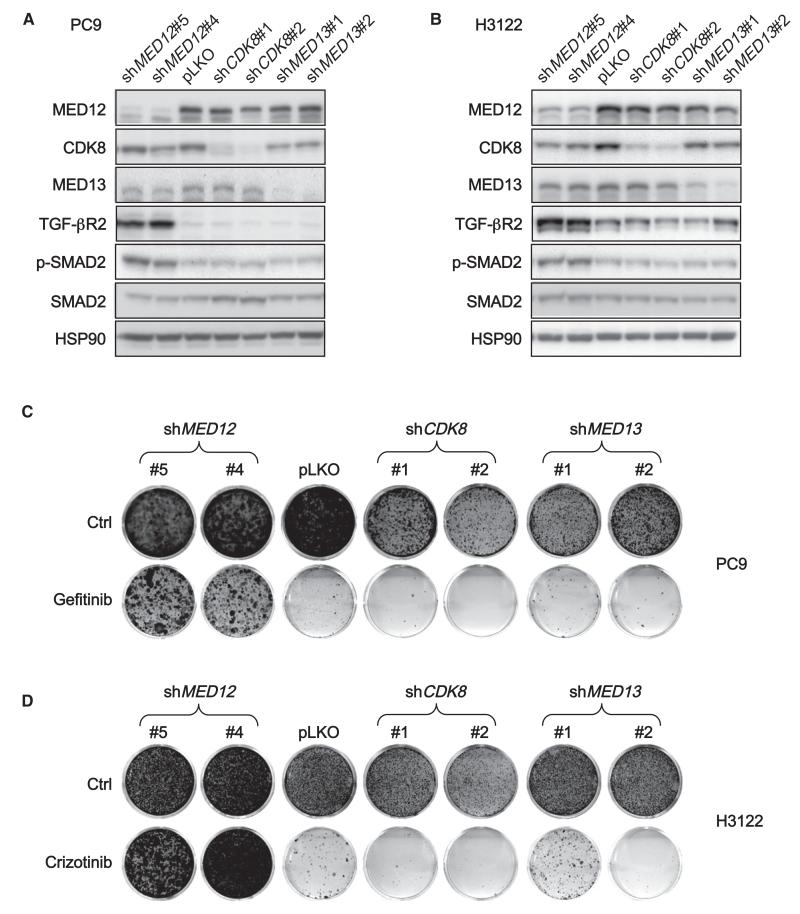
Because MED12 is part of the MEDIATOR transcriptional complex that functions in the nucleus, we assumed that MED12 would act on TGF-βR2 transcription. However, there was only a modest increase in TGF-βR2 mRNA upon MED12KD (Figure 4I). Moreover, we observed a progressive increase in TGF-βR2 protein levels in time after MED12KD. These results suggest that MED12 predominantly suppresses TGF-βR2 in a posttranscriptional manner. To investigate this, we determined the subcellular localization of MED12. We carried out nuclear and cytoplasmic fractionation of PC9 cells expressing control vector or shMED12 followed by western blotting (Figure 4K). Lamin A/C and SP1 were used as controls for nuclear fractions, α-TUBULIN and HSP90 for cytoplasmic fractions. Abundant nuclear MED12 was detected, consistent with its function in the MEDIATOR transcriptional complex. Unexpectedly, a significant quantity of MED12 was also present in the cytoplasmic fraction. Cytoplasmic MED12 was also seen in H3122 cells (Figure S5K). No cytoplasmic CDK8, another subunit of the MEDIATOR kinase module with which MED12 is known to associate closely, was detected. This suggests that cytoplasmic MED12 might have a second function distinct from its role in the MEDIATOR complex. Consistent with this, downregulation of other MEDIATOR subunits, such as CDK8 and MED13 in PC9 and H3122 cells, did not lead to upregulation of TGF-βR2 or activation of SMAD2 (Figures 5A and 5B) and failed to confer resistance to EGFR and ALK inhibitors (Figures 5C and 5D).
Figure 5. Downregulation of MED12, but Not of Other MEDIATOR Components, Activates TGF-β Signaling by Elevating TGF-βR2 Protein Levels and Confers Multidrug Resistance.
(A and B) Suppression of MED12, but not of other MEDIATOR components CDK8 or MED13, leads to strong induction of TGF-βR2 and elevated levels of p-SMAD2. Western blotting analysis of PC9 (A) and H3122 (B) cells expressing pLKO or shRNAs targeting MED12 CDK8 or MED13 is shown.
(C and D) MED12KD, but not knockdown of CDK8 or MED13, confers resistance to TKIs. (C) PC9 cells expressing pLKO or independent shRNAs targeting MED12 CDK8 or MED13 were cultured in 50 nM gefitinib. The cells were fixed, stained, and photographed after 10 (untreated) or 21 days (treated). (D) H3122 cells expressing pLKO or independent shRNAs targeting MED12, CDK8, or MED13 were cultured in 300 nM crizotinib. The cells were fixed, stained, and photographed after 14 (untreated) or 28 days (treated).
The unexpected cytoplasmic localization of MED12 prompted us to examine a potential physical interaction between MED12 and TGF-βR2. We first performed coimmunoprecipitation (coIP) experiments with Phoenix cells cotransfected with TGF-βR2 and MED12. As indicated in Figure 4L, TGF-βR2 coimmunoprecipitated with MED12, and conversely MED12 coimmunoprecipitated with TGF-βR2, indicating that MED12 interacts physically with TGF-βR2. Consistent with this, coIP experiments with the cytoplasmic fraction of untransfected PC9 cells indicate that endogenous TGF-βR2 interacts with endogenous MED12 (Figure 4M). As a second independent approach, we used a proximity ligation assay (PLA) to validate the TGF-βR2-MED12 interaction in situ. PLA technology allows sensitive detection of protein-protein interaction and requires two primary antibodies from different species against the proteins that are presumed to interact. Because our best antibodies against TGF-βR2 and MED12 were produced in rabbits, we generated MED12KD PC9 cells reconstituted with Flag-Med12 to be able to use PLA technology with mouse anti-Flag to detect Med12. These reconstituted cells expressed levels of MED12 and TGF-βR2 proteins similar to those in parental cells (Figure S6A). The results shown in Figure S6B indicate that there is a significant in situ interaction of TGF-βR2 and MED12 in the cytoplasm of PC9 cells, which is consistent with the data from the coIP experiments above.
The observation that MED12KD caused a strong increase of cell-surface TGF-βR2 (Figure S5H) suggests that MED12 could inhibit TGF-βR signaling by preventing the maturation of TGF-βR2. To test this, we performed coIP experiments with antibodies against HA tag and MED12 on Phoenix cells cotransfected with HA-TGF-βR2 and MED12 and incubated the immunoprecipitates with Endo H or PNGase F enzymes. Endo H removes oligosaccharides of glycoproteins in the endoplasmic reticulum (ER), but not the highly processed complex oligosaccharides processed in the Golgi. In contrast, PNGase F deglycosylates glycoproteins in both the ER and Golgi. As indicated in Figure S6C, in the TGF-βR2 immunoprecipitate, we observed three distinct forms of TGF-βR2: the 60 kDa form that was insensitive to both Endo H and PNGase F corresponding to unglycosylated TGF-βR2; the 70 kDa form that was sensitive to Endo H corresponding to the partially glycosylated TGF-βR2 in the ER; the smear from 80 to 100 kDa that was Endo H resistant but PNGase F sensitive corresponding to the fully glycosylated TGF-βR2. We found that only the nonprocessed and partially processed forms of TGF-βR2 coimmunoprecipitated with MED12. These data are consistent with a model in which MED12 interferes with the proper glycosylation of TGF-βR2 and hence blocks cell-surface expression of the receptor (Kim et al., 2012).
A MED12KD Gene Signature Has Features of EMT and Is Both Prognostic and Predictive
MED12 suppression leads to activation of TGF-β signaling and expression of mesenchymal markers, suggestive of a partial EMT-like process. Recently, EMT has been identified as a program in human CRC that correlates with poor prognosis (Loboda et al., 2011). We therefore asked whether MED12KD indeed induces an EMT-like process and whether the processes induced by MED12KD are likewise associated with poor prognosis in CRC. We first compared the 237 genes that are upregulated in the MED12KD signature (Table S2A) to the 229 genes upregulated in a more general EMT signature (see Extended Experimental Procedures; Table S2B). We found a significant overlap of 31 genes between both signatures (p = 8.9 × 10−23; Figure S7A and Table S2C). This further supports the notion that MED12 loss initiates a partial EMT. Next we asked whether genes that are deregulated after MED12KD predict survival in CRC. Hierarchical clustering of a set of 231 CRC tumor samples using the MED12KD signature genes led to the identification of two groups of patients that have significantly different disease-specific survival (DSS) (Figure S7B). The group with higher over-all expression of signature genes that are upregulated upon MED12KD had worse outcomes compared to the group with lower expression of the same genes. These results indicate that the processes induced by MED12KD are associated with a poor survival in CRC patients.
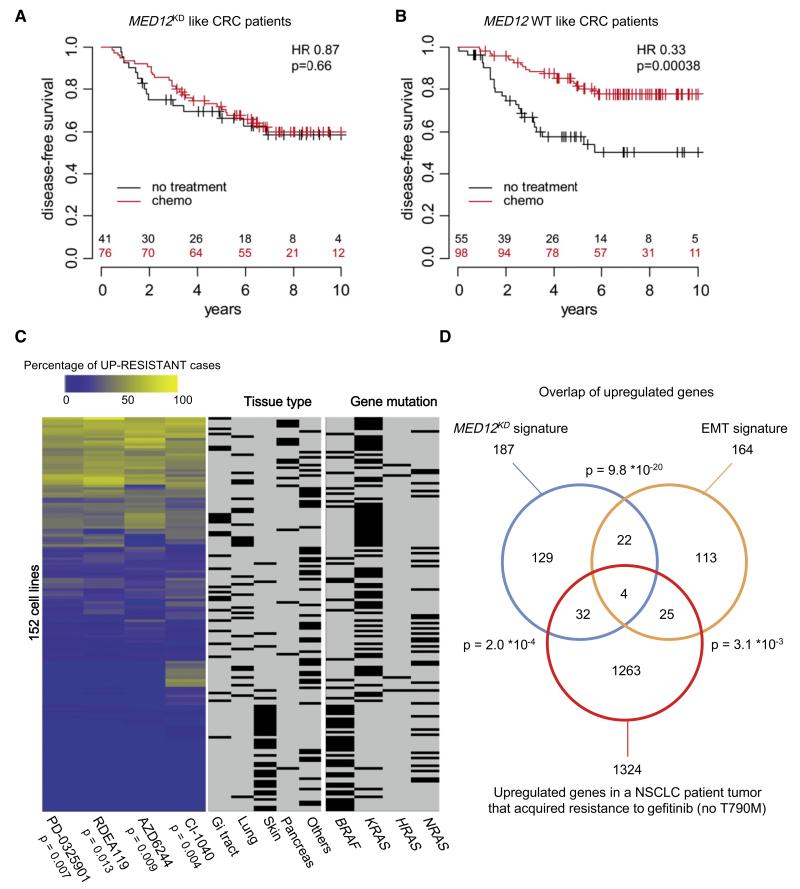
Next, we examined a second cohort of 270 stage III CRC patients, only some of whom were treated with 5-FU-based chemotherapy and whose responses to chemotherapy are known, to determine whether the MED12KD signature could also predict responses to chemotherapy in patients (Salazar et al., 2011; P.R., unpublished data). We used the MED12KD signature genes that were also present in the microarray previously used for the expression analysis of these tumors to classify patients as MED12KD like or MED12 wild-type (MED12WT) like (see Extended Experimental Procedures; Tables S2D and S2M). We found that chemotherapy did not lead to noticeable change in DSS of patients with MED12KD-like tumors (Figure 6A) whereas it did cause a significant increase in DSS of patients with MED12WT-like tumors (Figure 6B). These results indicate that the MED12KD signature predicts response to 5-FU-based chemotherapy in CRC patients, consistent with our finding that MED12KD confers resistance to 5-FU (Figure S2F).
Figure 6. MED12KD Signature Predicts Drug Responses to Cancer Therapies.
(A) Kaplan-Meier (KM) analysis of disease-specific survival (DSS) for the cohort of 117 CRC tumors with MED12KD-like gene signature. Patients with MED12KD-like tumors treated with chemotherapy did not show more significant DSS than untreated patients (red line, chemotherapy; black line, no treatment; HR = 0.87; p = 0.66). See Extended Experimental Procedures for details. HR = hazard ratio; p = p value.
(B) KM analysis of DSS for the cohort of 153 CRC tumors with MED12WT-like gene signature. Patients with MED12WT-like tumors treated with chemotherapy showed more significant DSS than untreated patients (red line, chemotherapy; black line, no treatment; HR = 0.33; p = 0.00038). See Extended Experimental Procedures for details. HR = hazard ratio; p = p value.
(C) MED12KD signature predicts drug responses to MEK inhibitors in 152 cell lines of different cancer types harboring the matching RAS or RAF mutations. High expression of subsets of genes upregulated in the MED12KD signature is significantly associated with higher IC50s for all four MEK inhibitors (AZD6244, p = 0.009; CI-1040, p = 0.004; PD-0325901, p = 0.007; RDEA119, p = 0.013). Across these gene sets, each cell line was scored for the percentage of times it had high expression of the gene as well as resistance to the inhibitor. The heatmap in the left panel of this figure depicts this percentage for each MEK inhibitor. The cell lines are sorted using hierarchical clustering for visualization. The middle and right panel depict the tissue type of the cell lines and their RAS/RAF mutation status.
(D) MED12KD signature genes may be associated with drug resistance to the EGFR inhibitor in NSCLC patients. Genes that are upregulated after acquisition of resistance to gefitinib in a NSCLC patient tumor (not harboring the EGFR T790M gatekeeper mutation) significantly overlap with both the upregulated genes in the MED12KD signature (p = 2.0 × 10−4) and the genes upregulated during EMT (p = 3.1 × 10−3). See Extended Experimental Procedures for details.
See also Figure S7 and Table S2.
To further substantiate our finding that MED12 suppression confers resistance to cancer drugs targeting the MEK-ERK pathway, we asked whether the MED12KD signature could predict response to MEK inhibitors in a large and heterogeneous panel of cancer cell lines of different tissue types. As MEK inhibitors are currently being evaluated to treat tumors that have activating mutations in RAS or BRAF, we focused on 152 tumor cell lines harboring either RAS or BRAF mutations for whom the IC50 values of four different MEK inhibitors and gene-expression patterns have been determined (Garnett et al., 2012) (seeExtended Experimental Procedures and Table S2F). Of the 237 genes that were upregulated by MED12KD as identified by RNA-Seq, we could read the expression levels for 170 genes in these 152 cell lines (Table S2E). We found that high expression of these 170 genes is significantly associated with higher IC50s for all four MEK inhibitors in these cell lines (AZD6244, p = 0.009; CI-1040, p = 0.004; PD-0325901, p = 0.007; RDEA119, p = 0.013; Figure 6C and Table S2E). The analysis of one of these genes, ZBED2, is shown as an example in Figure S7C. Thus the group of genes that is upregulated upon MED12KD predicts response to MEK inhibitors in a very heterogeneous panel of cancer cell lines, consistent with our finding that MED12 acts independently of cellular context to influence cancer drug response (Figure 6C).
Finally, we asked whether expression of MED12KD signature genes is associated with drug resistance to targeted agents in the clinic. We obtained pairs of tumor samples derived from three patients (cases 3, 6, and 10) that have NSCLC tumors with EGFR-activating mutations both before and after development of resistance to gefitinib (Uramoto et al., 2011). Two of the resistant tumors did have the EGFR T790M gatekeeper mutation (cases 3 and 6). RNA was isolated from these formalin-fixed tumor slides followed by transcriptome sequencing by RNA-Seq. For each pair, we selected genes that showed a greater than 2-fold upregulation after acquisition of gefitinib resistance and then asked whether these genes overlap with the MED12KD signature. For the tumor pair without the EGFR T790M mutation (case 10), we did observe a significant overlap of genes upregulated after acquisition of gefitinib resistance with the MED12KD signature genes (Figure 6D and Tables S2G-S2L), but not for the two tumor pairs with EGFR T790M mutation (cases 3 and 6) (Figure S7B and Table S3). This result indicates that in the patient of case 10, a gene-expression program was activated upon gefitinib resistance that resembles the program induced by MED12KD.
TGF-βR Inhibitor and TKIs Synergize to Suppress Proliferation of MED12KD NSCLC Cells
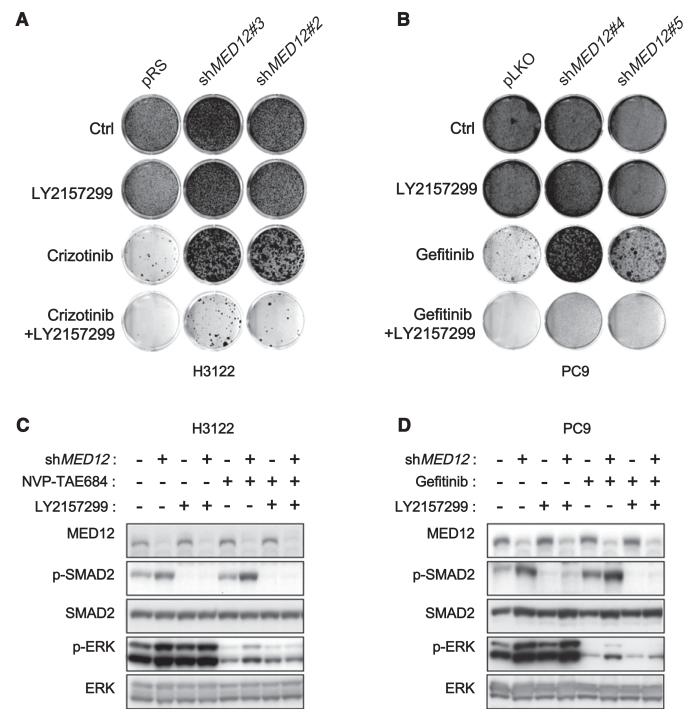
As inhibition of TGF-βR2 by RNAi resensitized MED12KD cells to tyrosine kinase inhibitors (TKIs), we reasoned that TGF-βR inhibitors should synergize with TKIs to inhibit proliferation in MED12KD cells. To test this, we cultured both parental and MED12KD H3122 cells in the absence and the presence of crizotinib, the TGF-βR inhibitor LY2157299, or the combination of both drugs. Crizotinib alone potently inhibited the growth of the control, but not of the MED12KD cells. LY2157299 alone had little effect on all cells. However, strong synergy was seen when crizotinib was combined with LY2157299 (Figure 7A). The same synergistic response was also obtained when LY2157299 was combined with gefitinib in MED12KD PC9 cells (Figure 7B). Moreover, the combination of LY2157299 with crizotinib or gefitinib suppressed the ERK activation driven by MED12KD in both H3122 and PC9 cells (Figures 7C and 7D). These biochemical data are in line with our previous RNAi results where TGF-βR2KD suppressed ERK activation in MED12KD cells (Figures S4D–S4F). Thus, the combination of TGF-βR inhibitors and TKIs might be a strategy for treating tumors with elevated TGF-β signaling.
Figure 7. TGF-βR Inhibitor and TKIs Synergize to Suppress Proliferation of MED12KDNSCLC Cells.
(A) Combination of TGF-βR and ALK inhibitors synergistically inhibits growth of MED12KD NSCLC cells harboring EML4-ALK translocation. H3122 cells expressing pRS or shMED12 vectors were cultured in the absence and the presence of 1 μM LY2157299, 300 nM crizotinib, or the combination of 1 μM LY2157299 and 300 nM crizotinib. The cells were fixed, stained, and photographed after 14 (untreated and LY2157299 alone) or 28 days (crizotinib alone and LY2157299 plus crizotinib).
(B) Combination of TGF-βR and EGFR inhibitors synergistically inhibits growth of MED12KD NSCLC cells harboring EGFR-activating mutation. PC9 cells expressing pLKO or shMED12 vectors were cultured in the absence and the presence of 1 μM LY2157299, 100 nM gefitinib, or the combination of 1 μM LY2157299 and 100 nM gefitinib. The cells were fixed, stained, and photographed after 10 (untreated and LY2157299 alone) or 28 days (gefitinib alone and LY2157299 plus gefitinib).
(C and D) Combination of LY2157299 with crizotinib or gefitinib suppressed the ERK activation driven by MED12KD in both H3122 and PC9 cells. (C) H3122 cells were grown in the absence or presence of 20 μM NVP-TAE684, 5 μM LY2157299, or the combination of 20 μM NVP-TAE684 and 5 μM LY2157299 for 6 hr, and the cell lysates were harvested for western blotting analysis. (D) PC9 cells were grown in the absence or presence of 25 nM gefitinib, 5 μM LY2157299, or the combination of 25 nM gefitinib and 5 μM LY2157299 for 6 hr, and the cell lysates were harvested for western blotting analysis.
See also Figure S4.
DISCUSSION
We identify here MED12 as a candidate biomarker of response to a range of cancer drugs in a variety of cancer types through a previously unappreciated role of this protein in TGF-β receptor signaling. MED12 is a component of the MEDIATOR transcriptional adaptor complex that serves as a molecular bridge between the basal transcription machinery and its upstream activators (Conaway et al., 2005). More specifically, MED12 is a subunit of the “kinase” module of the MEDIATOR complex, which also contains MED13, CYCLIN C, and CDK8, whose gene sequence is amplified in some 50% of colon cancers (Firestein et al., 2008). However, neither CDK8KD nor MED13KD caused upregulation of TGF-βR2 or conferred drug resistance, highlighting the unique role of MED12 in both TGF-βR2 activation and drug resistance. The involvement of MED12 in the response to TKIs was unexpected as most of the known genes that influence responses to TKIs involve components of signaling pathways that act downstream of or in parallel to these receptors. We reconcile this apparent discrepancy by demonstrating that part of MED12 resides in the cytosol, where it interacts with the immature forms of TGF-βR2 and inhibits its glycosylation, thereby preventing cell-surface expression (Kim et al., 2012). Consequently, MED12KD strongly enhances cell-surface expression of TGF-βR2 and activates TGF-β signaling. Activation of TGF-β signaling has also been linked to increased RAS-MEK-ERK signaling (reviewed by Zhang, 2009). Indeed we observed activation of ERK signaling by MED12 suppression, which persists in the presence of drugs like crizotinib, gefitinib, vemurafenib, seluteminib, and sorafenib (Figures 2 and S4), thus providing a rationale for why suppression of MED12 confers resistance to these drugs.
Our data indicate that MED12 suppression also induces an EMT-like phenotype and that this EMT-like phenotype induced by MED12KD is associated with chemotherapy resistance in both cell lines and patients. Our data are consistent with the findings of others who also witnessed resistance to EGFR inhibitors in cell lines undergoing EMT (Fuchs et al., 2008; Yao et al., 2010). In the clinic, EMT transdifferentiation was also seen in NSCLC patients who developed resistance to EGFR TKIs (Sequist et al., 2011; Uramoto et al., 2011). Consistent with this, we observed in a NSCLC patient who developed resistance to gefitinib without gatekeeper T790M mutation that a program of gene expression that resembled the one induced by MED12KD was activated (Figure 6D). It is at this point not clear whether patients that acquire EMT during drug resistance do so as a result of MED12 loss. This appears possible as MED12 is mutated in some 70% of uterine leiomyomas and in 5% of prostate cancers (Barbieri et al., 2012; Mäkinen et al., 2011). We note that these mutations are highly clustered, raising the possibility that these mutations are not null alleles. Consistent with this, we observe that MED12 suppression often confers a slow-growth phenotype to cancer cells and that near-complete suppression of MED12 is not tolerated by most cells. Thus suppression of MED12 may not confer a selective advantage in the absence of drug but may only become a benefit to the cancer cells when undergoing drug selection pressure. Consistent with this, we observed that PC9, NSCLC, A375, melanoma, and Huh-7 HCC cells are growth-inhibited by MED12KD, but this turns into a proliferative advantage when exposed to EGFR, BRAF, or MEK inhibitors or the multikinase inhibitor sorafenib. Therefore, MED12 suppression may not be a marker of intrinsic drug resistance as its constitutive suppression could well be disadvantageous to the cancer cell, but it may be acquired during drug selection to resist the therapy. That cancer cells can transiently assume a reversible drug-tolerant state was recently shown (Sharma et al., 2010).
Finally, our data demonstrate that inhibition of TGF-β signaling in MED12KD cells with small-molecule drugs can reverse resistance to targeted cancer drugs (Figure 7). This raises the possibility that EMT arising during drug-resistance development as seen in NSCLC (Sequist et al., 2011; Uramoto et al., 2011) may be countered by combination with a TGF-β antagonist, a notion that can readily be tested in the clinic.
EXPERIMENTAL PROCEDURES
shRNA Screens
The NKI shRNA library and the barcode screen are as described (Berns et al., 2004; Brummelkamp et al., 2006). Additional details can be found at http://screeninc.nki.nl/. See the Extended Experimental Procedures for details on the kinome “dropout” shRNA screen.
Cell Culture, Viral Transduction, and Long-Term Cell Proliferation Assays
Experiments were performed as described (Huang et al., 2009). See theExtended Experimental Procedures for details.
Gene-Expression and Statistical Analysis
Transcriptome sequencing analysis of cell lines was performed with RNA-Seq to generate the MED12KD gene signature, which was employed to hierarchically cluster a data set consisting of gene expression data for 231 CRC tumor samples for their outcome and to predict responses to chemotherapy in a second cohort of 270 CRC patients. Differences in DSS were determined using the Kaplan-Meier (KM) statistics.
See the Extended Experimental Procedures for details.
COSMIC Cell-Line Panel Analysis
Drug-response data (IC50 values) and gene-expression levels were obtained from Catalogue Of Somatic Mutations In Cancer (COSMIC) (Forbes et al., 2010). See the Extended Experimental Procedures for details for the analysis.
NSCLC Patient Samples
Tumor samples derived from three patients (cases 3, 6, and 10) that have NSCLC tumors with EGFR-activating mutations both before and after acquisition of resistance to gefitinib are as described (Uramoto et al., 2011). The institutional review board’s approved informed consent for the use of the tumor tissue specimens was obtained either from all the patients or from the patient’s legal guardians.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the NKI Genomics Core Facility, Maarten van Dinther, Jelle Wesseling, Ingrid Hofland, Ian Majewski, Kylie Greig, Johan Kuiken, and Erik Voets for technical support and discussion. We are grateful to Cinzia Pochet for support. This work was supported by grants from the Dutch Cancer Society, a European Research Council grant, The Cancer Systems Biology Center grant by NWO, The Netherlands Genomics Initiative (NGI), and a Spanish BAE FIS travel grant by Instituto Carlos III (to R.S.). P.R. and R.B. are employees and shareholders of Agendia Inc.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.10.035.
REFERENCES
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. BRIM-,3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al. ALK Lung Cancer Study Group. (2010). EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M, Tanabe KK. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391–2399. doi: 10.1158/0008-5472.CAN-07-2460. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Hölzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Laoukili J, Epping MT, Koster J, Hölzel M, Westerman BA, Nijkamp W, Hata A, Asgharzadeh S, Seeger RC, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328–340. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Kim YW, Park J, Lee HJ, Lee SY, Kim SJ. TGF-β sensitivity is determined by N-linked glycosylation of the type II TGF-β receptor. Biochem. J. 2012;445:403–411. doi: 10.1042/BJ20111923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A, Nebozhyn MV, James W, Watters JW, Carolyne A, Buser CA, Shaw PM, Huang PS, Van’t Veer LJ, Tollenaar RAEM, et al. EMT is the dominant program in human colon cancer. BMC Medical Genomics. 2011;2011:4–9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- Mäkinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, et al. MED12, the mediator complex subunit, 12 gene is mutated at high frequency in uterine leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, Zhou W, Choi HG, Smith SL, Dowell L, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J. Clin. Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Uramoto H, Shimokawa H, Hanagiri T, Kuwano M, Ono M. Expression of selected gene for acquired drug resistance to EGFR-TKI in lung adenocarcinoma. Lung Cancer. 2011;73:361–365. doi: 10.1016/j.lungcan.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc. Natl. Acad. Sci. USA. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.