Abstract
Genome-wide association studies into complex immune-mediated diseases have indicated that many genetic factors, each with individual low risk, contribute to overall disease. It is therefore timely and important to characterise how immune responses may be subtly modified by tissue context. Here we explore the role of tissue-derived molecules in influencing the function of T-cells, which, due to their migratory nature, come into contact with many different microenvironments through their lifespan. Hedgehog (Hh) proteins act as secreted morphogens, providing concentration-dependent positional and temporal cell-fate specification in solid tissues. Hh signalling is required for embryogenesis and is important in postnatal tissue renewal and in malignancy. However, the function of Hh in dynamic, fluid systems such as in mammalian immunity is largely unknown. Here we show that Hh-dependent transcription in T-cells promoted Th2 transcriptional programs and differentiation, exacerbating allergic pathology. Interestingly, expression of Sonic Hedgehog (Shh) increased in lung epithelial cells following the induction of allergic disease, and lung T-cells upregulated Hh-target gene expression, indicating that T-cells respond to locally-secreted Hh ligands in vivo. We show that Il4, the key Th2 cytokine, is a novel transcriptional target of Hh signals in T-cells, providing one mechanism for the role of Hh in Th differentiation. We propose that Hh, secreted from inflamed, remodelling or malignant tissue can modulate local T-cell function. Our data present an unexpected and novel role for tissue-derived morphogens in the regulation of fluid immune responses, with implications for allergy and tumour responses, suggesting new uses for anti-Hh therapeutics.
Keywords: Hedgehog, Morphogen, Gli2, T cells, Th2, Il4
Introduction
Hedgehog (Hh) family proteins, Sonic Hh (Shh), Indian Hh (Ihh) and Desert Hh (Dhh), are secreted inter-cellular signalling molecules, essential for embryogenesis and important in homeostasis of adult tissues (1). Morphogens such as Hh specify cell fate by establishing a concentration gradient, where the position of a target cell relative to the source of Hh determines the signal received. The Hh signalling pathway is initiated by ligand binding to the cell-surface receptor Patched 1 (Ptch1), relieving inhibition of Smoothened (Smo), resulting in signal transduction. At the end of the pathway are the Gli family of transcription factors (Gli1, Gli2 and Gli3) (2). Gli proteins bind DNA at consensus Gli-family binding sites and directly modulate target gene transcription. Gli2 is necessary to initiate the signal and acts mainly as an activator but can be processed to activate or repress transcription by post-translational modification (3). Strength and duration of the signal received, and so outcome, is determined by the balance of intracellular Gli-Repressor and Gli-Activator proteins.
Knockout and transgenic mouse models show that Hh signalling regulates thymocyte development (4-14). Peripheral T-cells express components of the Hh signalling pathway (15), which is involved in regulation of T-cell activation in vitro (11, 12, 15, 16). Hh family proteins are widely expressed in postnatal tissues, many of which harbour resident T-cells, including skin, lung, gut, bone marrow and spleen (17-21). Here we tested the hypothesis that Hh-dependent transcription modulates differentiation and effector function of peripheral CD4+ T-cells.
Naïve CD4+ T-cells can differentiate down several lineage pathways, with distinct T-helper (Th) functions (22). Th1 and Th2 cells are distinguished by their hallmark profiles of cytokine secretion, lineage-specific transcription factors and different cellular functions. Th1 cells express Tbet, produce interferon-gamma (IFNγ) and control immune responses against intracellular pathogens. Th2 cells express Gata3, secrete interleukin-4 (IL-4), IL-5, IL-9, IL-13 and IL-25 and are important for protection against extracellular parasites. Th2 cells are also involved in the pathogenesis of allergy and atopic disease (23). IL-4, the primary Th2 cytokine, is necessary for the generation of Th2-driven immune responses (24). Conditional deletion of Gata3 from naive T-cells blocked differentiation into functional IL-4-secreting cells (25). Expression of Gata3 and Il4 are closely linked: Gata3 can directly activate transcription of the Il4 gene (26), but for Th2 differentiation, naive T-cells require TCR and IL-4 signalling for strong induction of Gata3 (27). However, the complex cell-intrinsic and environmental mechanisms that induce upregulation of these key regulators of differentiation are incompletely understood. A large body of research has identified major regulators of Th differentiation including cytokines, transcription factors and other immune cell-derived molecules. However, little is known about the contribution of non-immune factors, including Hh proteins, which are secreted from the cells’ environment, in T-cell differentiation and plasticity. Genome-wide association studies (GWAS) into complex immune-mediated diseases have indicated that many genetic factors, each with individual low risk, contribute to overall disease. For asthma, GWAS have shown that multiple loci contribute to allergic pathology and drug responsiveness (28, 29) and that several factors may be involved in the communication between epithelial and immune cells in pathophysiology (29). It is therefore timely and important to characterise how immune responses may be subtly modified by environmental cues. Here we explore the role of tissue context in influencing the function of T-cells, which, due to lymphocyte trafficking, come into contact with many different microenvironments through their lifespan.
Materials and Methods
Mice
Lck-Gli2ΔN2 (11), Lck-Gli2ΔC2 (12), Dhh KO (30) mice and littermate/age-matched controls, all on C57BL/6 background, were used under UK Home Office regulations. The allergic airways model was as described (31).
Cell culture
Splenocytes were magnetically-purified using the EasySep mouse CD4+ cell negative selection kit (StemCell Technologies). T-cells were cultured at 5×106/ml in AIMV (Invitrogen). For microarray cells were activated for 6h with 0.01μg/ml soluble anti-CD3 and anti-CD28 (BD). For ChIP/qPCR/±anti-IL4 experiments, CD4+ cells were cultured for 48h with anti-CD3/anti-CD28-coated beads (1:1ratio, Invitrogen). 500ng/ml rmShh (R&D systems) or 5μg/ml 5E1 (anti-Hh mAb, DSHB, Iowa) were used. For Ic-cytokine staining, splenocytes were cultured for 3-4 hours with 50ng/ml PMA (Sigma), 500ng/ml Ionomycin (Sigma) and 3μg/ml Brefeldin A (eBiosciences). For Th skewing cultures, 5×105/ml CD4+ cells were cultured on 5μg/ml plate-bound anti-CD3 with 1μg/ml soluble anti-CD28 in complete RPMI+FCS (Invitrogen) for 6 days. For Th1 conditions, 5ng/ml rmIL-12 and 5μg/ml anti-mouse IL-4 were added. For Th2 conditions, 10ng/ml rmIL-4, 5μg/ml anti-mouse IFNγ, 5μg/ml anti-mouse IL-12 was added (antibodies/proteins: eBioscience).
Flow cytometry
Flow cytometry and reagents were as described (14).
ELISA
Th1/Th2 panel or Ready-Set-Go ELISA kits (eBioscience) were used. Shh ELISA was performed using Shh DuoSet kit (R&D Systems).
Histology
Lung tissue was fixed in formalin and embedded in paraffin wax for sectioning. Periodic acid Schiff stains were analysed by double-blinded scoring. Shh immunohistochemistry was as described (32).
Microarray and data analysis
UCL Genomics processed total RNA and acquired data from Affymetrix MOE430 2.0 mouse whole-genome arrays (dataset: GSE33156, available 01.01.2013, GEO depository). Data were normalised using mas5 of affy in each dataset. Differentially expressed genes (DEG) were identified by p<0.05 considering a false discovery rate by limma (Bioconductor). PCA was performed using the CRAN package. A novel application of canonical correspondence analysis (CCA) was used to compare our dataset relative to external data (33), where a Th1→Th2 axis was generated from GSE14308 (GEO dataset, Affymetrix, Th1/Th2-skewed mouse T-cells) using PCA, and used as a gradient for CCA of our own datasets.
Quantitative (q)RT-PCR
qPCR was carried out as described (12, 14). Samples were analysed in triplicate (13) following normalisation to Hprt expression and independently verified in 2-3 separate experiments.
Chromatin-Immunoprecipitation
SABiosciences ChampionChIP kit was used. Pre-cleared, sonicated chromatin was immunoprecipitated with anti-Gli2 (Santa Cruz), anti-RNA polymerase II (SABiosciences) or mIgG (SABiosciences). DNA was purified and used in PCR/qPCR using primers specific for HS2 region of Il4 (as primer pair 21 in (34)) and ChampionChIP Gata3 primer assays (SABiosciences). Results were validated in replicate experiments.
Data analysis
Statistical analyses were performed using Microsoft Excel or Prism 4 (Graph Pad) as stated in the text or Figure Legends. Significance was reached at p<0.05.
Results
Gli2 modulates Th-cell gene expression
Morphogens establish concentration gradients in solid tissues, but the immune system is fluid, with lymphocytes trafficking between blood, lymphoid organs and tissues. It is not known how morphogens such as Hh influence immune-cell biology. Therefore, to study the effect of Hh signalling on T-cells, independent of their history or migratory patterns, we established transgenic models where transcription by Gli2 is either constitutively activated or repressed in T-lineage cells. Lck-Gli2ΔN2 (Gli2A) mice carry a truncated form of Gli2 that acts as a permanent transcriptional activator of Hh target genes (11). Conversely, Lck-Gli2ΔC2 (Gli2R) mice express a repressor of Gli2-dependent transcription, which by binding to Gli-binding sites inhibits endogenous Hh-dependent transcription, and hence Hh signalling, in the cell (12). Thus, comparison of transcriptional profiles in WT and Gli2R cells identifies genes whose expression is regulated by physiological Hh signal transduction. Both transgenes are driven by the proximal lck promoter and as such are expressed peripheral T-cells (35, 36).
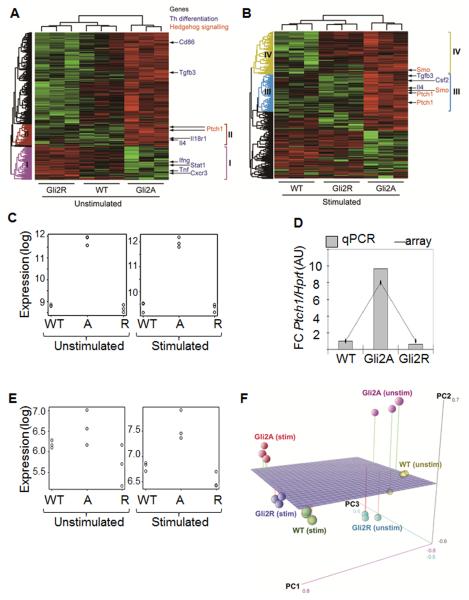
In order to define the transcriptional response of CD4+ T-cells to Hh pathway activation, we examined Hh-dependent gene expression in resting CD4+ cells (unstimulated) and CD4+ cells treated with anti-CD3/CD28 for 6h (stimulated) from WT, Gli2A and Gli2R spleen. Hundreds of differentially expressed genes were identified between WT and transgenic groups (GEO ref: GSE33156). Samples clustered according to genotype (Fig. 1A: unstimulated; B: stimulated), where Hh signalling/responsive genes, Ptch1 and Smo, were upregulated in Gli2A but not Gli2R (Fig. 1A, cluster II; 1B, clusters III and IV; 1B, 1C, 1D& 1E) as expected. We validated expression levels by qPCR using RNA from microarray experiments and independent cell-sorts (Fig. 1D). In agreement with microarray data, relative expression of Ptch1 was nine-fold higher in Gli2A cells than WT, but was downregulated two-fold in Gli2R cells compared to WT, indicating that active Hh signalling is taking place in T cells ex vivo.
Figure 1. Hedgehog-dependent transcription of genes involved in CD4+ T-cell differentiation.
(A, B) Gene expression heatmaps for (A) unstimulated and (B) stimulated CD4+ T-cells from WT, Gli2A and Gli2R spleen analysed by Affymetrix microarray. Th differentiation and Hh signalling genes are indicated. (C) Expression of Ptch1 in each sample. (D) qPCR (n=3, in triplicate) for Ptch1 relative to Hprt, normalised to WT samples compared to gene expression level by microarray. (E) Expression of Smo in each sample. (F) 3D-PCA showing sample relationships in PC1, PC2 and PC3.
In order to identify common effects of Hh signalling in resting and activated T-cells we applied principle component analysis (PCA) (Fig. 1F). Principle component 1 (PC1) showed that the largest difference in transcriptomes was between the unstimulated and stimulated samples, indicating that activation stimuli caused profound changes in transcription. PC2, the second largest measure of difference in gene expression, reflected differences between Gli2A and the other samples, especially Gli2R (Fig. 1F) and therefore reflected differences in Hh signalling.
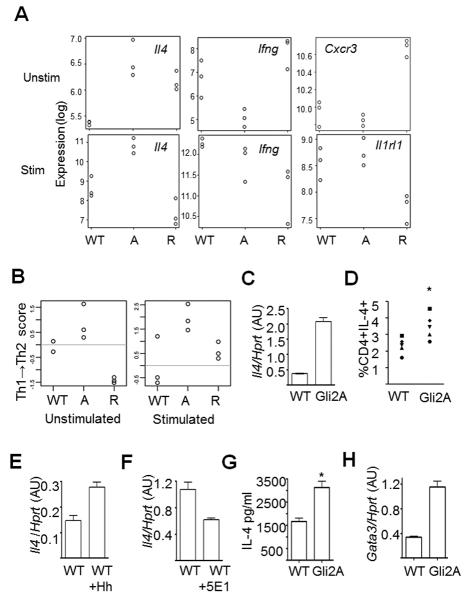
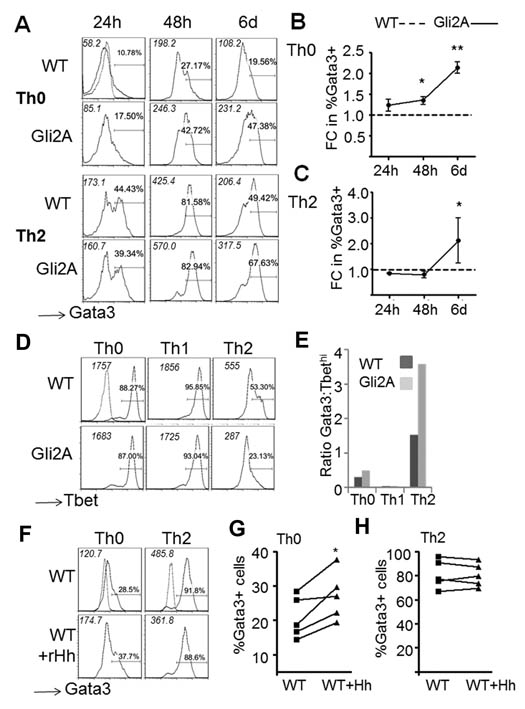
Interestingly, when we analysed gene lists generated by t-testing and by PCA, we found that genes involved in Th-cell differentiation were differentially expressed between groups. The Th2 cytokine, Il4, was upregulated in Gli2A unstimulated and stimulated cells (Fig. 2A) and belonged to the same clusters as Ptch1, a known Hh target gene (Fig. 1A-B, clusters II&III), suggesting that Il4 is downstream of Hh signalling in T-cells. In addition, Th1-related genes including Ifng and Cxcr3 were differentially regulated in Gli2A and Gli2R cells (Fig. 1A, cluster I, Fig. 2A). Stimulated Gli2R cells showed decreased expression of Il4 and Il1rl1, the IL-33 receptor, both important for Th2 function (Fig. 2A). Ptch1, Smo, and Il4 had high PC2 scores, indicating strong association with Gli2A, and therefore active Hh signalling. However, Th1-related genes showed negative PC2 scores, indicating that expression of these genes was increased in Gli2R and suppressed in Gli2A, confirming the results in Fig. 1A.
Figure 2. Hedgehog-dependent transcription favours a Th2 transcriptional profile.
(A) Gene expression in CD4+ T-cells from WT, Gli2A and Gli2R spleen (n=3/group) analysed by Affymetrix microarray. Expression of (A) Il4, Ifng, Cxcr3 and Il1rl1 in each sample. (B) Samples were measured by Th1→Th2 score, scale generated from publically-available Affymetrix datasets from in vitro-skewed Th1 and Th2 cells. On the scale 0 is the mean value of WT samples. Scale generated from publically-available microarray datasets from Th1 (negative) or Th2 (positive) samples. (C) Representative example qPCR measuring Il4 expression relative to Hprt in CD4+ splenocytes cultured with anti-CD3/CD28-coated beads. (D) %CD4+IL-4+ splenocytes directly ex vivo by intracellular cytokine staining, independent sets of experiments indicated by different shaped points (n=5 experiments, unpaired t-test: p<0.02). qPCR for Il4 as in (C), cells cultured with (E) rShh, or (F) anti-Shh (5E1). Mean±SD Il4 expression per independent sample was calculated relative to WT (unpaired t-tests: (C) n=3 p=0.04; (E) n=7 p=0.03; (F) n=4 p=0.02). (G) Mean±SD production of IL-4 by ELISA after 72h in culture with 5μg plate-bound anti-CD3/CD28 and 20U/ml rIL-2 (WT n=2, Gli2A n=3, unpaired t-test, p=0.02). (H) qPCR as in (C) to measure Gata3 relative expression. AU: arbitrary units.
Gli2A T-cells are Th2-like
These analyses suggested that Gli2A cells display transcriptional features similar to Th2 cells. To test this, we used a method with minimal assumptions to generate a scale of Th1/Th2 skewedness (Th1→Th2 score) based on publicly available whole-genome array data derived from Th-skewed cells (GEO database ref: GSE14308). We found Gli2A samples showed high, Th2-like scores when cells were either resting or stimulated (Fig. 2B). Interestingly, although the skewed reference data was obtained from T-cells that had been cultured for several days in skewing conditions, our transgenic T-cells showed a clear bias to Th2 (Gli2A) when prepared fresh from the mouse, compared to WT cells. After only six hours stimulation, Gli2A cells had further increased their Th2 score. Unstimulated Gli2R had a low score on the scale, suggesting a Th1 bias, supported by higher levels of Ifng and Cxcr3, and lower expression of Il1rl1 (Fig. 1A, cluster I, Fig. 2A). However, this trend in global gene expression disappeared upon stimulation (Fig. 2B).
Together, these analyses suggested that active Hh signalling in T-cells mediated transcriptional changes that promote Th2 differentiation. We therefore tested this hypothesis experimentally. We investigated the abundance of Il4 transcript by qPCR in activated CD4+ T-cells from WT and Gli2A spleen and found that Il4 expression in Gli2A cells was ~6-fold higher than WT (Fig. 2C). There was a consistent increase in the proportion of Gli2A cells producing IL-4 directly ex vivo compared to WT as assessed by intracellular cytokine staining on freshly isolated splenocytes (Fig. 2D). Expression of Il4 was also upregulated in activated WT T-cells following treatment with recombinant Shh (rShh), confirming that this effect is Hh-specific (Fig. 2E), and was repressed by the addition of neutralising anti-Hh mAb (5E1) to the cultures (Fig. 2F). Thus, physiological Hh signalling increases Il4 transcription in WT T-cells.
To confirm that Gli2A T-cells produce more IL-4 protein, we stimulated lymphocytes in vitro with anti-CD3/CD28 for 72h and assayed IL-4 secretion by ELISA. Gli2A cells produced on average twice as much IL-4 as WT (Fig. 2G).
The Th2 lineage-specific transcription factor Gata3 was not upregulated in Gli2A cells at 6h (GEO database ref: GSE33156). However, as Gli2A CD4+ cells readily upregulated Il4 mRNA expression and cytokine production, we tested whether Gata3 expression was increased in these cells after 48h activation. qPCR analysis showed that the presence of Gli2A led to a greater induction of Gata3 than in WT stimulated cells (Fig. 2H).
Gli2A enhances Th2-associated pathology
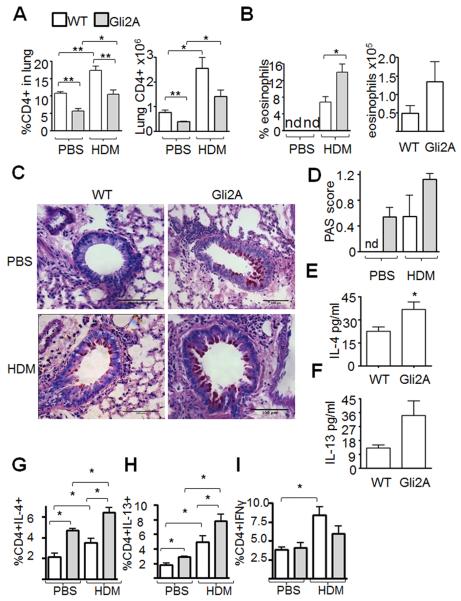
Our data suggest that activation of Hh signalling predisposes T-cells to become Th2-like upon stimulation. To test whether Hh-dependent transcription enhances Th2 differentiation and function in vivo, we used a murine model of mild allergic asthma, where dosing with house dust mite (HDM) extract elicits allergic pathology (31). After repeated intranasal administration of allergen/PBS bronchoalveolar lavage fluid (BAL), lung lobes and mediastinal lymph nodes (mLN) were collected from WT and Gli2A animals. For WT and Gli2A groups, the percentage of infiltrating lung CD4+ cells increased upon HDM-treatment (Fig. 3A), indicating T-cell recruitment during the response. The percentage and number of CD4+ cells in lung was reduced in Gli2A mice compared to WT (Fig. 3A), reflecting the decreased proportions of peripheral T-cells in these mice (11), but in both groups the percentage and number of CD4+ cells increased upon disease induction, reflecting T-cell recruitment to the lung. As expected, eosinophils were observed in BAL of HDM-treated mice only. There were more eosinophils in the airways in HDM-treated Gli2A mice compared to WT (Fig. 3B), indicating increased severity of the inflammatory phase of disease. Periodic acid Schiff staining showed that the prevalence of mucus-secreting cells in the bronchioles of Gli2A lungs was increased compared to WT (Fig. 3C,D). Interestingly, positive PAS staining was seen in PBS-treated Gli2A mice, possibly indicating some underlying airway pathology. In addition, BAL were analysed by ELISA for Th2 cytokines IL-4 and IL-13. There were increased levels of both Th2 cytokines in the HDM-treated Gli2A group compared to WT (Fig. 3E,F) despite the decreased percentage and number of CD4+ cells in lung tissue of transgenic animals (Fig. 3A).
Figure 3. Active Hh-dependent transcription enhances Th2-mediated disease in allergen-challenge allergic airways disease.
WT and Gli2A mice (n=5 in each group) underwent repeated intranasal challenge with PBS or 15μg HDM allergen in PBS as described. (A) Flow cytometric analysis of mean±SEM %CD4+ and number (×106) of CD4+ cells present in lung (*p=0.03, **p<0.006) and (B) percentage and number (×105) of eosinophils (CD11b+SiglecF+) in BAL 3wks after treatment (*p=0.02), nd: not detectable. (C,D) Bronchioles analysed using periodic acid shift (PAS) by histological scoring (bar=100μm) to identify mucus-secreting goblet cells (mean±SEM, non-parametric ANOVA: p=0.04). nd: not detectable. (E,F) ELISA analysis on BAL (2wks treatment) to quantify mean±SEM IL-4 (unpaired t-test: p=0.04) and IL-13 produced (p=0.08). (G-I) Ic-cytokine staining in CD4+ cells from lung for (G) IL-4, (H) IL-13 and (I) IFNγ. Statistically significant differences between group mean±SEM were identified by unpaired t-testing (*). WTvGli2A: IL-4 in PBS-treated p=0.0002, IL-4 in HDM-treated p=0.002; IL-13 in PBS-treated p=0.008, IL-13 in HDM-treated p<0.05. PBSvsHDM: IFNγ in WT (p=0.004), IFNγ in Gli2A (p=0.17); IL-4 in WT (p=0.04), IL-4 in Gli2A (p=0.006); IL-13 in WT (p=0.04), IL-13 in Gli2A (p=0.0008).
To characterise T-cells in the allergic airways model, we examined IL-4, IL-13 and IFNγ production on a per-cell basis by intracellular-cytokine staining in CD4+ cells. Allergen treatment induced production of Th2 cytokines IL-4 and IL-13 in WT and Gli2A mice as expected (Fig. 3G,H). Following HDM treatment, the proportions of CD4+ cells in lung expressing IL4 (Fig. 3G), and IL13 (Fig. 3H) were higher in Gli2A compared to WT. IFNγ was induced by HDM treatment in the WT T-cells, but not in the Gli2A group (Fig. 3I). Together this suggests a Th2-biased response in Gli2A animals. As expected (see Fig. 2), T-cells from Gli2A PBS-treated animals showed increased proportions of cells expressing Th2 cytokines compared to WT (Fig. 3G,H). Interestingly, goblet cell hyperplasia (Fig. 3C,D), was also observed in this group, suggesting that Hh-mediated transcription favours the Th2 phenotype even without allergen treatment. However, PBS-treated Gli2A mice did not recruit eosinophils into the airway (Fig. 3B), indicating that any basal Th2 inflammation is not equivalent to HDM-induced allergic response. This may indicate an uncoupling of goblet cell formation and IL4/IL-13 production from eosinophil recruitment.
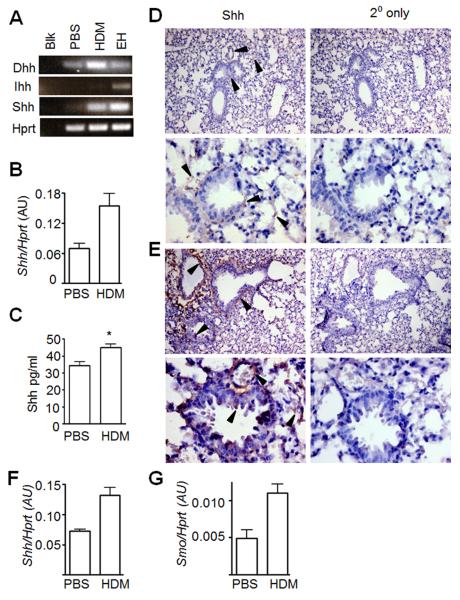
These data show that Gli-dependent transcription in T-cells exacerbates Th2-mediated allergen-induced pathology in vivo. Therefore, Hh proteins released from tissue may signal to local T-cells, resulting in enhanced Th2 differentiation/function. This would be of particular importance if Hh were released by inflamed tissue, as it would drive a positive feedback loop. To confirm that Hh family member proteins are present in non-transgenic, WT lung and investigate whether Hh expression is upregulated in allergic tissue, we assayed whole lung tissue homogenate from WT mice following PBS or HDM treatment. Dhh and Shh mRNA was detectable in lung samples by non-quantitative RT-PCR (Fig. 4A). We found that expression of Shh transcript (Fig. 4B, qPCR) and protein (Fig. 4C, ELISA) was significantly upregulated in lung homogenates from HDM-treated mice compared to PBS controls. Immunohistochemistry (IHC) showed that Shh protein is present at low levels in control lung (Fig. 4D), but is strongly expressed in HDM-treated WT lung, localised to basal stroma of airways, bronchial and alveolar epithelium, and in perivascular areas (Fig. 4E). To investigate further, we induced disease in WT mice and sorted epithelial (CD45-EpCAM+) and CD4+ T-cells (CD45+CD3+CD4+) from fresh lung by FACS and assessed gene expression by qPCR. In support of IHC data, expression of Shh was increased in epithelial cells from HDM-treated compared to PBS-treated WT mice (Fig. 4F). To determine whether lung T-cells were actively receiving Hh-signals in vivo, we measured expression of Smo, a reliable Hh-target gene in T-cells (Fig. 1E), in sorted lung CD4+ cells. Smo levels were raised in CD4+ cells from HDM-treated mice versus controls (Fig. 4G), indicating that lung T-cells transduced Hh signals in vivo, and that the pathway increased in activity upon induction of allergic disease.
Figure 4. Hh proteins are expressed in lung and lung-resident T-cells upregulate Hh target gene expression during allergic airways disease.
(A) Expression of Dhh, Ihh and Shh by non-quantitative RT-PCR in homogenised lung tissue from PBS- or HDM-treated WT BALB/c mice; Blk: water blank; EH: embryo head positive control. (B) Representative triplicate qPCR showing mean±SD expression Shh transcript in lung relative to Hprt (mean±SEM relative Shh expression in PBS- (n=4) compared to HDM-treated (n=6) WT mice: p=0.02). (C) Mean±SEM Shh protein concentration (ELISA, p=0.04) in homogenised lung of WT mice treated with PBS (n=4) or HDM (n=6). Immunohistochemical detection of Shh in (D) PBS- and (E) HDM-treated WT fixed paraffin-embedded lung sections at 100x (upper panels) and 400x (lower panels) magnification. Black arrows indicate Shh staining. (F, G) Lung epithelial cells (CD45-EpCAM+) and lung CD4+ T-cells (CD45+CD3+CD4+) were sorted by FACS from WT mice treated with PBS or HDM. Representative triplicate qPCR showing mean±SD expression of (F) Shh and transcript in lung epithelial cells, and (G) Smo in lung CD4+ T-cells, relative to Hprt. (mean±SEM relative Shh expression in PBS- (n=3) compared to HDM-treated (n=3) independent lung epithelial cell samples from WT mice: p=0.0095, for Smo in lung CD4+ cell samples: p=0.02).
Hh signals promote Th2 differentiation
Expression of Gata3 is induced and maintained by Th2-skewing conditions, drives production of Th2 cytokines, occurs over a course of hours-days (37), and can thus be used as an indicator of Th2 transcriptional identity. Therefore, to investigate the molecular mechanisms behind the exacerbation of allergic airways disease caused by Hh-mediated transcription in T-cells, we cultured WT and Gli2A CD4+ cells in Th-skewing conditions following activation, and measured commitment to Th2, by intracellular expression of Gata3 protein.
In neutral Th0 conditions, levels of Gata3 were always higher in Gli2A compared to WT CD4+ cells (Fig. 5A,B), suggesting an inherent bias towards Th2. In Th2 conditions, where exogenous IL-4 is added, WT cells displayed Gata3 levels similar to Gli2A cells at 24h and 48h, suggesting that enhanced expression of Gata3 by Th0-conditioned Gli2A cells is the result of their increased IL-4 production. However, after six days in culture, on average double the proportion of Gli2A cells expressed Gata3 than WT, showing enhanced commitment to the Th2 lineage (Fig. 5A,C). Th1 differentiation is controlled by Tbet, which antagonises the effects of Gata3 and vice versa (38). To test whether Gli2A cells displayed decreased Th1 potential compared to WT, we examined Tbet expression by staining in skewing cultures at 48h. Expression of Tbet was high and similar in Th0- or Th1-conditioned cultures, but in Th2 conditions was decreased in Gli2A cells compared to WT (Fig. 5D). This resulted in an increased ratio of Gata3:Tbet, particularly in Th2-skewed Gli2A CD4+ cultures (Fig. 5E), confirming that these cells are biased to a Gata3+ Th2 phenotype.
Figure 5. Active Hh-dependent transcription upregulates expression of Gata3 and skews cells towards a Th2 phenotype in vitro.
(A) Ic-Gata3 protein expression in WT and Gli2A CD4+ splenocytes after culture in Th-neutral (Th0) or Th2-skewing conditions. Mean±SEM (B) %Gata3+ cells as fold change (FC) of the WT cultures (n=3 independent experiments) in (B) Th0 (unpaired t-test, *p=0.01, **p=0.001) and (C) Th2 conditions (unpaired t-test, *p=0.02). (D) Ic-Tbet protein in Th0 or Th1/Th2-skewed cells. (E) Example Gata3:Tbet ratio at 48h. (F) WT CD4+ cells cultured for three days with a single dose of rShh at the start of the culture in (G) Th0 neutral (n=5, paired t-test: %Gata3+ p=0.04, MFI Gata3 p=0.03) and (H) Th2 conditions (n=5). Dotted overlays show example ic-isotype control staining, MFI of each histogram is shown in italics.
To verify that Hh signalling promotes Th2 differentiation of non-transgenic cells we cultured WT CD4+ cells in neutral Th0 conditions for 72h with a single dose of rShh. By 48h, Gata3 expression was higher in Hh-treated cells (Fig. 5F,G). To ask if the increase in Gata3 expression on rShh treatment was the result of increased IL-4 production, we cultured WT CD4+ cells in Th2 conditions in the presence of rShh. As expected, when IL-4 was added to the cultures, Gata3 expression was not affected by rShh (Fig. 5H), suggesting that strong IL-4 signals in the cultures (also in the presence of neutralising anti-IFNγ) are sufficient for maximal Th2 differentiation, so Shh treatment had no impact, or that IL-4 induction is downstream of Shh signalling.
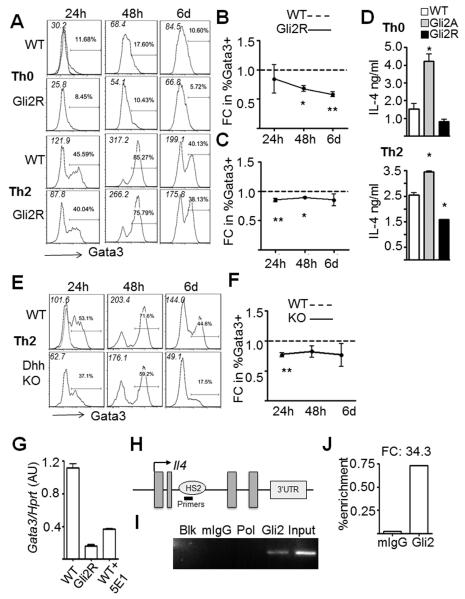
Repressing physiological Hh inhibits Th2
To test whether repression of physiological Hh-dependent transcription would impair Th2 potential and suppress Gata3 expression, we performed in vitro skewing experiments using Gli2R CD4+ cells. Gata3 did not reach WT levels over the time course in Th0 (Fig. 6A,B) or Th2 (Fig. 6A,C) conditions, indicating that repression of physiological Hh pathway activation in T-cells reduces their ability to maximally upregulate Gata3 and differentiate to the Th2-lineage. The fact that addition of exogenous IL-4 and anti-IFNγ (Th2 conditions) to Gli2R cultures was not sufficient to restore Gata3 expression to WT levels indicates that other factors in addition to these cytokines are involved in the mechanisms of Hh-mediated modulation of Th differentiation, and thus reflects our microarray data (Fig. 1,2).
Figure 6. Physiological Hh-signalling controls expression of Gata3 and Il4.
(A) Ic-Gata3 staining as Fig. 5 in WT and Gli2R CD4+ splenocytes in (B) Th0 (mean±SEM, n=3, *p=0.004, **p=0.009) and (C) Th2 conditions, (**p=0.005, *p<0.05). (D) IL-4 production per 105 cells measured by ELISA after 6 days in culture in Th0 (upper panel) or Th2 (lower panel) conditions (WT n=3, Gli2A n=2, Gli2R n=2, in triplicate *p<0.01). (E,F) Ic-Gata3 in Dhh KO and WT CD4+ cells (n=3, *p=0.0004) in Th2 conditions. Dotted overlays show control stain, MFI of each histogram is shown in italics. (G) Mean±SD Gata3 expression relative to Hprt in activated CD4+ splenocytes from Gli2R, or WT ±anti-Hh (5E1), triplicate qPCR representative of two independent experiments (WT vs. Gli2R, WT vs. WT ±anti-Hh: p<0.05). (H) The murine Il4 locus. (I,J) Sonicated chromatin from activated Gli2A CD4 splenocytes was immunoprecipitated with anti-Gli2, control IgG and anti-RNA-PolII and ChIP-PCR was performed on the intronic HS2 region of Il4. (I) End-point PCR showed a product in input (non-immunoprecipitated) and Gli2-precipitated fractions. (J) Fold enrichment in the Gli2-precipitated fraction was calculated based on input and control, quantified by qPCR.
To confirm that the activation or inhibition of Hh-dependent transcription altered IL-4 production by cells cultured in skewing conditions, we measured IL-4 protein concentration by ELISA after 6 days in culture (Fig. 6D). As expected, cultures of Gli2A cells contained on average 2.8-fold more IL-4 than WT cells, and Gli2R cells half the amount of IL-4 after culture in Th0 conditions (Fig. 6D upper panel). In Th2 conditions, compared to WT, Gli2A and Gli2R culture supernatants contained 1.5-fold and 0.5-fold IL-4 respectively (Fig. 6D lower panel). Similar levels of IL-4 were detected in supernatants from Th0 conditions as from equivalent Th2-skewed cultures. This has been observed before (39), and most likely reflects both the fact that Gli2A cells secrete high levels of endogenous IL-4, and the complex balance between the kinetics of IL-4 production, uptake and degradation during the prolonged six-day culture period in different conditions.
Physiological levels of Hh pathway activation in T-cells isolated fresh from the mouse spleen thus skew transcriptional processes to favour Th2 differentiation (Fig. 6). Given this, we examined the effect of reducing environmental Hh protein in the spleen on Th2 differentiation of non-transgenic CD4+ cells ex vivo. We used Dhh−/− knockout (KO) CD4 splenocytes, as Dhh is expressed by spleen stroma (40), and unlike Shh−/−, is not an embryonic-lethal mutation. We cultured Dhh KO and WT cells in Th2 skewing conditions and found that Dhh KO cells upregulate Gata3 less efficiently than WT after stimulation (Fig. 6E). This was statistically significant at 24h (Fig. 6F), suggesting that fresh KO cells are inherently impaired in their ability to undergo rapid Th2 differentiation, as a result of reduction in Hh signal from their environment.
Given that Gata3 protein expression was lowered by a reduction in Hh signalling, we measured Gata3 mRNA in Gli2R CD4 cells and in WT cells treated with anti-Hh mAb after 48h anti-CD3/CD28 stimulation. Gata3 transcription was reduced by inhibition of Hh signalling (Fig. 6G). Taken together, these experiments show that the physiological Hh signal can functionally control expression of this lineage-specifying transcription factor in CD4+ splenocytes.
Gli2 binds the Il4 gene
The rapid (6h) induction of Il4 (Fig. 1) and increased IL-4 production (Fig. 2, 6) by Gli2A cells, altered Gata3 induction (Figs. 5,6) and marked repression of Gata3 expression in Gli2R CD4+ T-cells and 5E1-treated WT cells (Fig. 6G) prompted us to investigate whether Gli2 could act directly to initiate or enhance transcription of Il4 and/or Gata3. We examined genomic sequences for suggested Gli consensus binding sequences and found several potential sites in Gata3 and Il4 genomic sequences. We therefore investigated whether Gli2 can directly bind Il4 and Gata3 by chromatin immunoprecipitation (ChIP). CD4+ cells from Gli2A mice were stimulated for 48h and then fragmented chromatin was immunoprecipitated with anti-Gli2 or control antibodies. DNA was purified from bound targets and PCR was performed to amplify regions of the Gata3 and Il4 genes identified as containing potential Gli binding sites. We found no enrichment of Gata3 by conventional PCR or qPCR assays which span 1kb up- and downstream of the TSS incorporating regions containing possible Gli binding sites. However, in the case of the Il4 locus (Fig. 6H), we observed significant binding to a region localising to an important enhancer element in intron 2 (DNAse-hypersensitivity site HS2 (41)) (Fig. 6I,J), but not to regions around the Il4 promoter (not shown). The HS2 region is critical for full IL-4-dependent Th2 responses (34), and necessary for chromatin remodelling essential for lineage-specific Il4 expression. Our data therefore identify Il4 as a novel target gene of Gli2-dependent Hh signalling and provide a mechanism for the role of Gli2 in skewing Th differentiation.
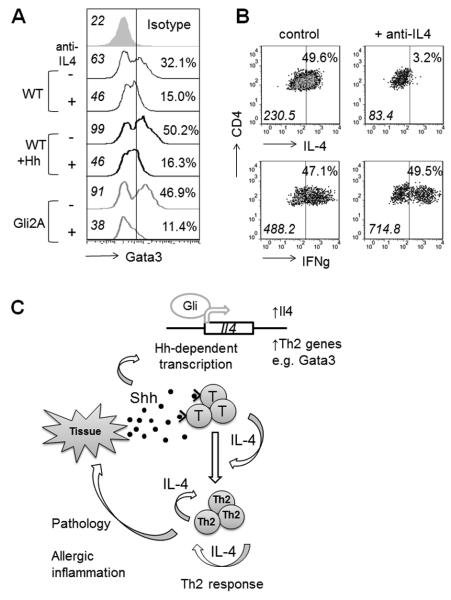
To test whether the Hh-dependent induction of Gata3 requires IL-4, we cultured CD4+ cells with anti-CD3/28-coated activator beads with or without anti-IL4 antibody and measured Gata3 expression after 48h. As expected, inhibition of IL-4 signalling in WT cultures markedly decreased Gata3 expression, confirming that IL-4 is required to support Th2 differentiation. Activation of Hh signalling enhanced Gata3 expression in CD4+ cells, but this effect was lost when anti-IL4 was present in the culture (Fig. 7A), confirming that IL-4 is required to induce Gata3 in WT cells and demonstrating that the Gli2A transgene does not activate Gata3 transcription independently of IL-4.
Figure 7. Hh regulates Th2 differentiation.
(A) Ic-Gata3 staining in CD4+ splenocytes after 48h in culture with anti-CD3/CD28-coated beads either with (+) or without (−) 5μg/ml anti-IL4 mAb. Histograms represent at least two independent experiments, MFI is shown in italics, percentages refer to proportion of cells to the right of the marker. (B) Ic-cytokines measured after 6 days in culture ±anti-IL-4 as described in (A). Marker on dot plots denote %cells with positive staining as assessed by isotype control (not shown), italics indicate MFI of population. (C) Schematic illustrating a proposed model where inflamed/injured/activated tissue releases Hh protein, signalling to local T-cells, causing upregulation of Il4 and influencing other Th genes by Gli-dependent transcription. IL-4 is released leading to enhanced Th2 differentiation/survival via Gata3 upregulation, and an increase in Th2 function, exacerbating pathology and tissue damage.
To test the impact of neutralisation of IL-4 on the cytokine production of Gli2A cells, we measured IL-4 expression by Ic-cytokine staining in cells after 6 days stimulation with activator beads. The presence of anti-IL-4 in the culture significantly blocked IL-4 expression (Fig. 7B), indicating that autocrine signalling is required for full IL-4 production. In contrast, the expression of IFNγ was unaffected by the presence of anti-IL4.
Discussion
Here we demonstrate that expression of Hh ligand increases in tissue following induction of Th2-mediated disease, and that Hh signalling in T-cells promotes Th2 function by skewing global transcription of T-cells towards a Th2-like profile, directly enhancing IL-4 production. In asthma, this process is pathological, but it may have evolved as an anti-inflammatory mechanism, or to enhance immunity to parasitic infection. Our data have important consequences for understanding how non-immune, tissue-derived factors influence T-cell immunity, particularly given the wide tissue expression of Hh family proteins. We demonstrate that morphogens can play a surprising role in the regulation of fluid, as well as solid organ systems. Highly migratory cells such as lymphocytes will traffic through many microenvironments in the course of their life, each of which therefore has the potential to alter the cells’ function. The role of context dependent, tissue-derived signals in influencing immunity in disease is an exciting emerging field. Understanding the impact of such events will be important for dissecting the mechanisms controlling complex diseases, which may have several sub-phenotypes, where multiple signals integrate to drive pathology.
In lung, we have shown that Hh proteins are present under normal conditions and that Shh expression is upregulated in lung epithelial cells during allergic inflammation. Further, CD4+ T-cells from lung express enhanced levels of Smo transcript after HDM-treatment, indicating active Hh signalling. Thus, inflamed tissue, including but probably not limited to epithelia, releases Shh, potentiating Hh signalling to local T-cells. This leads to transcriptional changes, increased IL-4 production, and enhanced Th2 responses, signalling to other immune effector cells, maintaining allergic inflammation and further aggravating disease (Fig. 7C). It will be of interest to measure Hh levels in BAL/lung from asthma sufferers. Importantly, recent GWAS have identified key SNPs in the hedgehog-sequestering molecule HHIP as important for lung capacity and function (spirometric measures) in human asthma (42). Our data demonstrate that Hh signalling is also involved in lymphocyte function in murine asthma. Therefore, it is timely to investigate the potential therapeutic use of small molecule inhibitors of Hh signalling in Th2-mediated disease.
Our study also investigated the influence of Hh proteins on normal T-cell differentiation. Hh signalling is involved adult stem cell renewal and in the pathogenesis of several cancers. To understand and develop strategies to treat Hh-dependent haematological and lymphoid malignancies (43, 44) it is clearly important to understand how Hh proteins function in healthy cells. We identified Il4 as a transcriptional target of Hh signalling in T-cells, and this is of interest not only because of its pivotal role in Th2 biology, but also because of the function of IL-4 in signalling to other immune cells. Increased IL-4 in tumour microenvironments can inhibit anti-tumour responses or promote tumour growth (45, 46). Interestingly, increased IL-4 has been observed in several cancers strongly associated with aberrant Hh signalling (47-49), and IL-4 derived from tumour-resident T-cells sustained growth of follicular B-lymphomas (50), tumours known to produce Hh ligands (51). Future work should therefore characterise the role of Hh in human allergic disease, and investigate whether Hh plays a role in modulating other Th-responses and tumour immune evasion.
Acknowledgements
We thank UCL Genomics for microarray services, JP Martinez-Barbera and N. Charolidi for guidance with ChIP, A. Eddaoudi/A. Camille for cell sorting and ICH/GOSH Histopathology for PAS staining.
Abbreviations
- Hh
Hedgehog
- Shh
Sonic Hh
- Ihh
Indian Hh
- Dhh
Desert Hh
- Th
T-helper
- WT
Wild-Type
Footnotes
Funding sources: Wellcome Trust, BBSRC, MRC; ALF is supported by GOSH/ICH BRC, funded by the Department of Health’s NIHR’s Biomedical Research Centres scheme; MO by a Human Frontier Science Program Long Term Fellowship; HS by a UCL CHRAT studentship.
- MIAME compliant microarray data are available at the GEO repository (http://www.ncbi.nlm.nih.gov/geo/) dataset: GSE33156.
- The authors have no conflicting interests.
References
- 1.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koebernick K, Pieler T. Gli-type zinc finger proteins as bipotential transducers of Hedgehog signaling. Differentiation. 2002;70:69–76. doi: 10.1046/j.1432-0436.2002.700201.x. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 4.Shah DK, Hager-Theodorides AL, Outram SV, Ross SE, Varas A, Crompton T. Reduced Thymocyte Development in Sonic Hedgehog Knockout Embryos. J Immunol. 2004;172:2296–2306. doi: 10.4049/jimmunol.172.4.2296. [DOI] [PubMed] [Google Scholar]
- 5.Hager-Theodorides AL, Dessens JT, Outram SV, Crompton T. The transcription factor Gli3 regulates differentiation of fetal CD4− CD8− double-negative thymocytes. Blood. 2005;106:1296–1304. doi: 10.1182/blood-2005-03-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Andaloussi A, Graves S, Meng F, Mandal M, Mashayekhi M, Aifantis I. Hedgehog signaling controls thymocyte progenitor homeostasis and differentiation in the thymus. Nat Immunol. 2006;7:418–426. doi: 10.1038/ni1313. [DOI] [PubMed] [Google Scholar]
- 7.Crompton T, Outram SV, Hager-Theodorides AL. Sonic hedgehog signalling in T-cell development and activation. Nat Rev Immunol. 2007;7:726–735. doi: 10.1038/nri2151. [DOI] [PubMed] [Google Scholar]
- 8.Rowbotham NJ, Hager-Theodorides AL, Furmanski AL, Ross SE, Outram SV, Dessens JT, Crompton T. Sonic hedgehog negatively regulates pre-TCR-induced differentiation by a Gli2-dependent mechanism. Blood. 2009;113:5144–5156. doi: 10.1182/blood-2008-10-185751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Outram SV, Hager-Theodorides AL, Shah DK, Rowbotham NJ, Drakopoulou E, Ross SE, Lanske B, Dessens JT, Crompton T. Indian hedgehog (Ihh) both promotes and restricts thymocyte differentiation. Blood. 2009;113:2217–2228. doi: 10.1182/blood-2008-03-144840. [DOI] [PubMed] [Google Scholar]
- 10.Outram SV, Varas A, Pepicelli CV, Crompton T. Hedgehog signaling regulates differentiation from double-negative to double-positive thymocyte. Immunity. 2000;13:187–197. doi: 10.1016/s1074-7613(00)00019-4. [DOI] [PubMed] [Google Scholar]
- 11.Rowbotham NJ, Hager-Theodorides AL, Cebecauer M, Shah DK, Drakopoulou E, Dyson J, Outram SV, Crompton T. Activation of the Hedgehog signaling pathway in T-lineage cells inhibits TCR repertoire selection in the thymus and peripheral T-cell activation. Blood. 2007;109:3757–3766. doi: 10.1182/blood-2006-07-037655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowbotham NJ, Furmanski AL, Hager-Theodorides AL, Ross SE, Drakopoulou E, Koufaris C, Outram SV, Crompton T. Repression of hedgehog signal transduction in T-lineage cells increases TCR-induced activation and proliferation. Cell Cycle. 2008;7:904–908. doi: 10.4161/cc.7.7.5628. [DOI] [PubMed] [Google Scholar]
- 13.Hager-Theodorides AL, Furmanski AL, Ross SE, Outram SV, Rowbotham NJ, Crompton T. The Gli3 transcription factor expressed in the thymus stroma controls thymocyte negative selection via Hedgehog-dependent and -independent mechanisms. J Immunol. 2009;183:3023–3032. doi: 10.4049/jimmunol.0900152. [DOI] [PubMed] [Google Scholar]
- 14.Furmanski AL, Saldana JI, Rowbotham NJ, Ross SE, Crompton T. Role of Hedgehog signalling at the transition from double positive to single positive thymocyte. European Journal of Immunology. 2012;42:489–499. doi: 10.1002/eji.201141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowrey JA, Stewart GA, Lindey S, Hoyne GF, Dallman MJ, Howie SE, Lamb JR. Sonic hedgehog promotes cell cycle progression in activated peripheral CD4(+) T lymphocytes. J Immunol. 2002;169:1869–1875. doi: 10.4049/jimmunol.169.4.1869. [DOI] [PubMed] [Google Scholar]
- 16.Stewart GA, Lowrey JA, Wakelin SJ, Fitch PM, Lindey S, Dallman MJ, Lamb JR, Howie SE. Sonic hedgehog signaling modulates activation of and cytokine production by human peripheral CD4+ T cells. J Immunol. 2002;169:5451–5457. doi: 10.4049/jimmunol.169.10.5451. [DOI] [PubMed] [Google Scholar]
- 17.Le H, Kleinerman R, Lerman OZ, Brown D, Galiano R, Gurtner GC, Warren SM, Levine JP, Saadeh PB. Hedgehog signaling is essential for normal wound healing. Wound Repair Regen. 2008;16:768–773. doi: 10.1111/j.1524-475X.2008.00430.x. [DOI] [PubMed] [Google Scholar]
- 18.Stewart GA, Hoyne GF, Ahmad SA, Jarman E, Wallace WA, Harrison DJ, Haslett C, Lamb JR, Howie SE. Expression of the developmental Sonic hedgehog (Shh) signalling pathway is up-regulated in chronic lung fibrosis and the Shh receptor patched 1 is present in circulating T lymphocytes. J Pathol. 2003;199:488–495. doi: 10.1002/path.1295. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen CM, Williams J, van den Brink GR, Lauwers GY, Roberts DJ. Hh pathway expression in human gut tissues and in inflammatory gut diseases. Lab Invest. 2004;84:1631–1642. doi: 10.1038/labinvest.3700197. [DOI] [PubMed] [Google Scholar]
- 20.Dierks C, Grbic J, Zirlik K, Beigi R, Englund NP, Guo GR, Veelken H, Engelhardt M, Mertelsmann R, Kelleher JF, Schultz P, Warmuth M. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nat Med. 2007;13:944–951. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 21.Sacedon R, Diez B, Nunez V, Hernandez-Lopez C, Gutierrez-Frias C, Cejalvo T, Outram SV, Crompton T, Zapata AG, Vicente A, Varas A. Sonic hedgehog is produced by follicular dendritic cells and protects germinal center B cells from apoptosis. J Immunol. 2005;174:1456–1461. doi: 10.4049/jimmunol.174.3.1456. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul WE. What determines Th2 differentiation, in vitro and in vivo? Immunol Cell Biol. 2010;88:236–239. doi: 10.1038/icb.2010.2. [DOI] [PubMed] [Google Scholar]
- 24.Kopf M, Le Gros G, Bachmann M, Lamers MC, Bluethmann H, Kohler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr., Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 26.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 27.Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol. 2002;168:4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- 28.Akhabir L, Sandford AJ. Genome-wide association studies for discovery of genes involved in asthma. Respirology. 2011;16:396–406. doi: 10.1111/j.1440-1843.2011.01939.x. [DOI] [PubMed] [Google Scholar]
- 29.Tamari M, Tomita K, Hirota T. Genome-wide association studies of asthma. Allergol Int. 2011;60:247–252. doi: 10.2332/allergolint.11-RAI-0320. [DOI] [PubMed] [Google Scholar]
- 30.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by Desert hedgehog regulates the male germline. Current biology: CB. 1996;6:298–304. doi: 10.1016/s0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 31.Gregory LG, Causton B, Murdoch JR, Mathie SA, O’Donnell V, Thomas CP, Priest FM, Quint DJ, Lloyd CM. Inhaled house dust mite induces pulmonary T helper 2 cytokine production. Clin Exp Allergy. 2009;39:1597–1610. doi: 10.1111/j.1365-2222.2009.03302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins D, Winyard PJ, Woolf AS. Immunohistochemical analysis of Sonic hedgehog signalling in normal human urinary tract development. J Anat. 2007;211:620–629. doi: 10.1111/j.1469-7580.2007.00808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, T R, Kano M, Sugiman T. Visualising the Cross-Level Relationships between Pathological and Physiological Processes and Gene Expression: Analyses of Haematological Diseases. PLoS ONE. 2013;8:e53544. doi: 10.1371/journal.pone.0053544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka S, Motomura Y, Suzuki Y, Yagi R, Inoue H, Miyatake S, Kubo M. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat Immunol. 2011;12:77–85. doi: 10.1038/ni.1966. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu C, Kawamoto H, Yamashita M, Kimura M, Kondou E, Kaneko Y, Okada S, Tokuhisa T, Yokoyama M, Taniguchi M, Katsura Y, Nakayama T. Progression of T cell lineage restriction in the earliest subpopulation of murine adult thymus visualized by the expression of lck proximal promoter activity. International immunology. 2001;13:105–117. doi: 10.1093/intimm/13.1.105. [DOI] [PubMed] [Google Scholar]
- 36.Buckland J, Pennington DJ, Bruno L, Owen MJ. Co-ordination of the expression of the protein tyrosine kinase p56(lck) with the pre-T cell receptor during thymocyte development. European Journal of Immunology. 2000;30:8–18. doi: 10.1002/1521-4141(200001)30:1<8::AID-IMMU8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 38.Chakir H, Wang H, Lefebvre DE, Webb J, Scott FW. T-bet/GATA-3 ratio as a measure of the Th1/Th2 cytokine profile in mixed cell populations: predominant role of GATA-3. J Immunol Methods. 2003;278:157–169. doi: 10.1016/s0022-1759(03)00200-x. [DOI] [PubMed] [Google Scholar]
- 39.Cameron SB, Stolte EH, Chow AW, Savelkoul HF. T helper cell polarisation as a measure of the maturation of the immune response. Mediators Inflamm. 2003;12:285–292. doi: 10.1080/0929350310001619744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry JM, Harandi OF, Porayette P, Hegde S, Kannan AK, Paulson RF. Maintenance of the BMP4-dependent stress erythropoiesis pathway in the murine spleen requires hedgehog signaling. Blood. 2009;113:911–918. doi: 10.1182/blood-2008-03-147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, Calhoun WJ, Castro M, Chung KF, Erzurum SC, Fitzpatrick AM, Gaston B, Israel E, Jarjour NN, Teague WG, Wenzel SE, Peters SP, Hawkins GA, Bleecker ER, Meyers DA. Importance of hedgehog interacting protein and other lung function genes in asthma. The Journal of allergy and clinical immunology. 2011;127:1457–1465. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, Munchhof M, VanArsdale T, Beachy PA, Reya T. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ok CY, Singh RR, Vega F. Aberrant activation of the hedgehog signaling pathway in malignant hematological neoplasms. Am J Pathol. 2012;180:2–11. doi: 10.1016/j.ajpath.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler A, Heidenreich R, Braumuller H, Wolburg H, Weidemann S, Mocikat R, Rocken M. EpCAM, a human tumor-associated antigen promotes Th2 development and tumor immune evasion. Blood. 2009;113:3494–3502. doi: 10.1182/blood-2008-08-175109. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Jiang J, Wang Z, Zhang J, Xiao M, Wang C, Lu Y, Qin Z. Endogenous interleukin-4 promotes tumor development by increasing tumor cell resistance to apoptosis. Cancer Res. 2008;68:8687–8694. doi: 10.1158/0008-5472.CAN-08-0449. [DOI] [PubMed] [Google Scholar]
- 47.Yamamura M, Modlin RL, Ohmen JD, Moy RL. Local expression of antiinflammatory cytokines in cancer. J Clin Invest. 1993;91:1005–1010. doi: 10.1172/JCI116256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prokopchuk O, Liu Y, Henne-Bruns D, Kornmann M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: evidence for autocrine and paracrine actions. Br J Cancer. 2005;92:921–928. doi: 10.1038/sj.bjc.6602416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zisakis A, Piperi C, Themistocleous MS, Korkolopoulou P, Boviatsis EI, Sakas DE, Patsouris E, Lea RW, Kalofoutis A. Comparative analysis of peripheral and localised cytokine secretion in glioblastoma patients. Cytokine. 2007;39:99–105. doi: 10.1016/j.cyto.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Ame-Thomas P, Le Priol J, Yssel H, Caron G, Pangault C, Jean R, Martin N, Marafioti T, Gaulard P, Lamy T, Fest T, Semana G, Tarte K. Characterization of intratumoral follicular helper T cells in follicular lymphoma: role in the survival of malignant B cells. Leukemia. 2011 doi: 10.1038/leu.2011.301. epub 2011/10/22 10.1038/leu.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh RR, Kim JE, Davuluri Y, Drakos E, Cho-Vega JH, Amin HM, Vega F. Hedgehog signaling pathway is activated in diffuse large B-cell lymphoma and contributes to tumor cell survival and proliferation. Leukemia. 2010;24:1025–1036. doi: 10.1038/leu.2010.35. [DOI] [PubMed] [Google Scholar]