Abstract
This study investigated pregnane X receptor polymorphisms in relation to unboosted atazanavir plasma concentrations in 2 cohorts of patients. The polymorphism 63396T→C predicted concentrations below the minimum effective concentration (150 ng/mL) with odds ratios of 18 (P = .008) and 5.13 (P = .02). Prospective studies determining potential clinical usefulness are now warranted.
Atazanavir is a protease inhibitor that can be administered at a dosage of 400 mg once per day (unboosted) or 300 mg with a 100-mg dose of ritonavir once per day (boosted). Although the use of unboosted atazanavir is currently not licensed in Europe, it remains the protease inhibitor of choice for patients who are unable to tolerate ritonavir and whose treatment options are limited. Atazanavir plasma concentrations correlate with treatment response, and a minimum effective concentration of 150 ng/mL has been proposed as a target in treatment guidelines [1]. This is particularly relevant for unboosted atazanavir, because mean trough concentration (Ctrough) values are lower for patients receiving unboosted atazanavir than they are for those receiving boosted atazanavir [2].
Plasma concentrations of atazanavir are influenced by many processes that are mediated by different transporters and enzymes. Atazanavir metabolism is effected mainly by cytochrome P450 3A4 (CYP3A4), and ritonavir inhibits CYP3A4, thereby increasing atazanavir plasma concentrations when coadministered [2]. The only single-nucleotide polymorphism (SNP) that substantially influences CYP3A4 expression and activity is the CYP3A4*1B (rs2740574), which is uncommon among white individuals [3]. Most protease inhibitors are also substrates of ABCB1, and a polymorphism in the ABCB1 gene (3435C→T; rs1045642) correlates with atazanavir plasma concentrations in patients treated with boosted and unboosted therapy [4, 5]. It must be noted that many studies have evaluated this polymorphism in relation to the disposition of other protease inhibitors, with conflicting results [6], which emphasizes the need to ensure that pharmacogenetic studies are underpinned by biologically plausible mechanisms and that findings are replicated in multiple cohorts.
The pregnane X receptor (PXR; NR1I2) regulates the expression of CYP3A4 [7] and ABCB1 [8] in response to endobiotics and xenobiotics [9]. Several studies have scanned for polymorphisms in the regulatory and coding regions of PXR [10–12]. Recently, 3 polymorphisms (44477T→C [rs1523130], 63396C→T [rs2472677], and 69789A→G [rs763645]) were reported in putative transcription factor binding sites of PXR regulatory regions [11]. Moreover, the T allele at position 63396 was associated with increased expression of PXR and increased activity of CYP3A4 in primary hepatocytes and liver [11]. Thus, it is likely that this polymorphism influences the expression of PXR, which in turn influences the expression and activity of CYP3A4.
We have previously hypothesized about the contribution of PXR polymorphisms to the disposition of antiretrovirals [13]. In the present study, the effect of these SNPs on atazanavir plasma concentrations in patients treated with an unboosted regimen was evaluated.
Methods
An analysis of 3 PXR SNPs (44477T→C, 63396C→T, and 69789A→G) was first conducted in a cohort of 47 patients (cohort A) from the Department of Infectious Diseases at the University of Turin (Turin, Italy). The genotyping for 63396C→T was then repeated in the Liverpool Therapeutic Drug Monitoring (TDM) Registry (62 patients; cohort B). Inclusion criteria for both cohorts were as follows: receipt of unboosted atazanavir (400 mg once per day), age >18 years, and no receipt of drugs known to alter plasma concentration of atazanavir (e.g., proton pump inhibitors or nonnucleoside reverse-transcriptase inhibitors). The following data were collected: age, sex, body weight, time after dose, concomitant antiretroviral drugs, and other medications. Cohort A patients had a good adherence record as assessed by analyses of clinical and pharmacy records (>95% adherence). Sampling was performed after written informed consent was obtained in accordance with local ethics committee indications. For cohort B, DNA was extracted from unlinked, anonymous plasma samples obtained from eligible patients. The TDM registry constitutes a database of TDM requests for HIV drugs from 1999 through 2007 (>18,000 requests) with archived plasma. Data collected include patient demographic information, all concomitant medications, drug dosage, and time since most recent dose. Ethical approval was obtained from the UK North West Multicentre Research Ethics Committee.
Genomic DNA was extracted with use of the QIAamp whole blood mini kit (Qiagen) according to the manufacturer’s instructions. Genotyping was conducted by real time–based allelic discrimination with use of standard methodology [14]. All primers, probes, and PCR conditions are available on request. Atazanavir plasma concentrations were quantified using internally validated liquid chromatography–mass spectrometry [15] and high-performance liquid chromatography UV methodology [16] for cohort A and cohort B, respectively. The lower limit of quantification for both methods was 25 ng/mL, and both methods are externally validated by a quality control program [17]. Ctrough was measured in samples collected 20–26 h after dosing. When samples were obtained 12–20 h after ingestion of the drug, Ctrough was derived according to a half-life of 4.6 h [2, 4].
All variants were tested for Hardy-Weinburg equilibrium by χ2 test of observed and predicted genotype frequencies. Results for categorical data are expressed as medians with interquartile ranges (IQRs). All data were assessed for normality using a Shapiro-Wilk test, and categorical data were compared using the Mann-Whitney U test. To investigate continuous data, a Pearson or Spearman correlation was used. Because none of the demographic factors were associated with atazanavir concentrations, multivariate analyses were not conducted. A Bonferoni correction was applied to data from cohort A to correct for multiple comparisons. P values <.05 were considered to be statistically significant.
Results
A total of 47 white patients met the inclusion criteria and were included in cohort A. Of these, 28 patients (60%) were male, the median age was 45 years (IQR, 39–50 years), the median weight was 65 kg (IQR, 56–76 kg), and the median body mass index (calculated as weight in kilograms divided by the square of height in meters) was 23.7 (IQR, 20.4–26.0). No associations between demographic or physical characteristics and the atazanavir Ctrough were observed. Sex did not influence median atazanavir Ctrough values (men, 55 ng/mL [IQR, 25–179 ng/mL); women, 82 ng/mL [IQR, 41–248]; P = .13). Similarly, atazanavir plasma concentrations were not correlated with age (ρ, −0.01; P = .96), weight (ρ, −0.05; P = .75), or body mass index (ρ, 0.11; P = .47). Median atazanavir Ctrough did not differ between patients treated with tenofovir and patients who were not treated with tenofovir (median Ctrough, 50 ng/mL [IQR, 25–163 ng/mL] vs. 68 ng/mL [IQR, 29–235 ng/mL]; P = .42).
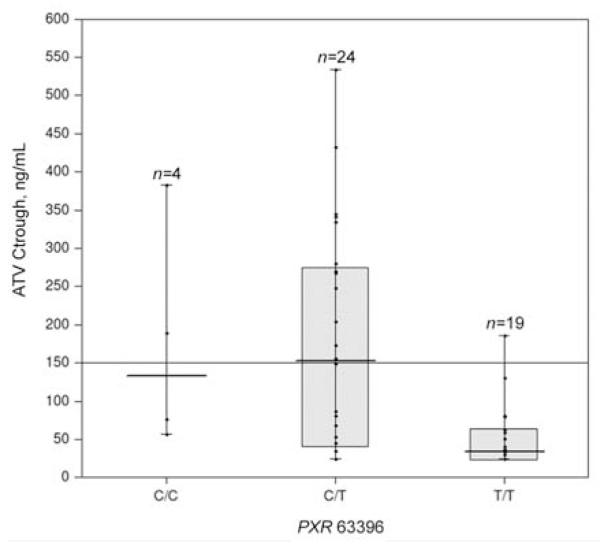
The allele frequencies for 44477C, 63396T, and 69789G were 55%, 66%, and 37%, respectively. All SNPs were in Hardy-Weinburg equilibrium. Median Ctrough was lower for individuals characterized by PXR 63396T homozygosity, compared with the other 2 groups (C allele homozygotes and heterozygotes) (34 ng/mL [IQR, 25–63 ng/mL] vs. 152 ng/mL (IQR, 47–388 ng/mL; P = .001) (figure 1). PXR 63396T homozygosity was associated with atazanavir concentrations of <150 ng/mL, with an OR of 18 (95% CI, 2.1–153.9; P = .008) (table 1).
Figure 1.
Atazanavir (ATV) trough plasma concentrations (Ctrough) in a cohort of 47 patients from the Department of Infectious Diseases at the University of Turin (Turin, Italy) that are associated with the PXR 63396 genotype. Median values (horizontal thick line), interquartile ranges (bars), patient values (black dots), highest and lowest values (whiskers), and the minimum effective concentration (150 ng/mL; horizontal thin line) are shown. CC, C allele homozygotes; CT, heterozygotes; TT, T allele homozygotes.
Table 1.
Factors correlated with subtherapeutic concentrations of atazanavir.
| Cohort A |
Cohort B |
|||
|---|---|---|---|---|
| Variable | OR (95% Cl) | P | OR (95% Cl) | P |
| Female sex | 0.5 (0.1–1.6) | .22 | 1.2 (0.4–3.8) | .88 |
| Median age, per year increase | 1.0 (1.0–1.1) | .92 | 1.0 (1.0–1.1) | .67 |
| Median weight, per kg increase | 1.0 (1.0–1.1) | .57 | 1.0 (0.9–1.1) | .31 |
| Administration of tenofovir | 1.5 (0.4–5.2) | .53 | 1.1 (0.4–3.2) | .80 |
| Homozygous for PXR 44477C | 0.6 (0.2–2.1) | .42 | … | … |
| Homozygous for PXR 63396T | 18.0 (2.1–153.9) | .008 | 5.1 (1.3–20.8) | .02 |
| Homozygous for PXR 69789G | 0.7 (0.1–4.5) | .68 | … | … |
A total of 62 patients were included in cohort B. Of these, 47 (76%) were male, and the median age was 40 years (IQR, 33–45 years). The median weight was 72 kg (IQR, 65–84.5 kg). No associations between atazanavir Ctrough and sex (median Ctrough, 151 ng/mL [IQR, 64–224 ng/mL] for male patients vs. 83 ng/mL [IQR, 50–247 ng/mL] for female patients; P = .66), age (ρ, 0.02; P = .86), or weight (ρ, 0.10; P = .45) were observed. Ethnicity data were not available. Atazanavir Ctrough did not differ between patients treated with tenofovir (median Ctrough value, 130 ng/mL [IQR, 61–235 ng/mL]) and patients who were not treated with tenofovir (median Ctrough value, 154 ng/mL [IQR, 53–216 ng/mL]; P = .99).
The allele frequency for 63396T was 44%, and the SNP was in Hardy-Weinburg equilibrium. Homozygosity for the PXR 63396T allele was confirmed to be a predictor of atazanavir concentrations <150 ng/mL, with an OR of 5.1 (95% CI, 1.3–20.8; P = .02) (table 1).
Discussion
Previous studies have shown that up to 40% of subjects who received atazanavir at a dosage of 400 mg once per day had a Ctrough value that was below the minimum effective concentration (i.e., <150 ng/mL), with considerable interindividual variability [2]. The plasma concentrations of atazanavir are likely to be influenced by the action of multiple enzymes and transporter systems that affect their absorption, metabolism, distribution, and elimination. Many of these proteins are themselves regulated by nuclear receptors, such as PXR. Concentration of atazanavir was strongly associated with homozygosity for the PXR 63396T allele, which was a predictor of concentrations <150 ng/mL in both cohorts. Because we have shown elsewhere that PXR is also expressed in PBMCs [18], further studies are now required to assess the impact of this SNP on intracellular accumulation of atazanavir.
TDM cohorts, such as cohort B, have inherent limitations that result from selection bias and a lack of information on factors such as ethnicity. Nevertheless, they represent a potentially valuable resource for pharmacogenetic studies, provided that these limitations are understood. Indeed, a strength of the present study is that the findings in the first cohort were replicated in a second cohort that involved different patients who were observed at a different time and employed a different methodology for drug analysis. These data also replicate the findings of Lamba et al. [11] by confirming a functional role of this polymorphism in vivo. Pharmacogenetic effects exerted by polymorphisms in PXR may be most apparent in patients who receive unboosted atazanavir, because potent inhibition of CYP3A4 and ABCB1 by ritonavir may mask these effects [19, 20]. Atazanavir concentrations were not correlated with any demographic factors, which is in line with the findings of a previous report [2].
In summary, we report a novel association between a SNP in PXR and atazanavir Ctrough, which suggests that PXR is important in the regulation of disposition of this drug. If this finding is confirmed in other cohorts, a prospective clinical study that assesses the usefulness of genetic tests for patients who request unboosted atazanavir is warranted.
Acknowledgments
Financial support. UK Department of Health Biomedical Research Centre for Microbial Diseases.
Footnotes
Potential conflicts of interest. S.K., D.J.B., and A.O. have received research funding from Boehringer Ingelheim, GlaxoSmithKline, Abbott Laboratories, Pfizer, AstraZeneca, Tibotec, Merck, and Roche Pharmaceuticals. G.D.P. has received research funding from Abbott Laboratories, GlaxoSmithKline, Pfizer, Merck, Janssen Cilag, Roche Pharmaceuticals, and Schering Plough. All other authors: no conflicts.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents . Guidelines for the use of antiretroviral agents in HIV-1–infected adults and adolescents. Departmetn of Health and Human Services; [Accessed 19 September 2008]. Jan 29, 2008. Available at: http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. [Google Scholar]
- 2.Colombo S, Buclin T, Cavassini M, et al. Population pharmacokinetics of atazanavir in patients with human immunodeficiency virus infection. Antimicrob Agents Chemother. 2006;50:3801–8. doi: 10.1128/AAC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–94. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez Novoa S, Barreiro P, Rendon A, et al. Plasma levels of atazanavir and the risk of hyperbilirubinemia are predicted by the 3435C→T polymorphism at the multidrug resistance gene 1. Clin Infect Dis. 2006;42:291–5. doi: 10.1086/499056. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Novoa S, Martin-Carbonero L, Barreiro P, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS. 2007;21:41–6. doi: 10.1097/QAD.0b013e328011d7c1. [DOI] [PubMed] [Google Scholar]
- 6.Owen A, Pirmohamed M, Khoo SH, Back DJ. Pharmacogenetics of HIV therapy. Pharmacogenet Genomics. 2006;16:693–703. doi: 10.1097/01.fpc.0000236338.41799.57. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem. 2001;276:14581–7. doi: 10.1074/jbc.M010173200. [DOI] [PubMed] [Google Scholar]
- 9.Martin P, Riley R, Back DJ, Owen A. Comparison of the induction profile for drug disposition proteins by typical nuclear receptor activators in human hepatic and intestinal cells. Brit J Pharmacol. 2008;153:805–19. doi: 10.1038/sj.bjp.0707601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hustert E, Zibat A, Presecan-Siedel E, et al. Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab Dispos. 2001;29:1454–9. [PubMed] [Google Scholar]
- 11.Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab Dispos. 2008;36:169–81. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J, Kuehl P, Green ED, et al. The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics. 2001;11:555–72. doi: 10.1097/00008571-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Owen A. The impact of host pharmacogenetics on antiretroviral drug disposition. Curr Infect Dis Rep. 2006;8:401–8. doi: 10.1007/s11908-006-0052-2. [DOI] [PubMed] [Google Scholar]
- 14.Wyen C, Hendra H, Vogel M, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61:914–8. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford J, Boffito M, Maitland D, et al. Influence of atazanavir 200 mg on the intracellular and plasma pharmacokinetics of saquinavir and ritonavir 1600/100 mg administered once daily in HIV-infected patients. J Antimicrob Chemother. 2006;58:1009–16. doi: 10.1093/jac/dkl379. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez de Requena D, Bonora S, Canta F, et al. Atazanavir trough is associated with efficacy and safety: definition of therapeutic range [abstract 645. Program and abstracts of the 12th Conference on Retroviruses and Opportunistic Infections; Boston, MA. Alexandria, VA: Foundation for Retrovirology and Human Health; 2005. [Google Scholar]
- 17. [Accessed 19 September 2008];Association for Quality Assessment in Therapeutic Drug Monitoring and Clinical Technology Web page. Available at: http://www.kkgt.nl.
- 18.Owen A, Chandler B, Back DJ, Khoo SH. Expression of pregnane-X-receptor transcript in peripheral blood mononuclear cells and correlation with MDR1 mRNA. Antiviral Therapy. 2004;9:819–21. [PubMed] [Google Scholar]
- 19.Eagling VA, Back DJ, Barry MG. Differential inhibition of cytochrome P450 isoforms by the protease inhibitors, ritonavir, saquinavir and indinavir. Br J Clin Pharmacol. 1997;44:190–4. doi: 10.1046/j.1365-2125.1997.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drewe J, Gutmann H, Fricker G, Torok M, Beglinger C, Huwyler J. HIV protease inhibitor ritonavir: a more potent inhibitor of P-glycoprotein than the cyclosporine analog SDZ PSC 833. Biochem Pharmacol. 1999;57:1147–52. doi: 10.1016/s0006-2952(99)00026-x. [DOI] [PubMed] [Google Scholar]