Abstract
Cell-to-cell heterogeneity in ganglioside catabolism was determined by profiling fluorescent tetramethylrhodamine-labeled GM1 (TMR-GM1) breakdown in individual primary neurons and glia from the rat cerebellum. Cells isolated from 5–6 day old rat cerebella were cultured for 7 days, and then incubated for 14 h with TMR-GM1. Intact cells were recovered from cultures by mild proteolysis, paraformaldehyde fixed, and subjected to single cell analysis. Individual cells were captured in a capillary, lysed, and the released single-cell contents subjected to capillary electrophoresis with quantitative laser-induced fluorescent detection of the catabolic products. Non-neuronal cells on average took up much more exogenous TMR-GM1 than neuronal cells, and catabolized it more extensively. After 14 h of incubation, non-neuronal cells retained only 14% of the TMR products as GM1 and GM2, compared to >50% for neurons. On average, non-neuronal cells contained 74% of TMR-labeled product as TMR-ceramide, compared to only 42% for neurons. Non-neuronal cells retained seven times as much TMR-GM3 (7%) compared to neuronal cells (1%). To confirm the observed single cell metabolomics, we lysed and compared TMR-GM1 catabolic profiles from mixed neuron/glial cell cultures and from cultures depleted of non-neuronal cells by treatment with the antimitotic agent cytosine arabinoside. The whole culture catabolic profiles were consistent with the average profiles of single neurons and glia. We conclude that the ultrasensitive analytic methods described accurately reflect single cell ganglioside catabolism in different cell populations from the brain.
Introduction
Gangliosides, glycosphingolipids bearing one or more sialic acid residue(s), are found on all mammalian tissues and cells, and are major cell surface determinants in the brain [1–3]. They vary among tissues and cell types in their glycan structural complexity and expression levels [4]. Gangliosides function in physiological and pathological processes by binding to lectins or toxins, and by associating laterally and modifying the signaling activity of cell surface receptors [5,6]. Their improper catabolism is a hallmark of several lipid storage diseases including Tay-Sachs and Niemann-Pick disease [7,8]. The expression and metabolism of gangliosides in the brain and in isolated neural cell populations have been well documented [9,10]. Here, we extend those analyses to the single cell level, using primary cultured rat cerebellar granule (CG) cells.
CG cultures were chosen for single cell ganglioside metabolomic analysis for several reasons. Isolated from young (5–6 day old) rats, primary CG neurons are relatively homogeneous (typically >90%), plentiful, and characteristically small (<10 µm diameter), which allows them to be distinguished morphologically from glia in primary cultures both before and after collection for single cell analysis [11]. Their ganglioside and lipid compositions have been thoroughly documented as they develop in vitro [10]. They express relatively high levels of ganglioside (~2% of total lipid) including the major mammalian brain gangliosides GM1, GD1a, GD1b, and GT1b.
Technologies designed to measure metabolism in single cells must meet several criteria. The sensitivity must provide for detection of the minute amount of product found in the sub-picoliter volume of a small neuron. The selectivity must allow detection of expected and unexpected components. The dynamic range must be large enough to allow discrimination between the starting material and both highly expressed and low abundance metabolic products. The term chemical cytometry was coined to refer to analytical techniques meeting these criteria, which include electrochemical, mass spectrometric, or fluorescence detection schemes [12].
Here, we report the catabolism of TMR-labeled ganglioside GM1 in single primary cerebellar granule neurons and primary non-neuronal cells (glia) in the same cell cultures using capillary electrophoresis with laser induced fluorescence detection (CE LIF). This technique provides very high sensitivity in the 100 yoctomole (1 ymol = 10−24 mole) range corresponding to ~70 molecules [13–15], highly efficient separations with hundreds of thousands of theoretical plates [16], and an unprecedented dynamic range of nine orders of magnitude [17].
Materials and Methods
Cerebellar cells
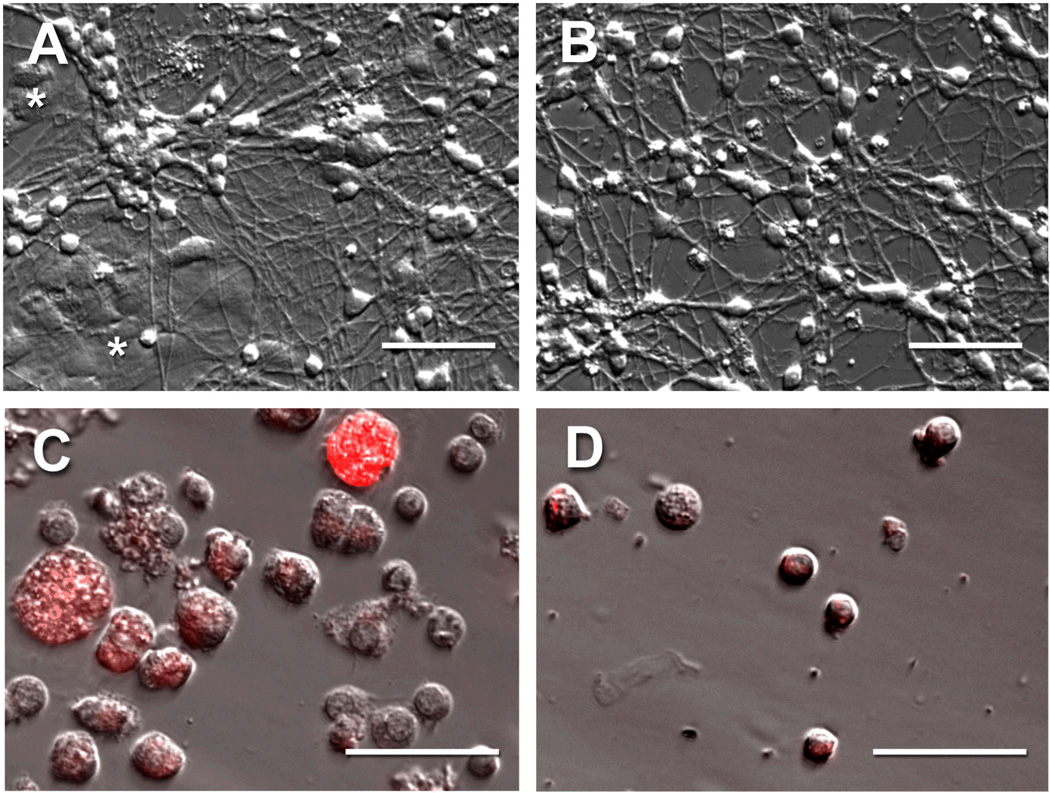
Primary cells from the rat cerebellum were isolated and cultured as described previously [18], except that a supplement (NS21) was used in place of serum [19]. Briefly, cerebella were dissected from 5–6 day old rats, meninges were removed and the tissue was treated with papain. Tissue was dissociated into single cells by triturating, the cells were collected and washed by centrifugation and then re-suspended in growth medium (Neurobasal supplemented to 25 mM KCl, 2 mM glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin). A mixture of neuron-supportive factors (NS21) was added to the growth medium as described previously [19]. Cells were plated on polylysine-coated tissue culture dishes at a density of 2 million cells in 2 ml NS21-containing medium on each 35-mm dish, and then were incubated at 37°C, 5% CO2 in a humidified incubator for 7 d, at which time the neurons had formed a dense meshwork of axons (Fig. 1A,B). As indicated, 10 µM cytosine-β-D-arabinofuranoside (AraC, Sigma) was added to some cultures 18 h after plating and kept in the medium thereafter to deplete dividing (non-neuronal) cells. The medium was changed every 2 d for all plates. Cultures and dissociated cells were imaged on a Nikon T2000 microscope using Hoffman Modulation Contrast and epi-fluorescence optics with images collected using a CCD camera.
Fig. 1.
Neuronal and non-neuronal cells in cultures of dissociated rat cerebellum. Cells dissociated from the cerebella of 5–6 day old rats were cultured for 7 d and imaged by Hoffman Modulation Contrast Microscopy (A). These cultures contained small highly refractile neurons with long axons growing on and around characteristically flattened (*) non-neuronal cells. AraC was added to some cultures 18 h after plating and kept in the medium thereafter to deplete non-neuronal cells leaving primarily neurons and their axons (B). After 7 d in culture, 5 µM TMR-GM1 (complexed with BSA) was added and the cells cultured an additional 14 h prior to collection by mild trypsinization followed by fixation. The resulting single cells (C, D) were characterized by fluorescence and Hoffman Modulation Contrast Microscopy (overlay images shown). Primary cultures lacking AraC were characterized by large heavily fluorescent non-neuronal cells and small lightly fluorescent neurons (C), whereas cells collected from cultures grown in the presence of AraC were predominantly neurons (D). Scale bars, 50 µm.
Single cell metabolomics
Ganglioside GM1 with tetramethylrhodamine in its lipid moiety was synthesized as described [20]. Equimolar amounts of TMR-GM1 and fatty acid free bovine serum albumin (BSA) were pre-complexed at 0.17 mM in an ethanol-aqueous mixture (2:1) and then diluted to 5 µM in NS21-free growth medium. Seven-day cell cultures were washed with growth medium without NS21, and then incubated for 14 h with 5 µM TMR-GM1/BSA complex in the same medium. In some experiments, cultures were then washed once with medium, and further incubated for 45 min with 2.5 µM of Cell Tracker Green (5-chloromethylfluorescein diacetate, CMFDA, Invitrogen). The medium was replaced by fresh medium with neither TMR-GM1 nor CMFDA, followed by 30 min incubation to allow for CMFDA incorporation. Cells were then removed from the culture dish using phosphate buffer containing 2.5 mg/ml trypsin and 0.9 mM EDTA (Invitrogen), followed by 2.5 mg/ml soybean trypsin inhibitor. The released cells were washed with phosphate-buffered saline (PBS) by centrifugation, fixed by suspension in 4% paraformaldehyde in PBS, and then suspended in PBS containing 10 mM glycine until analysis. In some experiments, for comparison with single cell metabolomic studies, after washing with PBS the collected cells were lysed in 0.5 ml of 1% sodium dodecyl sulfate in water without fixation.
Capillary electrophoresis
The instrument used for the current analysis is an improved version from the instrument described in previous reports [21–25]. One of the key components is the use of a sheath-flow cuvette allowing post-column laser-induced fluorescence detection. Typically, a capillary filled with conductive buffer is held between a buffer or sample reservoir which is also the injection end, and the sheath-flow cuvette which is the detection end. The application of a high electric field separates fluorescently tagged analytes into zones across the capillary. As the analytes exit the capillary, the sheath buffer focuses the sample stream, which intersects with a focused laser beam, and fluorescence is gathered at 90° using collection optics. The improvement in the current instrument is in the wide linear dynamic range achieved (9 orders of magnitude) as reported previously [17]. In short, a cascade of four fiber optics beam-splitters (Timbercon, Inc.) is used to divide the collected fluorescence into five channels, each monitored by a single-photon counting avalanche photodiode with a 55 ns dead-time. This cascade of detectors allows monitoring of highly intense metabolite signals without sacrificing detection of trace amounts of other products.
For single cell analyses, individual cells were identified as neuronal or non-neuronal by size ([26], see Fig. 1C,D) and then captured in a capillary [27]. For the larger non-neuronal cells, a 31 µm ID, 150 µm OD, and 39 cm long bare fused silica capillary (Polymicro, Phoenix, AZ) was used, whereas for smaller neurons a 20 µm ID, 150 µm OD and 40 cm long capillary was used. Single cell capture was performed by applying a 1 s negative pressure on the sheath flow at the terminus of the capillary, causing siphoning and thus injection of a single cell into the capillary. Each cell was sandwiched between 2 plugs of a 0.4% solution of Triton X-100 detergent to ensure lysis. The electrophoresis buffer consisted of 10 mM sodium tetraborate, 35 mM sodium deoxycholate, and 5 mM methyl-β-cyclodextrin, pH 9.2. A 10 mW diode-pumped solid-state laser provided excitation of the sample at 532 nm, and fluorescence was collected by a 0.7 NA microscope objective, passed through a 580 DF30 bandpass filter, and imaged onto a GRIN lens coupled fiber optic. This fiber optic was connected to the first beam-splitter in the above-mentioned fiber optic cascade.
The data were acquired by a custom Labview program, using a National Instruments card (NI, TX), at a rate of 50 Hz. The signal was recorded in a PC, corrected for dead-time using a second order correction factor [28], and passed through a five point median filter to remove spikes from the signal. The data were then convoluted with a Gaussian function with a five-point standard deviation before plotting. Peak area was estimated by using a nonlinear least squares regression analysis to fit a Gaussian function to the data in the vicinity of each peak.
Results
Single cell TMR-GM1 catabolism
Cultured cells from the rat cerebellum were chosen for single cell metabolomic studies because they provide an abundant and relatively uniform population of primary nerve cells [11,26]. After 7 d, cultures were dominated by uniform small and refractile nerve cell soma on a dense mat of axons (Fig. 1A,B). Non-neuronal cells (glia) were also found in these cultures as larger round or flattened cells that were depleted when cultures were grown in the presence of the antimitotic agent AraC. This provided an opportunity to compare single cell GM1 metabolomics in different cell types isolated from the brain.
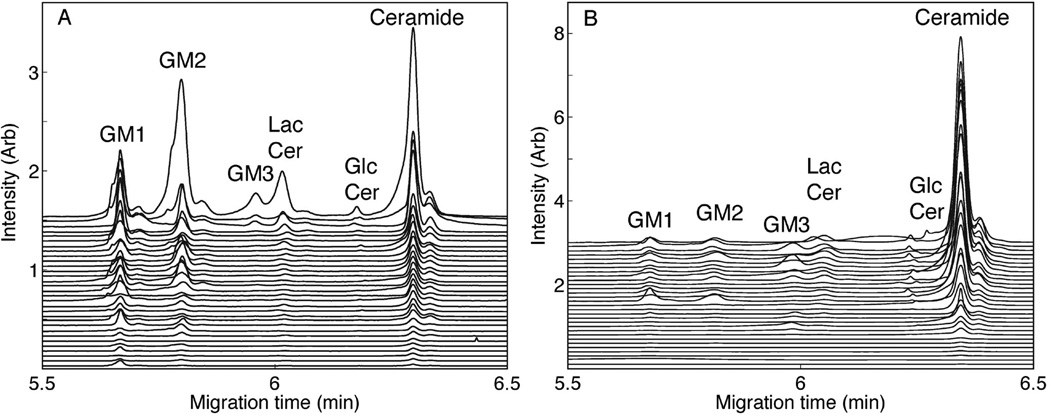
Cells were dissociated from cultures by mild proteolysis, resulting in small (<11 µm) nerve cells and larger (>15 µm) non-neuronal cells (Fig. 1C,D). The vital dye Cell Tracker Green revealed that the great majority of cells were viable upon collection and retained the intracellular vital dye at the time of analysis (data not shown). Neurons and non-neuronal cells were distinguished by size microscopically and individual cells were captured in capillaries then lysed with detergent. Single cell contents were then resolved by capillary electrophoresis into quantifiable metabolic products. TMR GM1 entered the catabolic pathway in individual primary neurons and non-neuronal cells, generating the sequential breakdown products: GM1 → GM2 → GM3 → lactosylceramide (LacCer) → glucosylceramide (GlcCer) → ceramide (Fig. 2). Each TMR-labeled catabolite was identified by its co-migration with synthetic TMR standards [20]. Labeled metabolic products more complex than GM1 were not detected (see Discussion).
Fig. 2.
Capillary electrophoresis of single cells. Cells from 7-day cerebellar cultures were labeled with TMR-GM1, incubated for 14 h, then collected and subjected to single cell analyses as described in the text. Fluorescence intensities as a function of CE migration time of TMR-labeled GM1 catabolites of individual cells are plotted in descending amplitude of the ceramide peak. The migration positions of standard TMR-labeled catabolites [20] are indicated. (A) 31 individual neurons were analyzed by CE-LIF. (B) 30 individual non-neuronal cells were analyzed by CE-LIF.
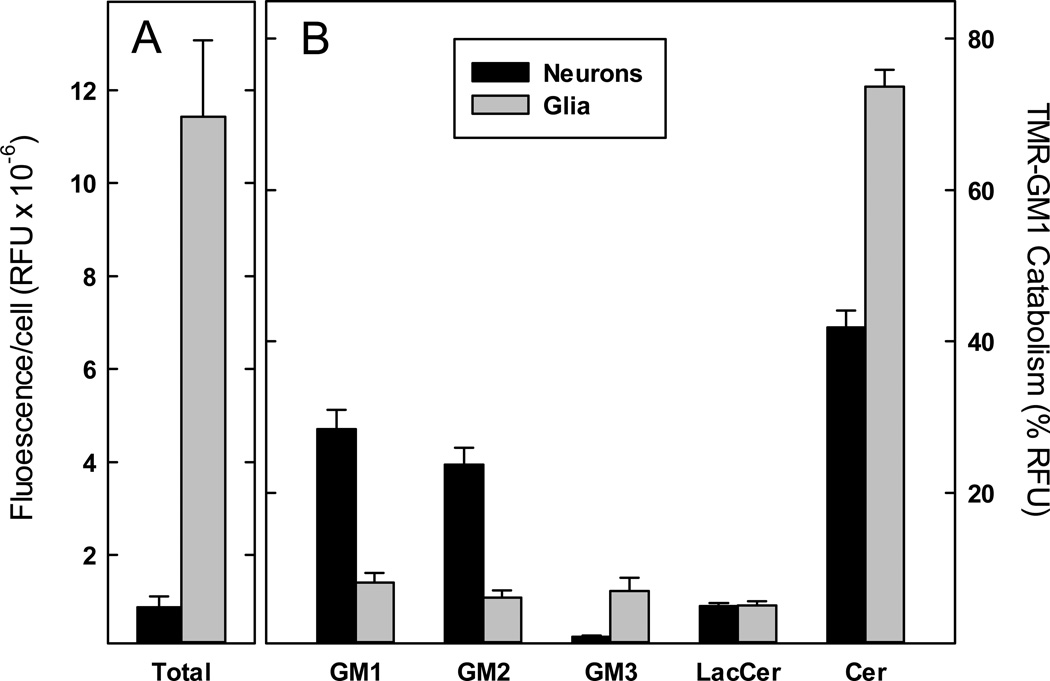
After 14 h of incubation with TMR-GM1, non-neuronal cells catabolized the precursor primarily (74%) to TMR-ceramide with the balance distributed approximately equally among TMR GM1 and three catabolic intermediates (each ≤8%, Fig. 3). In contrast, neurons retained >50% of the label as equivalent amounts of TMR-GM1 and TMR-GM2, with only 42% catabolized to ceramide. Neurons, on average, retained little of the label as TMR-GM3 (1%) compared to a significantly higher amount of TMR-GM3 in non-neuronal cells (7%). On average, non-neuronal cells accumulated more TMR label than neurons (Fig. 1C & Fig. 3). Both cell populations were heterogeneous in total TMR-lipid uptake and metabolism. Nevertheless, the broad dynamic range of our instrument allowed accurate quantification of the metabolic products in all cells. These data demonstrate the ability to segregate and analyze ganglioside metabolism in individual brain cells, and to associate differential ganglioside catabolism with different brain cell populations.
Fig. 3.
Aggregated single cell analyses of GM1 catabolism. Cells from 7-day cerebellar cultures were labeled with TMR-GM1, incubated for 14 h, then collected and subjected to single cell analysis as described in Fig. 2. Data were aggregated from 31 individual neurons and 30 individual non-neuronal cells, identified by size. (A) The fluorescence intensity of TMR-GM1 and all of its catabolites (areas under the curve in relative fluorescence units (RFU)) was summed for each neuron and non-neuronal cell analyzed in Fig. 2. Average fluorescence per cell for each class of cells is presented as mean ± SEM. (B) The average fluorescence intensity of TMR-GM1 and each of its catabolites, expressed as a percent of total fluorescence, is presented for neurons and non-neuronal cells. Despite cell-to-cell variation in cell fluorescence (panel A), the large dynamic range of our analytical system allows reproducible quantification of TMR-GM1 and its catabolites (mean ± SEM) for each cell class.
Comparison of TMR-GM1 catabolism in single cells to that in cell lysates
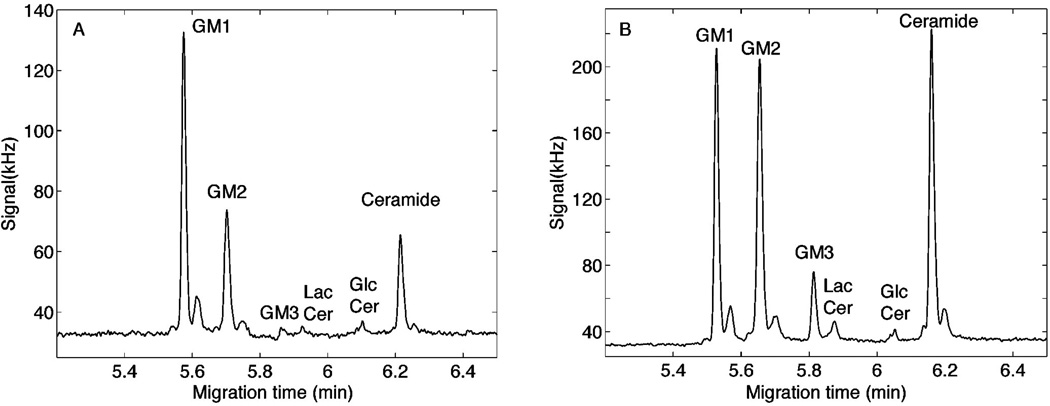
We observed preferential uptake and metabolism of TMR-GM1 by non-neuronal cells in mixed neuron/glia cultures from the rat cerebellum (Figs. 1–3). To compare population metabolism to single cell metabolism, the TMR-GM1 labeled cells collected from entire culture dishes (~500,000 cells), lysed in detergent and the metabolic products of the culture population determined on a nanoliter aliquot using the same capillary electrophoresis methods as used for single cells. Some cultures were grown in the presence of the antimitotic agent AraC to deplete the cultures of non-neuronal (dividing) cells (Fig. 1B,D). The results (Fig. 4) are in agreement with the single cell data (Fig. 3). TMR ceramide was higher in the control cultures compared to the AraC-treated culture, whereas TMR-GM1 dominated the AraC-treated cultures. Also similar to the single cell data, TMR GM3 was expressed at much higher concentration in the control cultures than in the AraC-treated cultures where glia were depleted.
Fig. 4.
Whole cerebellar cell culture analyses. Cells from 7-day cerebellar cultures were labeled with TMR-GM1, incubated for 14 h, collected by centrifugation and lysed in detergent for whole culture catabolite analysis. Fluorescence intensities of whole culture aliquots are shown, with the migration positions of standards [20] indicated. (A) Cerebellar cultures grown in the presence of AraC to deplete non-neuronal cells. (B) Control cerebellar cultures grown in the absence of AraC, which include both neurons and non-neuronal cells.
Discussion
Gangliosides are major cell surface determinants in the outer leaflet of the plasma membranes of nerve cells, and are well positioned to act as recognition and regulation molecules in physiological and pathological processes in the brain and elsewhere [1–3]. Knowledge about the functions of gangliosides in cell-cell recognition, in the regulation of cell surface receptor function, and as pathogen targets continues to emerge from biochemical, genetic and clinical studies [5,6]. Basic knowledge about ganglioside expression and metabolism is the basis for understanding their function. Although the study of ganglioside metabolism in tissues and cell populations is enlightening [9,10], the ability to track ganglioside metabolism in single neurons may provide additional insights.
The present study reports differences in the metabolic fate of exogenous TMR GM1 taken up by neurons and non-neuronal (glial) cells (based on size [26]) in the same culture. We were successful in investigating cellular subsets – one cell at a time – within a primary cell population from the brain. Non-neuronal cells metabolized TMR-GM1 more efficiently to the end product of this derivative, TMR-ceramide, than did neurons. Catabolic intermediates between GM1 and ceramide were present at significant but much lower abundance in glia, but notably included much more GM3 than in neurons. These results are consistent with glial vs. neuronal ganglioside metabolism, since proliferating glia are more catabolically active than post-mitotic resting neurons and GM3 is the major ganglioside in (for example) rodent brain astrocytes [29]. In contrast, in cultured cerebellar neurons metabolically labeled with sphingosine, GM3 was not readily detected [30]. Analyses of TMR-GM1 catabolic products in cultures grown with and without the mitotic inhibitor AraC support the single cell analyses. Cultures depleted of proliferating cells convert less TMR-GM1 to TMR-ceramide and express less GM3, consistent with the neuronal metabolism of this derivative.
A goal of single cell metabolomics is to survey both anabolic and catabolic pathways. In the current studies, biosynthetic conversion of TMR-GM1 to higher-order gangliosides, such as the major cerebellar neuronal gangliosides GD1a and GT1b, was not observed. Based on the cellular distribution of TMR-GM1, we suspect that it is selectively sampling catabolic pathways, and that its sub-cellular distribution in catabolic compartments is in part due to the TMR moiety. This would be consistent with prior studies showing differential uptake of ganglioside GM1 with different lipid moiety structures [31]. Additional studies with different fluorescent moieties and with molecules at different steps in the metabolic pathways will be required to test this hypothesis and expand single cell ganglioside metabolism to anabolic as well as catabolic pathways. Encouragement for these studies comes from the observation that [3H]GM1 labeled in the sphingosine 3-hydroxyl is converted to higher-order as well as catabolic products [32]. Improvements in resolution of higher-order gangliosides by CE may also enhance detection of anabolic products. Together, exploration of the power and limitations of exogenously added fluorescent gangliosides and their analyses may lead to enhanced single cell ganglioside metabolomics in various brain cell populations. The current findings comprise a platform for these enhancements, demonstrating the ability to analyze ganglioside metabolism cell-by-cell in different brain cell populations.
Acknowledgments
We gratefully acknowledge funding from the National Institutes of Health (R01NS061767).
Contributor Information
David C. Essaka, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN. 46556 USA and Department of Chemistry, University of Washington, Box 351700, Seattle, WA. 98195 USA
Jillian Prendergast, Departments of Pharmacology and Neuroscience, Johns Hopkins University School of Medicine, 725 N. Wolfe Street, Baltimore, MD 21205 USA.
Richard B. Keithley, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN. 46556 USA and Department of Chemistry, University of Washington, Box 351700, Seattle, WA. 98195 USA
Ole Hindsgaul, Carlsberg Laboratory, Gamle Carlsberg Vej 10, DK-2500, Valby, Copenhagen, Denmark.
Monica M. Palcic, Carlsberg Laboratory, Gamle Carlsberg Vej 10, DK-2500, Valby, Copenhagen, Denmark
Ronald L. Schnaar, Email: schnaar@jhu.edu, Departments of Pharmacology and Neuroscience, Johns Hopkins University School of Medicine, 725 N. Wolfe Street, Baltimore, MD 21205 USA.
Norman J. Dovichi, Email: Norman.Dovichi.1@nd.edu, Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN. 46556 USA.
References
- 1.Yu RK, Tsai YT, Ariga T, et al. Structures, Biosynthesis, and Functions of Gangliosides-an Overview. J Oleo Sci. 2011;60:537–544. doi: 10.5650/jos.60.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonnino S, Prinetti A. Gangliosides as regulators of cell membrane organization and functions. Adv Exp Med Biol. 2010;688:165–184. doi: 10.1007/978-1-4419-6741-1_12. [DOI] [PubMed] [Google Scholar]
- 3.Schnaar RL. Brain gangliosides in axon-myelin stability and axon regeneration. FEBS Lett. 2010;584:1741–1747. doi: 10.1016/j.febslet.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledeen RW, Yu RK. Gangliosides: structure, isolation, and analysis. Methods Enzymol. 1982;83:139–191. doi: 10.1016/0076-6879(82)83012-7. [DOI] [PubMed] [Google Scholar]
- 5.Todeschini AR, Hakomori SI. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim Biophys Acta. 2008;1780:421–433. doi: 10.1016/j.bbagen.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez PH, Schnaar RL. Gangliosides in cell recognition and membrane protein regulation. Curr Opin Struct Biol. 2009;19:549–557. doi: 10.1016/j.sbi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolter T, Sandhoff K. Glycosphingolipid degradation and animal models of GM2-gangliosidoses. J Inherit Metab Dis. 1998;21:548–563. doi: 10.1023/a:1005419122018. [DOI] [PubMed] [Google Scholar]
- 8.Vitner EB, Platt FM, Futerman AH. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Echten G, Sandhoff K. Ganglioside metabolism. Enzymology, topology, and regulation. J Biol Chem. 1993;268:5341–5344. [PubMed] [Google Scholar]
- 10.Prinetti A, Chigorno V, Prioni S, et al. Changes in the lipid turnover, composition, and organization, as sphingolipid-enriched membrane domains, in rat cerebellar granule cells developing in vitro. J Biol Chem. 2001;276:21136–21145. doi: 10.1074/jbc.M010666200. [DOI] [PubMed] [Google Scholar]
- 11.Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dovichi NJ, Hu S. Chemical cytometry. Curr Opin Chem Biol. 2003;7:603–608. doi: 10.1016/j.cbpa.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Whitmore CD, Olsson U, Larsson EA, et al. Yoctomole analysis of ganglioside metabolism in PC12 cellular homogenates. Electrophoresis. 2007;28:3100–3104. doi: 10.1002/elps.200700202. [DOI] [PubMed] [Google Scholar]
- 14.Turner EH, Lauterbach K, Pugsley HR, et al. Detection of green fluorescent protein in a single bacterium by capillary electrophoresis with laser-induced fluorescence. Anal Chem. 2007;79:778–781. doi: 10.1021/ac061778r. [DOI] [PubMed] [Google Scholar]
- 15.Chen DY, Dovichi NJ. Yoctomole detection limit by laser-induced fluorescence in capillary electrophoresis. J Chromatogr B Biomed Appl. 1994;657:265–269. doi: 10.1016/0378-4347(94)00014-x. [DOI] [PubMed] [Google Scholar]
- 16.Essaka DC, White J, Rathod P, et al. Monitoring the uptake of glycosphingolipids in Plasmodium falciparum-infected erythrocytes using both fluorescence microscopy and capillary electrophoresis with laser-induced fluorescence detection. Anal Chem. 2010;82:9955–9958. doi: 10.1021/ac1021776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dada OO, Essaka DC, Hindsgaul O, et al. Nine orders of magnitude dynamic range: picomolar to millimolar concentration measurement in capillary electrophoresis with laser induced fluorescence detection employing cascaded avalanche photodiode photon counters. Anal Chem. 2011;83:2748–2753. doi: 10.1021/ac103374x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehta NR, Lopez PH, Vyas AA, et al. Gangliosides and Nogo receptors independently mediate myelin-associated glycoprotein inhibition of neurite outgrowth in different nerve cells. J Biol Chem. 2007;282:27875–27886. doi: 10.1074/jbc.M704055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Stevens B, Chang J, et al. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson EA, Olsson U, Whitmore CD, et al. Synthesis of reference standards to enable single cell metabolomic studies of tetramethylrhodamine-labeled ganglioside GM1. Carbohydr Res. 2007;342:482–489. doi: 10.1016/j.carres.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsay LM, Dickerson JA, Dovichi NJ. Attomole protein analysis by CIEF with LIF detection. Electrophoresis. 2009;30:297–302. doi: 10.1002/elps.200800498. [DOI] [PubMed] [Google Scholar]
- 22.Sobhani K, Michels DA, Dovichi NJ. Sheath-flow cuvette for high-sensitivity laser-induced fluorescence detection in capillary electrophoresis. Appl Spectrosc. 2007;61:777–779. doi: 10.1366/000370207781393208. [DOI] [PubMed] [Google Scholar]
- 23.Zhao JY, Diedrich P, Zhang Y, et al. Separation of aminated monosaccharides by capillary zone electrophoresis with laser-induced fluorescence detection. J Chromatogr B Biomed Appl. 1994;657:307–313. doi: 10.1016/0378-4347(94)00015-8. [DOI] [PubMed] [Google Scholar]
- 24.Cheng YF, Dovichi NJ. Subattomole amino acid analysis by capillary zone electrophoresis and laser-induced fluorescence. Science. 1988;242:562–564. doi: 10.1126/science.3140381. [DOI] [PubMed] [Google Scholar]
- 25.Whitmore CD, Essaka D, Dovichi NJ. Six orders of magnitude dynamic range in capillary electrophoresis with ultrasensitive laser-induced fluorescence detection. Talanta. 2009;80:744–748. doi: 10.1016/j.talanta.2009.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce IA, Cambray-Deakin MA, Burgoyne RD. Glutamate acting on NMDA receptors stimulates neurite outgrowth from cerebellar granule cells. FEBS Lett. 1987;223:143–147. doi: 10.1016/0014-5793(87)80525-2. [DOI] [PubMed] [Google Scholar]
- 27.Krylov SN, Starke DA, Arriaga EA, et al. Instrumentation for chemical cytometry. Anal Chem. 2000;72:872–877. doi: 10.1021/ac991096m. [DOI] [PubMed] [Google Scholar]
- 28.Madsen MT, Nickles RJ. A precise method for correcting count-rate losses in scintillation cameras. Med Phys. 1986;13:344–349. doi: 10.1118/1.595874. [DOI] [PubMed] [Google Scholar]
- 29.Akasako Y, Nara K, Nagai Y, et al. Inhibition of ganglioside synthesis reduces the neuronal survival activity of astrocytes. Neurosci Lett. 2011;488:199–203. doi: 10.1016/j.neulet.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 30.Prinetti A, Chigorno V, Tettamanti G, et al. Sphingolipid-enriched membrane domains from rat cerebellar granule cells differentiated in culture. A compositional study. J Biol Chem. 2000;275:11658–11665. doi: 10.1074/jbc.275.16.11658. [DOI] [PubMed] [Google Scholar]
- 31.Bassi R, Sonnino S. The role of the ganglioside lipid moiety in the process of ganglioside-cell interactions. Chem Phys Lipids. 1992;62:1–9. doi: 10.1016/0009-3084(92)90048-t. [DOI] [PubMed] [Google Scholar]
- 32.Chigorno V, Valsecchi M, Sonnino S, et al. Formation of tritium-labeled polysialylated gangliosides in the cytosol of rat cerebellar granule cells in culture following administration of [3H]GM1 ganglioside. FEBS Lett. 1990;277:164–166. doi: 10.1016/0014-5793(90)80834-6. [DOI] [PubMed] [Google Scholar]