Abstract
Prostate cancer is the second most common cancer in men worldwide and causes over 250,000 deaths each year1. Overtreatment of indolent disease also results in significant morbidity2. Common genetic alterations in prostate cancer include losses of NKX3.1 (8p21)3,4 and PTEN (10q23)5,6, gains of the androgen receptor gene (AR)7,8 and fusion of ETS-family transcription factor genes with androgen-responsive promoters9–11. Recurrent somatic base-pair substitutions are believed to be less contributory in prostate tumorigenesis12,13 but have not been systematically analyzed in large cohorts. Here we sequenced the exomes of 112 prostate tumor/normal pairs. Novel recurrent mutations were identified in multiple genes, including MED12 and FOXA1. SPOP was the most frequently mutated gene, with mutations involving the SPOP substrate binding cleft in 6–15% of tumors across multiple independent cohorts. SPOP-mutant prostate cancers lacked ETS rearrangements and exhibited a distinct pattern of genomic alterations. Thus, SPOP mutations may define a new molecular subtype of prostate cancer.
We performed exome capture followed by paired-end, massively parallel sequencing on genomic DNA from 112 prostate adenocarcinomas and matched normal samples. We focused on treatment-naïve radical prostatectomy specimens from American and Australian patients that spanned a range of grades, stages, and risk of recurrence (Supplementary Table 1) (details in Materials and Methods). The exon capture baits targeted 98.2% of genes in the Consensus CDS database (http://www.ncbi.nlm.nih.gov/CCDS). A mean coverage depth of 118× per sample was achieved, with 89.2% of targets covered at ≥ 20× depth (Supplementary Fig. 1 and Supplementary Table 2). Tumor and normal DNA were also analyzed by Affymetrix SNP 6.0 arrays to detect somatic copy number alterations. In addition, transcriptome sequencing (“RNA-seq”) was performed on 22 exome-sequenced tumors and 41 independent samples (Supplementary Fig. 2).
We identified 5,764 somatic mutations that were present in tumor DNA but absent in peripheral blood or non-cancerous prostate (Supplementary Table 3). Of these, 997 variants occurred in a single tumor that harbored a frame-shift mutation of the mismatch-repair gene MSH6 (Supplementary Fig. 3). After excluding this highly-mutated sample, the remaining tumors harbored a median of 10 silent and 30 non-silent mutations (range 10 to 105 total mutations) or ~1.4 per Mb covered (Supplementary Fig. 3). Analysis of 229 non-silent mutations by mass-spectrometric genotyping validated 95.6% of variants with allelic fraction ≥ 0.2 (C.I. 92–98%) (Supplementary Table 3). The mutation rate of this cohort exceeded that of seven published prostate tumor genomes (0.9 mutations per Mb)14, perhaps because the increased exome sequence coverage improved detection of variants present at lower allelic fractions.
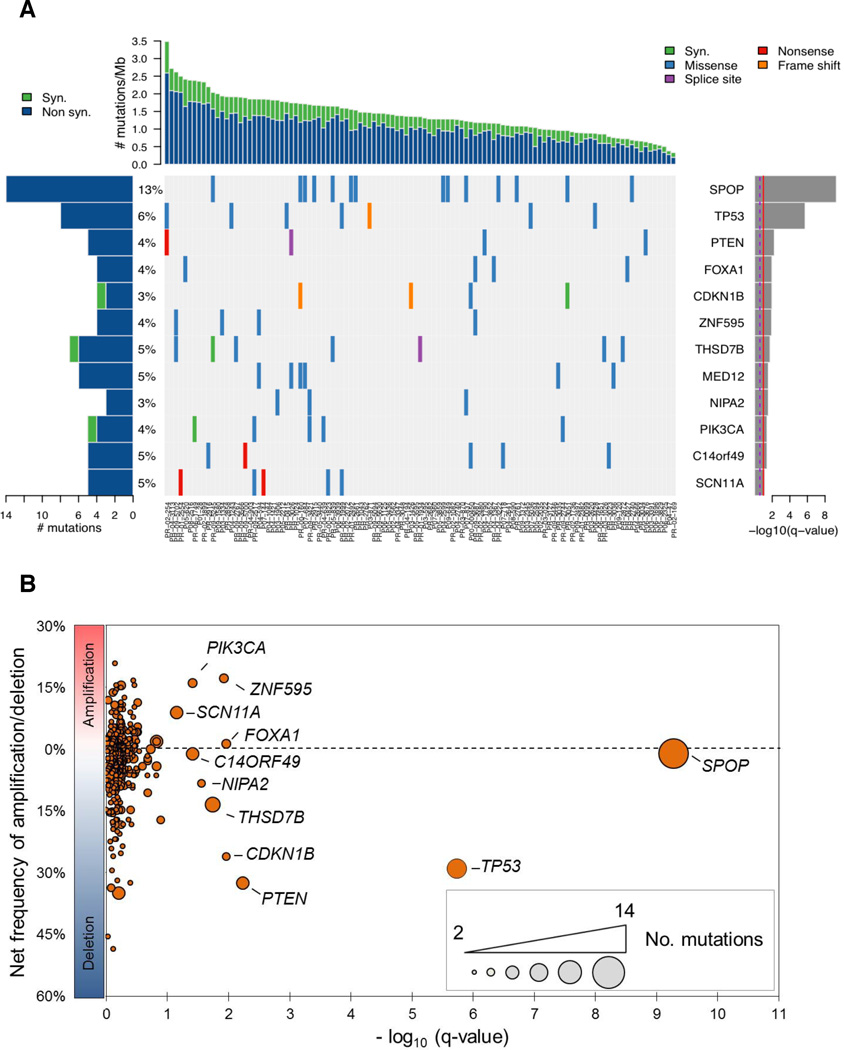
We searched for genes that harbored more non-synonymous mutations than expected by chance given gene size, sequence context and the frequency of mutations for each tumor (Fig. 1A and Supplementary Table 4). Twelve genes were enriched for mutations at q-value < 0.1, the majority of which are highly expressed at the transcript level in prostate tumors (Supplementary Fig. 4). The identification of PIK3CA, TP53 and PTEN confirmed that our approach detected alterations known to promote tumorigenesis in prostate cancer and other malignancies. We also found evidence of enrichment for mutations in the PTEN pathway, cell cycle regulatory machinery, and other gene sets (Supplementary Table 5)12.
Fig 1. Significantly mutated genes in aggressive primary prostate cancer.
(A) (Top) A cohort of 111 primary prostate tumors is ordered by number of mutations per Mb sequenced. (Center) Mutations in significantly mutated genes, colored by the coding consequence of the mutation. Each column represents a tumor and each row a gene. (Left) Number and percentage of tumors with mutations in a given gene. (Right) The negative log of the q-values for the significance level of mutated genes is shown (for all genes with q < 0.1). (B) Net frequency of gene deletion/amplification across 169 copy number-profiled tumors. Significantly mutated genes are indicated. Only autosomal genes with two or more mutations are shown.
The most frequently mutated gene was SPOP (13% of cases; Fig. 1), which encodes the substrate-binding subunit of a Cullin-based E3 ubiquitin ligase15,16. Although isolated SPOP mutations have been reported in prostate cancer14,17, this gene has not previously been found significantly mutated in any malignancy. Several novel genes not previously known to undergo somatic alteration in prostate cancer were enriched for mutations, including FOXA1, MED12, THSD7B, SCN11A and ZNF595. The p27Kip1 gene CDKN1B was somatically mutated in three samples and deleted in sixteen others (Fig. 1B). p27Kip1 constrains prostate tumor growth in mice18 and harbors a germline prostate cancer risk allele19, but somatic substitutions have not previously been observed in this cell cycle regulatory protein. Infrequent mutations were also detected in multiple proto-oncogenes, tumor suppressors, and chromatin-modifying enzymes (Supplementary Results and Discussion, Supplementary Table 4).
The Forkhead transcription factor gene FOXA1 harbored nonsilent mutations in 4 of 111 exomes and 4 of 41 independent RNA-seq samples. FOXA1 is required for epithelial cell differentiation in the murine prostate20 and promotes cell cycle progression in castration-resistant prostate cancer21. Notably, FOXA1 modulates AR-driven transcription22 and activates expression of CDKN1B23. Mutations strictly affected residues in the Forkhead domain (Supplementary Table 4) that reside near the DNA binding surface (Fig. 2A)24. The clustered nature of these mutations suggests that they may disrupt binding of FOXA1 DNA targets.
Fig 2. Recurrent somatic mutations in FOXA1 and MED12.
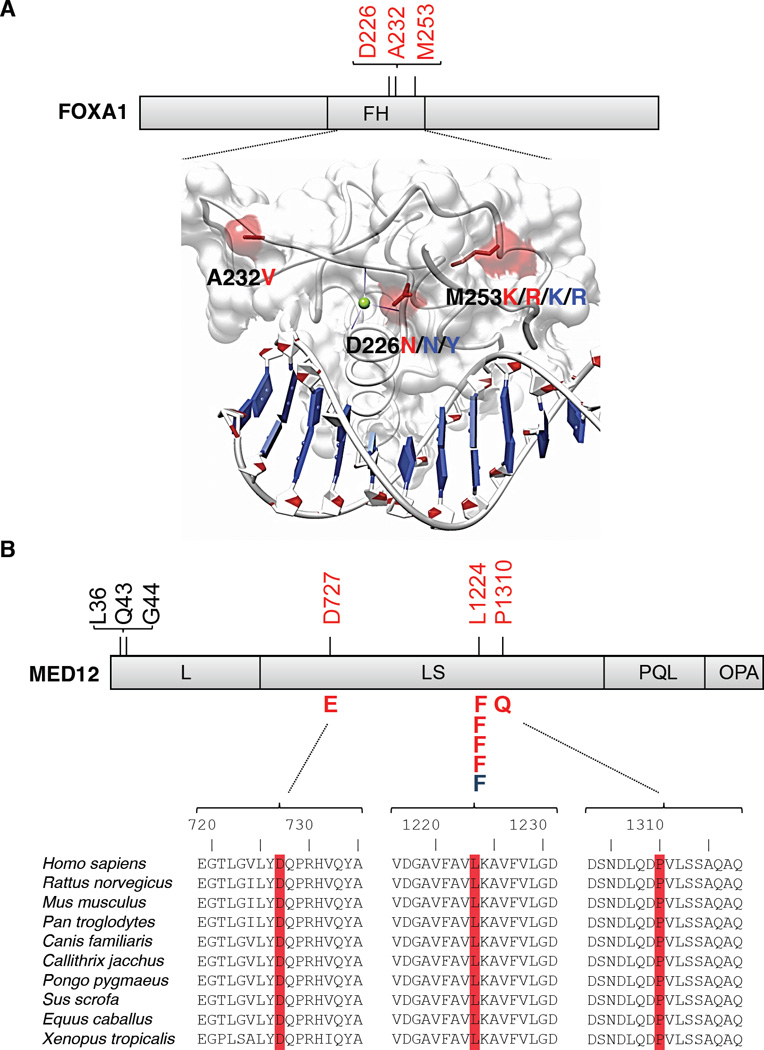
Mutations detected by exome sequencing are depicted (red), as are variants from non-overlapping transcriptome sequencing data (blue). (A) Structural analysis of mutations in FOXA1. Mutated residues are mapped to the structure of the HNF3γ fork-head domain from coordinate file 1VTN.pdb (www.pdb.org)24 and highlighted in red. FH, Fork-head domain. (B) Recurrent MED12 mutations in prostate cancer (red, blue) are distinct from those reported in uterine leiomyeoma (shown in black)28. Domains of MED12 are denoted as in Zhou et al.25. Multispecies conservation of the mutated sites is shown below the mutation.
Mutations affecting MED12 were observed in 6 out of 111 exomes, with a recurrent F1224L mutation in five samples (Fig. 2B and Supplementary Table 4). MED12 encodes a subunit of the mediator complex and the Cyclin-dependent kinase 8 (CDK8) sub-complex that regulates basal and stimulus-specific transcriptional programs25–27. Recently, MED12 mutations were reported in 70% of uterine leiomyomas28, benign stromal tumors of the smooth muscle lineage. Mutations in prostate cancer affected distinct codons from those in leiomyoma and occurred in epithelial cells rather than stroma as determined by laser-capture microdissection (LCM) (Supplementary Fig. 5). Conceivably, MED12 mutations may perturb CDK8-dependent modulation of transcriptional programs linked to p53 and androgen signaling26,27.
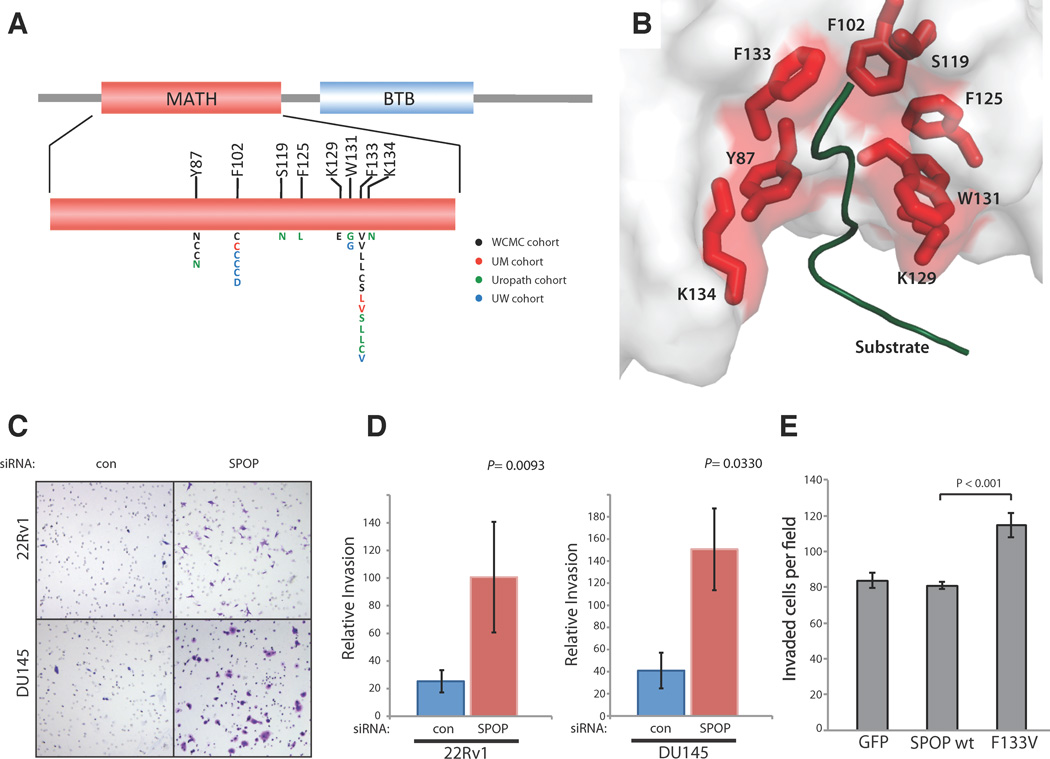
Although SPOP mutations were originally reported in genomic studies of prostate cancer (Supplementary Table 6)14,17, their prevalence and functional relevance remained unknown. We therefore sequenced this gene in multiple additional cohorts comprising over 300 primary tumors and metastases from the US and Europe. Using RNA-seq and Sanger sequencing of tumor and matched germline DNA, recurrent heterozygous SPOP substitutions were identified in 6–13% of primary prostate adenocarcinomas (Figs. 3A and Supplementary Fig. 6, Supplementary Table 7). No mutations were identified in 36 benign prostate tissue samples, prostate stroma, or 6 common prostate cell lines. SPOP mutations were also found in 6 of 41 patients with metastatic disease (14.5%) (Fig. 3A, Supplementary Table 7). Thus, SPOP mutations occur at a 6 to 15% frequency across localized and advanced prostate tumors.
Fig 3. Structural and functional studies of recurrent SPOP mutations in prostate cancer.
(A) Positional distribution of somatic mutations in SPOP across the Weill Cornell Medical College (WCMC), University of Michigan (UM), Uropath, and University of Washington (UW) prostate tumor cohorts. (B) Mutated residues in the crystal structure of the SPOP MATH domain bound to substrate (PDB 3IVV). (C) Representative images of invasive 22Rv1 and DU145 cells transfected with control and SPOP siRNA in Matrigel invasion assays. (D) Quantitation of invaded cells transfected with SPOP siRNA. (E) Quantitation of invaded DU145 cells transfected with GFP, SPOP wt, and SPOP F133V. Error bars depict standard deviation.
All SPOP mutations affected conserved residues in the structurally-defined substrate binding cleft (Fig. 3B, Supplementary Fig. 7)16. Several recurrently mutated residues exert key substrate-interacting roles; moreover, mutation of Y87, W131, and F133 disrupts substrate binding in vitro16. These results strongly suggest that prostate cancer SPOP mutations are biologically significant. To test this hypothesis, we examined the consequences of mutant SPOP protein expression or SPOP knockdown on tumorigenic phenotypes in vitro. Prostate cancer cells transfected with the most common SPOP mutant (F133V) or SPOP siRNA showed increased invasion compared to controls (Fig. 3C–E, Supplementary Fig. 8), but cell growth and viability were largely unaffected (Supplementary Fig. 9). The SPOP-CUL3 complex affects a variety of substrates that impact multiple pathways, including hedgehog, JNK, and steroid receptor signaling cascades29–31. SPOP undergoes amplification in other malignancies and may be overexpressed in renal cell carcinoma29; however multiple prostate cancer cohorts showed no evidence of SPOP amplification or upregulation (Fig. 1B, Supplementary Figs. 10, 11). Conceivably, prostate cancer-associated SPOP mutations exert de novo gain of function alterations (e.g., a distinct substrate profile), dominant negative effects, or more subtle alterations in substrate specificity. Further studies are necessary to determine the specific ubiquitin ligase functions and cellular pathways deregulated by SPOP mutation in prostate cancer.
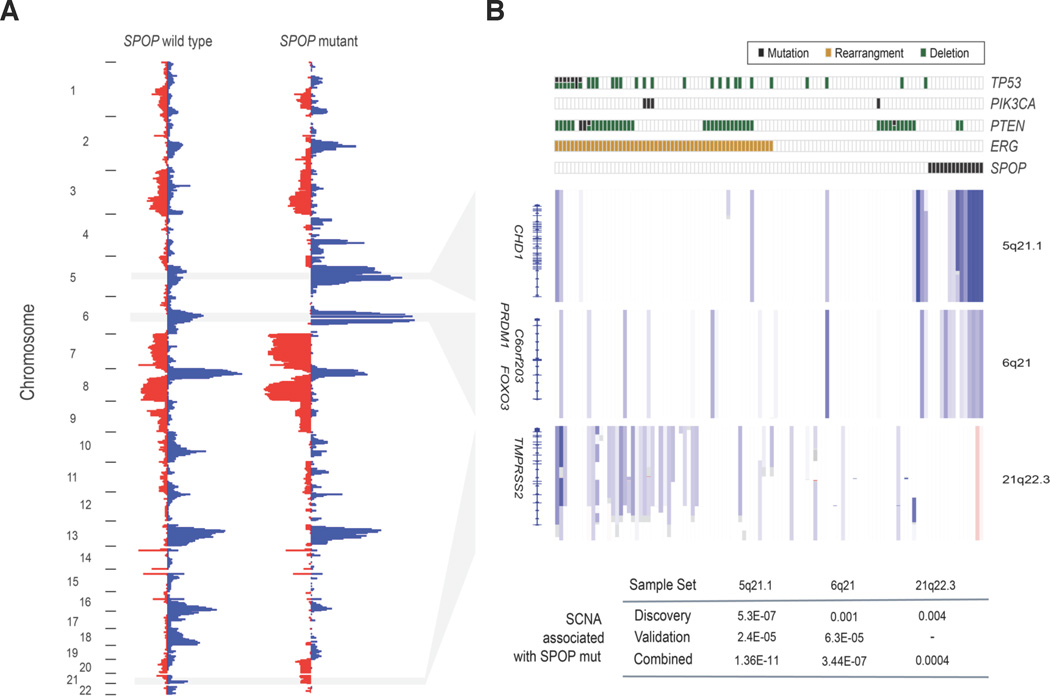
Strikingly, all exomes with SPOP mutations lacked the TMPRSS2-ERG fusion or other ETS rearrangements (Fig. 4, Supplementary Fig. 12), present in up to 50% of prostate cancers11,32. This mutually exclusive relationship between SPOP mutation and ERG rearrangement (P < 0.001, Fisher’s exact test) was confirmed in evaluable samples across all five cohorts tested (Supplementary Fig. 12), even within an individual prostate tumor (Supplementary Fig. 13). Thus, SPOP mutation and ETS fusions may represent early and divergent driver events in prostate carcinogenesis. SPOP mutations were identified in LCM-analyzed high-grade intraepithelial neoplasia (HG-PIN) adjacent to invasive adenocarcinoma, further strengthening the premise that SPOP mutation comprises an early event in prostate tumorigenesis (Supplementary Fig. 14).
Fig 4. SPOP mutation defines a distinct genetic subclass of prostate cancer.
(Left) Frequency of genomic copy number alterations in SPOP-mutant and SPOP-wildtype tumors. Length of bars reflects the frequency of copy number loss (blue) or gain (red). (Right) Heatmap showing selected recurrent somatic copy number aberrations (SCNA). Each row represents a single prostate cancer sample. Samples are annotated for mutations in SPOP, PTEN, PIK3CA, and TP53, deletions of PTEN, and ERG rearrangements. Deletions positively correlated (5q21, 6q21) or inversely correlated (21q22.3) with SPOP mutation are shown. P-values of peak association with SPOP mutation in both discovery and validation cohorts are displayed at bottom (Fisher’s exact test). Regions are not to scale; full coordinates available in Supplementary Table 8.
In light of prior studies suggesting that prostate cancer may be classified by co-occurring genomic alterations12,33,34 we investigated whether SPOP-mutant tumors were enriched for other genomic lesions (Fig 4). Recurrent somatic deletions at 5q21 and 6q21 were enriched in SPOP-mutant tumors (P = 1.4×10−11 and P = 3.4×10−7, respectively, Fisher’s exact test) both in the whole-exome cohort and an independent prostate cancer collection (Fig. 4, Supplementary Table 8). Thus, loss of tumor-suppressor genes in these regions may collaborate with SPOP mutation to promote tumorigenesis. The relevant 5q21 locus contains CHD1, which encodes a chromatin-modifying enzyme that also undergoes disruptive rearrangements in prostate cancer14. The 6q21 region encompasses several genes including FOXO3, a FOXA1 homologue that has previously been implicated in prostate carcinogenesis and progression35, and PRDM1, a tumor suppressor in lymphoma36. In contrast, TP53 lesions were generally absent in SPOP-mutant tumors (P = 0.015, Fisher’s exact test), despite the fact that this tumor suppressor was recurrently mutated and deleted (Fig. 1). SPOP mutations also trended inversely with point mutations and/or copy number loss involving the PTEN locus in primary tumors (P = 0.044, Fisher’s exact test) (Fig. 4); this pattern was supported by FISH analysis for PTEN deletion (Supplementary Fig. 15). SPOP-mutant tumors also lacked PIK3CA mutations (Fig. 4). Although the inverse relationship between SPOP mutations and PTEN/PIK3CA alterations was evident in primary tumors (P = 0.041, Fisher’s exact test), these events co-occurred more frequently in metastatic tumors (Supplementary Fig. 15). Further studies are needed to determine if these genetic relationships also occur in other patient populations, and to elucidate the biological interactions that may underlie this phenomenon. Taken together, these results suggest that SPOP mutations may anchor a distinct genetic subtype of ETS-negative cancers.
In summary, whole-exome sequencing has identified genes that are recurrently mutated in prostate cancer. These efforts have also revealed a distinct ETS fusion-negative subclass of prostate cancer characterized by recurrent SPOP mutations and enriched for both 5q21 and 6q21 deletions. In the future, this expanded genetic framework may articulate new mechanisms of carcinogenesis that inform both disease modeling and patient stratification for clinical trials of experimental agents. Together with additional comprehensive analyses of the prostate cancer genome, epigenome, and transcriptome, these systematic approaches should illuminate the landscape of alterations that underlie disease biology and therapeutic vulnerability in this common and clinically heterogeneous malignancy.
Online Methods
DNA extraction and exome sequencing
H&E slides were cut from all frozen tissue blocks and examined by a board-certified pathologist to select for high-density cancer foci with <10% stroma or other noncancerous material to ensure high purity of cancer DNA. Biopsy cores were then taken from the corresponding frozen tissue block for DNA extraction. From each sample, 25–30mg of tissue was homogenized. DNA was extracted from homogenate and quantified using Picogreen dsDNA Quantitation Reagent (Invitrogen, Carlsbad, CA). Samples were qualified on an agarose gel (E-Gel, Invitrogen) to assess structural integrity. All DNA samples were stored at −20°C.
Whole-exome capture libraries were constructed from 100ng of tumor and normal DNA following shearing, end repair, phosphorylation and ligation to barcoded sequencing adapters37. Ligated DNA was size-selected for lengths between 200–350bp and subjected to exonic hybrid capture using SureSelect v2 Exome bait (Agilent). Samples were multiplexed and sequenced on multiple Illumina HiSeq flowcells to average target exome coverage of 118×.
Copy number analysis
Tumor and normal DNA were analyzed by Affymetrix SNP 6.0 arrays to detect regions of somatic copy number alteration. Quality control, segmentation and copy number analysis were performed as previously described33 with one additional step aimed at diminishing the number of recurrent lesions possibly caused by germline signal: we applied the same detection pipeline to the normal DNA samples alone. All peaks detected in both analyses were excluded from the list of recurrent somatic copy number aberrations. The remaining lesions with GISTIC q-value < 0.1 were included in analysis of copy number alterations associated with mutated genes. Two-tail Fisher Exact Test was applied for all association tests.
Sequence data processing and quality control
Exome sequence data processing and analysis were performed using Broad Institute pipelines14,38,39. A BAM file aligned to the hg19 human genome build was generated from Illumina sequencing reads for each tumor and normal sample by the “Picard” pipeline. The “Firehose” pipeline was used to manage input and output files and submit analyses for execution by GenePattern40.
Quality control modules in Firehose were used to compare genotypes derived from Affymetrix arrays and sequencing data to ensure concordance. Genotypes from SNP arrays were also used to monitor for low levels of cross-contamination between samples from different individuals in sequencing data using the ContEst algorithm41.
Mutation calling and identification of significantly mutated genes
The MuTect algorithm from the Broad Institute Genome Analysis Toolkit was used to identify SSNVs in targeted exons39,42. MuTect identifies candidate SSNVs by Bayesian statistical analysis of bases and their qualities in the tumor and normal BAMs at a given genomic locus. We required a minimum of 14 reads covering a site in the tumor and 8 in the normal for mutation calling. We determined the lowest allelic fraction at which SSNVs could be detected on a per-sample basis using estimates of cross-contamination from the ContEst pipeline41. Small somatic insertions and deletions were detected using the Indelocator algorithm after local realignment of tumor and normal sequences42.
The MutSig algorithm from the Broad Institute was applied to identify genes or gene sets that were significantly enriched for mutations given sequence context and genomic territory14,39,42. For each gene, we calculated the probability of detecting the observed constellation of mutations or a more extreme one, given the background mutation rates calculated across the dataset. This p-value was then adjusted for multiple hypotheses testing (Benjamini-Hochberg procedure) to obtain a q-value. Mutations in significantly mutated genes and others described in the text were manually reviewed by examination of BAM files in the Integrative Genomics Viewer (IGV)43.
Validation of selected mutations by mass spectrometric genotyping
We chose 240 non-silent mutations (231 SSNVs and 9 indels) across 48 T/N pairs to validate by mass spectrometric genotyping using the iPLEX platform (Sequenom, San Diego, CA). We targeted 74 mutations in significantly-mutated genes or gene sets with a q-value <0.1 and mutations reported in COSMIC. The remaining 166 non-silent mutations were chosen at random. Because the rate of validation using this technology falls significantly when the mutant allele is present at low allelic fraction14,38, we attempted to validate only mutations with allelic fraction ≥ 0.2 (i.e., where 20% of sequence reads from the tumor contain the mutation).
Of the 240 assays attempted, 228 gave successful genotype calls and 218 somatic mutations were confirmed (listed in Supplementary Table 3). All events called in the tumor were absent from the corresponding normal. The overall accuracy for mutation calling was 95.6% (CI: 92%–98%; Clopper-Pearson 95% confidence interval), in close agreement with previous studies14,38,39.
Mutation annotation
Somatic point mutations and indels were annotated using Oncotator (Ramos et al, submitted), which integrates information from publicly available databases including UCSC Genome Browser’s UCSC Genes track44, miRBase release 1545, dbSNP build 13246, UCSC Genome Browser’s ORegAnno track47, UniProt release 2011_0348 and COSMIC v5149.
Mutations were classified as heterozygous or homozygous using the ABSOLUTE algorithm (Carter et al., in press). ABSOLUTE integrates genome-wide copy number data from SNP arrays and the allelic fraction values of somatic mutations to model gene and mutation copy number in a tumor. Only a subset of tumors contained sufficient copy number alterations to permit analysis. Mutations in these tumors were annotated as heterozygous or homozygous and clonal or subclonal. Mutation multiplicity, equal to the average number of copies of a mutation per cancer cell, was also calculated.
RNA extraction, RNA-seq sample prep, sequencing, and processing
RNA was extracted from the frozen cancer tissue using TRIzol (Invitrogen) according to the manufacturer's protocol. Total RNA was prepared in accordance with Illumina's sample preparation protocol for paired-end sequencing of mRNA as previously described50. Paired-end reads were aligned to the human genome (hg18) using ELAND as previously described50. Data were visualized using the Integrated Genomics Viewer43, and candidate mutations were identified in SPOP coding regions.
DNA extraction and SPOP Genotyping
DNA was extracted using phenol-chloroform and purified by ethanol precipitation method as previously described14. Direct Sanger sequencing of putative SPOP somatic mutations in all tumor-blood pairs was performed by standard methods following PCR amplification using specific primers. Sequences of the primers used for amplifying and sequencing SPOP are given in Supplementary Table 9.
Laser Capture Microdissection (LCM)
5µm-thick tissue sections were cut, fixed and stained on membrane-coated slides followed by dissection with the ArcturusXT™ LCM Instrument (Life Technologies Corporation, California, USA). Tissue staining and Laser Capture Mircodissection (LCM) were performed by M.B. and K.P. as described by Espina et al.51. A combined IR capture and UV laser cutting was carried out to best recover a precise subset of cells. DNA was amplified with the Whole Genome Amplification kit (WGA4) as suggested by the manufacturer for the single cell approach (Sigma Aldrich, St. Louis, MO, USA). Standard PCR was used for targeted enrichment of SPOP exons 6 and 7 followed by Sanger sequencing.
FISH
The ETS rearrangement status and PTEN deletion status was assessed on tissue slides from the same tumor nodule used for RNA and DNA extraction. Methods for fluorescence in situ hybridization (FISH) for TMPRSS2-ETS gene fusion have been previously described9,11. We used ERG, ETV1, ETV4, and ETV5 break-apart FISH assays to confirm gene rearrangement on the DNA level52. To assess the status of PTEN, we used a locus specific probe and a reference probe as previously described14. All FISH probes are listed in Supplementary Table 10.
Quantitative RT-PCR
RNA was extracted using the TRIzol reagent (Invitrogen), subjected to DNase treatment (DNA-free kit; Applied Biosystems) according to the manufacturer's instructions, and used in quantitative RT-PCR. Quantitative RT-PCR was performed using the ABI 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer's RNA-to-CT 1-step protocol. Each target was run in triplicate, and expression levels relative to the housekeeping gene GAPDH were determined on the basis of the comparative threshold cycle CT method (2−ΔΔCT). The primer sequences used in these experiments are given in Supplementary Table 9. All experiments were run in triplicate; results are representative of three independent experiments.
Immunohistochemistry
ERG rearrangement status was confirmed by immunohistochemistry as previously described53. Briefly, primary rabbit monoclonal antibody was obtained from Epitomics (Burlingame, CA). Antigen recovery was conducted using heat retrieval and CC1 standard, a high pH Tris/borate/EDTA buffer (VMSI, catalog no. 950-124). Slides were incubated with 1:100 of the ERG primary antibody for 1 hour at room temperature. Primary antibody was detected using the ChromoMap DAB detection kit (VMSI, catalog no. 760-159) and UltraMap anti-Rb HRP (VMSI, catalog no. 760-4315). The anti-Rb HRP secondary antibody was applied for 16 minutes at room temperature. Slides were counterstained with Hematoxylin II (VMSI, catalog no. 790-2208) for 8 minutes followed by Bluing Reagent (VMSI, catalog no. 760-2037) for 4 minutes at 37°C. Subjective evaluation of ERG protein expression was scored as positive or negative by study pathologists (K.P., J-M.M., M.A.R.)
SPOP wt and mutant plasmids
Wild-type SPOP was obtained from Origene (Rockville, MD) with C-terminal myc and FLAG tags in a mammalian expression vector. SPOP-F133V was created by using the QuikChange II site-directed mutagenesis kit (Agilent). All plasmids were confirmed with Sanger sequencing, and protein expression was confirmed with Western blot using antibodies to SPOP, myc, and FLAG.
Cell Culture and Transfection
The human prostate cancer cell lines 22Rv1, and DU145 and the benign prostate cell line RWPE were obtained from the American Type Culture Collection (Manassas, VA). 22Rv1 and DU145Cells were maintained in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. RWPE cells were maintained in Keratinocyte-SFM (Invitrogen) supplemented with human recombinant Epidermal Growth Factor and Bovine Pituitary Extract (BPE).
For siRNA transfection, RWPE (2.5 × 105 per well), 22Rv1 (4 × 105 per well), and DU145 (2 × 105 per well), cells were seeded on 6-well tissue culture plates. The next day, cells were transfected with 100 nM SPOP or nontargeting (control) siRNAs (ON-TARGETplus; Thermo Scientific) using Dharmfect 2 reagent (Invitrogen) according to the manufacturer’s instructions. For plasmid transfection, DU145 (4 × 105 per well), cells were seeded on 6-well tissue culture plates. The next day, cells were transfected with 4 ug of pCMV6-WT SPOP or pCMV-SPOP-F133V using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Cell viability and proliferation assays
22Rv1 (2 × 103 per well) and DU145 (1 × 103 per well) cells transfected with control or SPOP siRNA or SPOP plasmids were seeded in 96-well tissue culture plates. Cell viability and growth was determined by performing WST-1 assay (Roche) reading absorbance at 450 nm according to the manufacturer's instructions. Values from three wells were obtained for each treatment and timepoint. Results are representative of three independent experiments.
Invasion assays
For invasion assays, 7.5 × 104 22Rv1 and 5 × 104 DU145 cells transfected with control or SPOP siRNA or SPOP plasmids were resuspended in 0.5 mL of RPMI-1640 medium containing 1% FBS and placed into the top chamber of Matrigel-coated 8-µm Transwell inserts (BD Falcon). The bottom wells contained RPMI supplemented with 5–10% FBS. After 24h (DU145) or 48h (22Rv1), the filters were fixed and stained with Crystal Violet 0.5% for 30 min, and cells on the upper surface of the filters were removed with a cotton swab. Migrated cells were quantified by counting the numbers of cells that penetrated the membrane in four microscopic fields (at 20X objective magnification) per filter. All experiments were run in triplicate; results are representative of three independent experiments.
Supplementary Material
Acknowledgements
This work was supported by the National Human Genome Research Institute (NHGRI) Large Scale Sequencing Program, Grant U54 HG003067 to the Broad Institute (E.S.L.), the Kohlberg Foundation (L.A.G.), the Starr Cancer Consortium (M.A.R., F.D., L.A.G), the Prostate Cancer Foundation (M.A.R.), Department of Defense Synergy Award PC101020 (F.D., L.A.G., M.A.R.), Dana-Farber / Harvard Cancer Center Prostate Cancer SPORE grant NIH P50 CA090381, New Investigator Award PC094516 (F.D.), the National Cancer Institute, Early Detection Research Network U01CA111275, NCI EDRN (F.D., M.A.R.), National Cancer Institute R01 CA125612 (F.D., M.A.R.), the PNW Prostate Cancer SPORE P50CA097186 (C.M., P.S.N.), the Department of Defense Synergy Award PC093372 (T.W., P.S.N.), the Swiss Science Foundation grant PASMP3_134379/1 (J.-P.T.) and the NIH Director’s New Innovator Award (DP2OD002750) (L.A.G.). S.C.B. is supported by a Medical Scientist Training Program (MSTP) grant from the United States National Institute of Health. C.E.B. is supported by a Prostate Cancer Foundation Young Investigator Award. We are grateful for the assistance of members of the Broad Institute Biological Samples Platform, Genetic Analysis Platform and Genome Sequencing Platform. We thank Samprit Banerjee for computational assistance, Robert Kim and Robert Leung for their critical contributions to the Weill Cornell Prostate Cancer Tumor Bank, Peter Schraml, Susanne Dettwiler and Martina Storz for assistance with the UHZ cohort and biobank, and the members of the University of Washington Rapid Autopsy Program. We thank the University of Michigan Prostate Cancer SPORE (P.I. Ken Pienta) for sample contribution to this study. We are also grateful to the patients and families who contributed to these studies.
Footnotes
Data Access: The binary Sequence Alignment/Map (BAM) files and SNP array data were deposited in dbGaP under accession no. (phs000447.v1.p1).
Author Contributions
S.C.B., M.S.L., P. S., W.M.H., E.V.A., N.S., K.C., A.S., S.L.C., G.S., D.V. and A.H.R. performed computational analyses. M.B., J.P.T., T.A.W., S.S.C., K.S., G.B., T.Y.M., K.P., and T.V. designed and performed experiments. E.N., D.A., R.C.O., C.G., W.W., M.C.R., K.A. and S.B.G. processed samples and supervised exome sequencing. J.M.M., K.P., N.R., P.J.R., H.M., C.M., and P.S.N. coordinated sample acquisition, processing, pathologic review, and analysis. C.E.B, S.C.B., F.D., P.W.K., T.R.G., M.M., E.S.L., G.G., M.A.R. and L.A.G. designed the study. C.E.B, S.C.B., F.D., G.G., M.A.R. and L.A.G analyzed the data and wrote the manuscript.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Daskivich TJ, et al. Overtreatment of men with low-risk prostate cancer and significant comorbidity. Cancer. 2011;117:2058–2066. doi: 10.1002/cncr.25751. [DOI] [PubMed] [Google Scholar]
- 3.He WW, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia-Gaur R, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Cairns P, et al. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 7.Linja MJ, Visakorpi T. Alterations of androgen receptor in prostate cancer. J. Steroid Biochem. Mol. Biol. 2004;92:255–264. doi: 10.1016/j.jsbmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Visakorpi T, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat. Genet. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 9.Perner S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 10.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 11.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 12.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar a, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 2011;108 doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai Y, et al. Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 1997;418:23–26. doi: 10.1016/s0014-5793(97)01340-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 18.Majumder PK, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibel AS, et al. CDKN1A and CDKN1B polymorphisms and risk of advanced prostate carcinoma. Cancer Res. 2003;63:2033–2036. [PubMed] [Google Scholar]
- 20.Gao N, et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–3443. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, et al. Definition of a FoxA1 Cistrome That Is Crucial for G1 to S-Phase Cell-Cycle Transit in Castration-Resistant Prostate Cancer. Cancer Res. 2011;71:6738–6748. doi: 10.1158/0008-5472.CAN-11-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao N, et al. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 23.Williamson Ea, et al. BRCA1 and FOXA1 proteins coregulate the expression of the cell cycle-dependent kinase inhibitor p27(Kip1) Oncogene. 2006;25:1391–1399. doi: 10.1038/sj.onc.1209170. [DOI] [PubMed] [Google Scholar]
- 24.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhou R, et al. SOX9 interacts with a component of the human thyroid hormone receptor-associated protein complex. Nucleic Acids Res. 2002;30:3245–3252. doi: 10.1093/nar/gkf443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 27.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makinen N, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science. 2011;252 doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, et al. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science. 2009;323:1218–1222. doi: 10.1126/science.1157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011 doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosquera JM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin. Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demichelis F, et al. Distinct genomic aberrations associated with ERG rearranged prostate cancer. Genes Chromosomes Cancer. 2009;48:366–380. doi: 10.1002/gcc.20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapointe J, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 35.Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15:752–757. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandelbaum J, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 41.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffith OL, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium U. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pflueger D, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Espina V, et al. Laser-capture microdissection. Nat. Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 52.Svensson MA, et al. Testing mutual exclusivity of ETS rearranged prostate cancer. Lab. Invest. 2011;91:404–412. doi: 10.1038/labinvest.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park K, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.