Abstract
India is home to approximately 60 per cent of the world's remaining wild tigers, a species that has declined in the last few centuries to occupy less than 7 per cent of its former geographical range. While Indian tiger numbers have somewhat stabilized in recent years, they remain low and populations are highly fragmented. Therefore, the application of evidence-based demographic and genetic management to enhance the remaining populations is a priority. In this context, and using genetic data from historical and modern tigers, we investigated anthropogenic impacts on genetic variation in Indian tigers using mitochondrial and nuclear genetic markers. We found a very high number of historical mitochondrial DNA variants, 93 per cent of which are not detected in modern populations. Population differentiation was higher in modern tigers. Simulations incorporating historical data support population decline, and suggest high population structure in extant populations. Decreased connectivity and habitat loss as a result of ongoing fragmentation in the Indian subcontinent has therefore resulted in a loss of genetic variants and increased genetic differentiation among tiger populations. These results highlight that anthropogenic fragmentation and species-specific demographic processes can interact to alter the partitioning of genetic variation over very short time scales. We conclude that ongoing strategies to maximize the size of some tiger populations, at the expense of losing others, is an inadequate conservation strategy, as it could result in a loss of genetic diversity that may be of adaptive significance for this emblematic species.
Keywords: Panthera tigris, genetic variation, historical samples, population decline
1. Introduction
The Indian subcontinent is a globally important region for biodiversity, and its tiger populations (Panthera tigris) represent 50–60 per cent of all those remaining [1]. The tiger's range has declined more than 50 per cent during the last three generations [2,3]; it now occurs in less than 7 per cent of its historical distribution. Historical records [4] and genetic data [5] show that the recent demographic history of Indian tigers is characterized by population collapse owing to anthropogenic habitat destruction and hunting. Tigers now persist in relatively small populations (20–120 individuals) in India, which are often spatially isolated [6]. These populations harbour 60–70 per cent of the species’s extant genetic diversity [5], emphasizing their importance for future species recovery.
While mitochondrial genetic diversity and population differentiation in modern Indian tigers seems high [5,7], the variation lost owing to population decline in this region is unknown. In subspecies such as the Amur tiger, human-induced population decline has left modern populations with very low genetic variation [8,9]. Other subspecies, such as the Caspian tiger, possessed low genetic variation even in historical times [10]. Studies on other apex predators have also revealed significant loss of genetic variation as a result of acute population decline (e.g. wolf [11,12]; cheetah [13–15]; puma [16,17]; Tasmanian tiger [18]). Additionally, historical presence/absence models indicate a much higher degree of tiger habitat connectivity historically compared with modern times [19]. This loss of habitat continuity is expected to result in demographic isolation, an increased genetic differentiation and a reduction in genetic diversity owing to genetic drift.
Loss of genetic variation, while rarely posing the immediate threats associated with demographic loss and environmental stochasticity [20–23], can nevertheless ultimately compromise populations and may lead to extinction [24–26]. Further, since genetic variation is often geographically structured and can be modified by species-specific behavioural patterns [27,28], the effects of local extinctions on range-wide genetic variation can exceed that predicted by population size change alone, resulting in a non-additive loss of unique genetic variants and an increase in genetic structure [29–31]. However, to understand the impact of habitat loss not only on genetic variation, but also on its partitioning, knowledge of the time period through which changes in habitat took place is important. Historical collections provide important temporal and geographically referenced samples for such studies [32]. In the case of Indian tigers, populations have undergone their strongest declines over the last 200 years [5], and therefore museum samples collected during the British colonial period in India are relevant to study these processes.
A recent tiger survey led by the government of India has asserted that the species is in recovery because numbers have increased from 1400 to 1700 between 2009 and 2011 [33,34]. However, while populations may increase under certain circumstances (e.g. the cessation or control of hunting), demographic recovery is only one indicator of long-term population viability, and management goals that only aim to increase numbers within modified landscapes and do not consider gene flow are often inadequate [35]. Here, by analysing both maternally and bi-parentally inherited marker data, we investigated (i) whether Indian tigers have lost genetic variation through recent historical time, (ii) whether this loss of variation can be explained by a decrease in population size alone and (iii) whether the geographical partitioning of genetic variation has changed during this period. Since we compared historical and modern populations, our modern samples included most extant tiger populations in the Indian subcontinent, while our historical samples include populations where tigers have been locally extirpated.
2. Material and methods
(a). Sampling
Historical Indian tigers were sampled from the Natural History Museum, London and National Museum of Scotland, Edinburgh, and predominantly comprised specimens hunted prior to 1950. Samples were mainly available from geographical regions where tigers are now extinct (for example, Afghanistan, Rajputana, Gujarat; see electronic supplementary material, table S1 and figure S1). Fifty-three samples with associated geolocation and dates (36 skulls and 17 skins) were analysed. Dried tissue, nasal bones (turbinals) or small pieces of skin were collected from skulls and skins. Each specimen was handled wearing gloves, carefully sampled with separate sterile blades and stored in a sterile vial. Samples were stored at −20°C until processed.
For contemporary tigers, we used mitochondrial (mt)DNA sequences (1263 bp, comprising up to four fragments) and microsatellite data (10 loci) from all Indian samples analysed by Mondol et al. [5] (GenBank accession numbers: EU661609–EU661691). Twenty-four additional faecal samples were collected during 2009–2010 from five protected areas (see the electronic supplementary material, tables S2 and S3). Faecal samples were processed as by Mondol et al. [36]. Seventy-seven mitochondrial and 112 microsatellite genotypes from individual tigers were used for comparison with the historical samples in this study (see the electronic supplementary material, tables S2 and S3).
(b). DNA extraction, PCR amplification and data quality control
All DNA extractions were performed in an ancient DNA laboratory exclusively dedicated to the handling of museum samples (Cardiff University), where no previous tiger work had been conducted (details are provided in the electronic supplementary material). To control for contamination issues commonly associated with museum samples [37,38], stringent PCR amplification and quality control measures were used. We amplified four mitochondrial fragments (NADH5—482 bp, Cytb—450 bp, NADH2—131 bp and control region—200 bp). Ten microsatellite loci were amplified. Details on PCRs and data quality control are provided in the electronic supplementary material.
(c). Data analysis
We used Microchecker v. 2.2.3 [39] to identify possible null alleles, large allele dropout and scoring errors owing to stutter peaks. We quantified the genotyping error rate in our dataset (both modern and historical) using the algorithm developed by Zhan et al. [40]. This approach quantifies error by using the entire dataset.
All related individuals were removed before Structure analysis (relatedness estimated in ML-Relate [41]).
Bayesian clustering was implemented using Structure v. 2.3.3 [42,43] and details are provided in the electronic supplementary material. We assumed that the combined samples (historical and modern) would reveal historical population structure. Admixture was estimated using the Q-values (estimated proportions of ancestry) calculated in Structure. We used Q > 0.75 as threshold for assigning individuals to populations [44].
Summary diversity statistics and indices of genetic differentiation (pairwise Fst for both mtDNA and microsatellites, and AMOVA) were estimated using Arlequin v. 3.1 [45], dividing tiger populations according to cluster assignment results from Structure. A median-joining network based on 1263 bp mtDNA sequences for all modern and historical tiger samples was created using Network [46]. Additionally, TempNet [47] was used to create a parsimony network to display sequence data in a heterochronous context. Subsampling was conducted to investigate effects of sample size (details are provided in the electronic supplementary material).
Coalescent simulations [48] allow the modelling of population genetic data and have been extensively used to test hypotheses of demographic history. In association with approximate Bayesian computation (ABC) approaches, they have been used to estimate parameters such as population size before and after demographic bottlenecks and the timing of such declines. Simulations based on modern samples alone necessarily assume that populations in the past and those present today at a specific location are synonymous, and hence ignore the possibility of population extinction or replacement. Serial coalescent models [49], however, allow incorporation of genetic data from different time points in the past. Such models result in a better ability to detect population decline [50].
We used serial coalescent simulations and an ABC approach (BayeSSC [51]) to estimate the time of onset of population decline for Indian tigers, and aimed to estimate three parameters: current (modern) effective population size, time of population decline and extent of population decline. The extent of decline is quantified as a ratio of the historical effective size to the current effective size. The ABC framework consists of simulating thousands of gene genealogies according to values chosen from the prior distributions for the parameters of interest. The observed and simulated data were compared against each other by determining the distance between a vector of summary statistics calculated from each dataset. The 0.001 percentile of the simulated parameters with the smallest distances were used to estimate the posterior distribution of the parameters of interest. Details of simulations (prior distributions, summary statistics used and rejection criteria) are provided in electronic supplementary material for both mtDNA and microsatellites. For the mitochondrial DNA dataset, we ran simulations for two datasets: (i) Indian subcontinent-wide tigers and (ii) north Indian (Terai and semi-arid regions) tigers (see figure 1; electronic supplementary material, tables S1 and S2 for sample details). Rejection was based on observed summary statistics for mtDNA data from the Indian subcontinent and the north Indian region, respectively.
Figure 1.
Forest cover map with modern and historical sample locations from different countries used in this study. The map also shows the inferred biogeographic tiger habitats, as found in this study.
For the microsatellite data, we conducted ABC analyses only for summary statistics based on modern and historical north Indian tigers (see figure 1; electronic supplementary material, tables S1 and S2 for sample details), because the sampling for historical samples from peninsular India was relatively poor.
3. Results
(a). Amplification success
Of the 53 museum samples collected during this study, we could generate mtDNA (1263 bp) and microsatellite data (eight loci) from 36 and 25 samples, respectively (see the electronic supplementary material, table S1). Sequences were submitted to GenBank (accession numbers: JN786601–JN786684). Individual amplification success for mtDNA and microsatellites are provided in detail in the electronic supplementary material (tables S5 and S6). Loci FCA230 and FCA232 had low amplification success on museum samples and were removed from the analysis. Although no null alleles were present in our modern samples, FCA090 and FCA628 showed evidence for null alleles in the historical samples. Analysis of genotyping error rates [40] revealed a 0.5 and 1.7 per cent allelic dropout rate (with no false alleles) for modern and historical samples, respectively (see the electronic supplementary material, table S4). Overall, our analysis showed a mean per-genotype error rate of 5 × 10–6 and 1 × 10–3 from modern and historical samples, respectively, corresponding to less than one erroneous multilocus genotype for both museum (0.8) and modern (0.01) microsatellites.
(b). Historical genetic variation
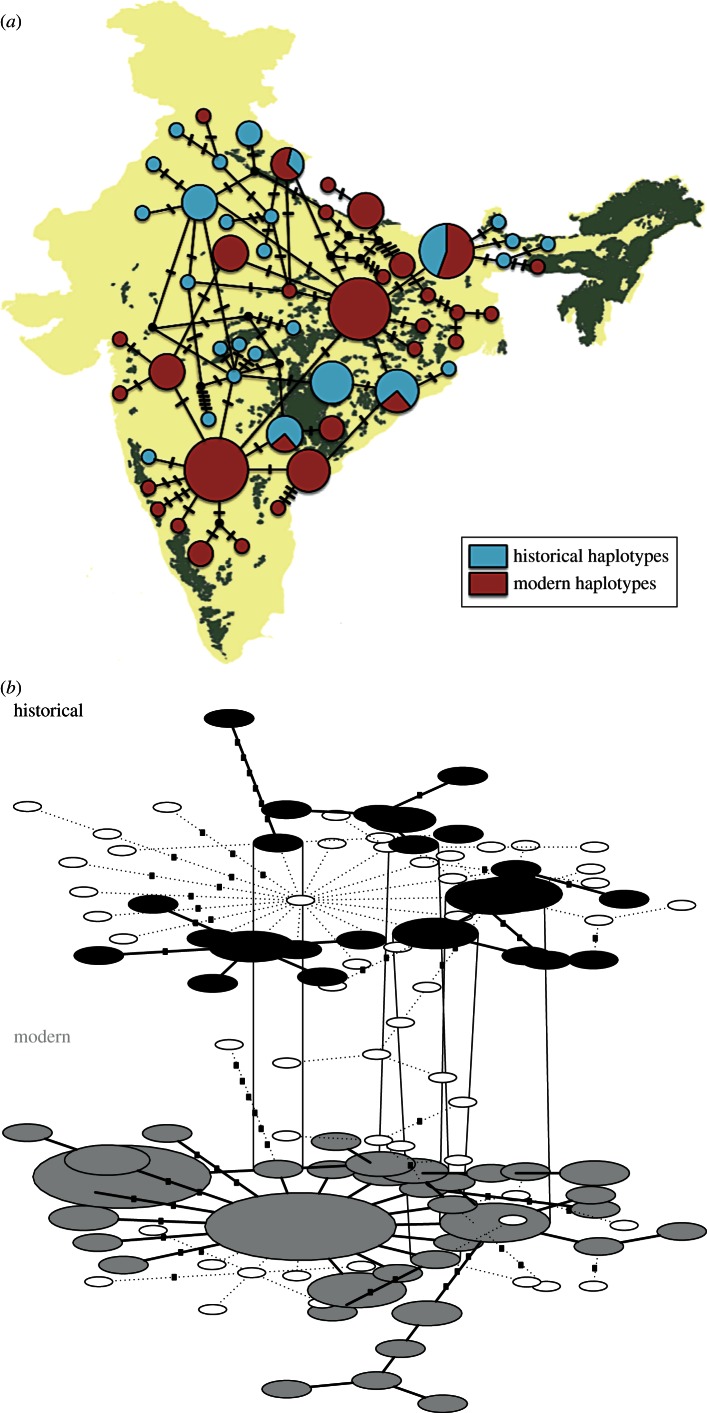
For mitochondrial DNA, our results reveal that Indian tigers historically possessed a substantial number of previously undetected haplotypes when compared with modern samples (figure 2a). Historical samples showed 25 haplotypes compared with 36 haplotypes found in modern tigers (figure 2a, table 1), with only four historical haplotypes being found in modern tigers (figure 2b), indicating a substantial loss and change of variation through time. Resampling confirmed that this inference was robust to lower historical sample sizes (table 1).
Figure 2.
(a) Distribution of modern and historical haplotypes based on 1263 bp of mtDNA data. (b) A statistical parsimony network from modern and historical samples created by the program TempNet [47] reveals shared haplotypes between different time periods. Open ellipses represent missing haplotypes. (Online version in colour.)
Table 1.
Genetic diversity in modern and historical samples.
| modern samples | historical samples | resampling simulation (average of 1000 runs) | ||
|---|---|---|---|---|
| mtDNA (1263 bp) | number of haplotypes | 36 | 25 | 17 (13,20) |
| microsatellites (eight loci) [six loci] | no. alleles (s.d.) | 15.37 (2.774) [15.16 (2.6)] | 6.25 (1.66) [6.66 (1.3)] | 11.827 (8.95–15.04) |
| allelic size range (s.d.) | 37 (7.17) [38.66 (6.2)] | 16.75 (8.06) [19.33 (6.07)] | 29.45 (26.5–32.5) | |
| expected heterozygosity (s.d.) | 0.85 (0.03) [0.84 (0.03)] | 0.748 (0.07) [0.73 (0.07)] | 0.85 (0.851–0.856) |
Analyses with eight nuclear microsatellites did not recapitulate the loss of mitochondrial genetic variation. When combined, all modern samples showed a higher number of alleles (NAModern = 15.37 (2.774), NAHistorical = 6.25 (1.66)), allelic size range (SizeModern = 37 (7.17), SizeHistorical = 16.75 (8.06)) and observed heterozygosity (Ho Modern = 0.53 (0.08), Ho Historical = 0.43 (0.08)), when compared with historical samples (table 1). Resampling confirmed that lower genetic variation in historical samples was not simply owing to lower sample size, and the removal of the loci with inferred null alleles from the dataset did not change this pattern (see table 1). Because historical and modern sampling was more comparable for the north Indian region, we also compared summary statistics for this region. This comparison revealed that historical and modern north Indian tigers had similar numbers of alleles (NAModern-NI = 7.28 (1.76), NAHistorical-NI = 6 (1.35)) and observed heterozygosity (Ho Modern-NI = 0.668 (0.11), Ho Historical-NI = 0.732 (0.09)) for seven loci.
(c). Population structure in current and historical tigers
To explore changes in population structure between historical and modern tigers, we analysed structure in historical and modern tigers versus modern tigers alone. Structure Harvester indicated that K = 3 was the most probable number of clusters for modern samples. Peninsular India (comprising Western Ghats and Deccan Plateau) and northeast India samples formed one cluster, the semi-arid region formed a second cluster and the Terai landscape formed the third. Surprisingly, when modern and historical samples were taken together two clusters were inferred. Peninsular India and northeast India samples formed one cluster, while north Indian samples formed the second, suggesting a recent increase in genetic differentiation (figure 3). Inference of three clusters for modern samples and two clusters for the modern and historical samples did not change when we omitted northeast (modern and historical) samples from this analysis.
Figure 3.
Clusters indicated by the program Structure, based on eight microsatellite loci, for (a) modern (n = 57) and (b) modern and historical (n = 82) tigers. (Online version in colour.)
Analysis of molecular variance (table 2) revealed that population differentiation has approximately doubled for both mitochondrial DNA and microsatellites in modern tigers, suggesting a substantial increase in genetic differentiation in recent times. Including northeast samples did not change the results.
Table 2.
AMOVA analysis with microsatellites and mitochondrial DNA based on combined (historical and modern) and modern samples with different combinations of populations.
| genetic population | sample | percentage of variation (mtDNA—1263 bp) |
percentage of variation (microsatellites—8 loci) |
overall Fst |
||||
|---|---|---|---|---|---|---|---|---|
| among populations | within populations | among populations | among individuals within populations | within individuals | mtDNA (1263 bp) | microsatellites (8 loci) | ||
| peninsular India, semi-arid, Terai landscape (three populations without northeast) | modern and historical | 9.04 | 90.96 | 9.15 | 28.05 | 62.8 | 0.09a | 0.09a |
| modern | 20.6 | 79.4 | 20.66 | 19.38 | 59.97 | 0.2a | 0.2a | |
| peninsular India versus north India (two populations without northeast) | modern and historical | 7.95 | 92.05 | 6.52 | 30.21 | 63.21 | 0.07a | 0.06a |
| modern | 17.79 | 82.21 | 8.12 | 26.43 | 65.45 | 0.17a | 0.08a | |
| peninsular India, semi-arid, Terai landscape (three populations with northeast) | modern and historical | 8.03 | 91.97 | 7.78 | 27.05 | 65.17 | 0.08a | 0.08a |
| modern | 18.47 | 81.53 | 19.02 | 21.24 | 59.74 | 0.18a | 0.19a | |
| peninsular India versus north India (two populations with northeast) | modern and historical | 6.7 | 93.3 | 4.5 | 30.03 | 65.47 | 0.06a | 0.04a |
| modern | 15.28 | 84.72 | 5.35 | 27.52 | 67.13 | 0.15a | 0.05a | |
aSignificant.
(d). Simulating demographic history of Indian tigers
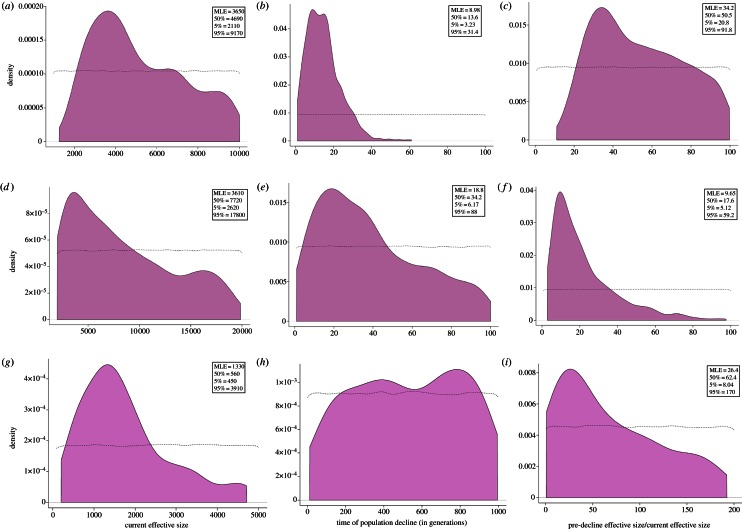
Our simulations support a recent population decline (MLEIndian subcontinent = 45 years (16, –160; 5–95th percentile), MLEsemi-arid Terai = 100 years (30–440)) of high magnitude (MLEIndian subcontinent = 34-fold (21–91), MLEsemi-arid Terai = 10-fold (5–59); figure 4a–f). In both Indian subcontinent-wide and regional analyses, the current effective size was estimated to be relatively high (MLEIndian subcontinent = 3650 (2110–9170), MLEsemi-arid Terai = 3610 (2620–17 800); figure 4a–f). We ran the final simulations at a mutation rate of 0.015/bp/106 years since initial simulations indicated this to be the most probable estimate. Overall, the simulations fitted the observed data better for the north Indian tiger dataset (delta = 0.563) compared with the Indian subcontinent-wide dataset (delta = 1.495).
Figure 4.
The posterior distributions for (a–c) all mitochondrial genetic data (modern and historical), (d–f) mitochondrial data from the semi-arid and Terai region (modern and historical), and (g–i) microsatellite data from the semi-arid and Terai region (modern and historical). In all cases, we include posterior distributions for current effective size, time of population decline and the magnitude of the population decline. The microsatellite results are based on the average-over-loci approach. (Online version in colour.)
Rejection analyses and posterior distributions for the microsatellite data based on an average-across-loci approach resulted in lower delta values (0.054927; figure 4i), indicating better fit than when posteriors were combined across loci (delta between 0.1 and 0.2; electronic supplementary material, figure S1).
Figure 4g–i reveals a signature of population decline, as indicated by the microsatellite data for tigers from the semi-arid and Terai regions. The magnitude of decline (MLEsemi-arid Terai = 26.5-fold (8–170)) was higher than that suggested by mitochondrial DNA (MLEsemi-arid Terai = 10-fold), while the current effective size (MLEsemi-arid Terai = 1330 (450–3910)) estimate was lower (MLEsemi-arid Terai = 3610). The microsatellite data did not, however, have sufficient power to detect the timing of this decline. MLE for mutation rate was 3 × 10−4. Analysis based on separate loci and then combining posteriors revealed qualitatively similar results (see the electronic supplementary material, figure S1), with a relatively high magnitude of population decline (MLE = 17.0-fold), while the effective size (MLE = 700) was lower than that estimated above.
4. Discussion
(a). Tigers have recently lost mitochondrial, but not nuclear genetic variation
Here, we assessed historical genetic variation for Indian tigers based on samples from areas where tigers have now been extirpated. Comparison of mitochondrial DNA for historical and modern samples reveal that a large number of haplotypes present historically in India have been lost. Apart from a decrease in genetic variation per se, only 7 per cent of the historical mitochondrial haplotypes are represented in the contemporary samples (4 of 57; figure 2b). MtDNA is maternally inherited, and ecological studies on tigers reveal that females have smaller home ranges to males and that dispersing females tend to establish their territory close to their mothers [52]. The apparent loss of mitochondrial haplotypes could be caused by a combination of sex-biased philopatry and our historical sampling (mainly in regions where tigers are now extinct) since we would expect regions to feature maternal genotypes that characterize them. Loss of habitat would be expected to disproportionately affect females (and their haplotypes) that represented those extirpated regions. While several studies suggest a decline in mitochondrial haplotype diversity in other taxa (e.g. birds: loggerhead shrike [53]; mammals: Scandinavian wolf [11], Arctic fox [54]), we could not find examples of studies where apparent haplotype turnover between historical and modern samples is so high. In addition to sampling effects, this could be because we sequenced a relatively long mitochondrial segment, revealing high haplotype diversity in historical as well as modern samples.
In contrast, across the Indian subcontinent, we found a different pattern for microsatellites. Modern tigers showed higher genetic variation than historical tigers. One explanation could be an inherent dissociation in genetic diversity between nuclear and mitochondrial DNA in tigers. However, this is unlikely because AMOVA indicates that genetic structure has approximately doubled for both marker types in modern samples. Yet, because our sampling included relatively few historical samples from peninsular India, inclusion of more individuals from this region could change the result. Museums skins are often sampled from regions under the heaviest hunting pressure [38], and a corollary of this observation is that historical sampling from extant populations can be much lower—as was the case for the Natural History Museum's tiger skin collection. However, while our modern and historical sample sizes are not comparable for peninsular India, they are comparable for north India (see figure 1, semi-arid and Terai regions), and here genetic variation (measured as heterozygosity and allelic variance) was lower for modern tigers. The results for the north Indian region therefore suggest that increased historical sampling from peninsular India might change our results. Additionally, several studies on historical genetic variation reveal that despite major bottlenecks, genetic variation is often comparable between modern and historical samples (e.g. fisher [55]; otter [56]; right whale [57]), while some studies reveal a relatively moderate loss of genetic variation (Scandinavian wolf [11]; puma [16,17]).
(b). Tiger population structure has increased
Our most striking result is an increase in population structure in Indian tigers since historical times. This inference is supported by structure analyses (figure 3) and AMOVA (table 2), which infer the existence of fewer populations when historical and modern samples are combined (for microsatellites) and an increase in mitochondrial structure for modern populations. The difference between AMOVA patterns for mtDNA and microsatellites could reflect the lower effective size for mtDNA, allowing differentiation to accumulate more rapidly [27]. Conflicting patterns for mtDNA and nuclear DNA are common for mammals with female philopatry and male-biased dispersal [28]. In contrast to females, male tigers have large home ranges and are capable of dispersing up to 280 km [58].
Changes in population structure have been observed over time scales of thousands of years for large species such as brown bears [59] and other mega-herbivores during the Pleistocene [60] at continental scales. A few recent studies [17,55,61] have demonstrated changes in population structure between historical and modern populations. However, all have been relatively limited in their geographical scale, and have only investigated microsatellite genetic variation. Our results are especially striking given the magnitude of increase in population structure evidenced both from mitochondrial and nuclear markers, given the short time scale over which these effects are observed.
(c). Both mitochondrial and nuclear simulations suggest tiger population size decline
Our coalescent simulations for mitochondrial DNA support a relatively recent population decline (figure 4a–f). The relatively high estimates of current effective size could be owing to the high population structure within both the Indian subcontinent and north India. Estimates of the timing of population decline, however, appear to be slightly older for north Indian tigers and could suggest a relatively older human impact in north India. Although historical reports indicate massive commercial forest logging by the British Empire for railway expansion [62] and post-Independence deforestation for agricultural lands [63] during the last 200 years, human densities have always been high in the fertile Gangetic plains and Terai landscape. Historical hunting records also suggest large-scale tiger hunting by Mughal rulers 500 years ago [4] in this landscape. Overall, the timing of decline as suggested by mitochondrial DNA simulations that incorporate historical data are more recent than those suggested by earlier results (based only on modern microsatellite data from peninsular India) [5]. This could be (a) because the current models assume a population bottleneck, while earlier models assumed gradual decline [5] (for a similar-sized population decline, population bottleneck scenarios will appear more recent when compared with gradual decline scenarios), or (b) because our historical samples are mostly more recent than 100 years ago. As a result, our statistical power to investigate demographic change is highest for the ‘time window’ corresponding to the last century [32].
Additionally, ABC and serial coalescent simulations for microsatellite markers reveal a population decline for north India (semi-arid and Terai region; figure 4g–i). A previous study of tiger demographic history [5] suggested a population decline in peninsular tigers based on microsatellite data. Rejection algorithm parameter estimation based on our north India data suggests that the magnitude of population decline might have been higher in the semi-arid and Terai regions (26-fold versus 10-fold). Using historical samples did not improve our ability to date this population decline. It is possible that there were multiple declines, and our approach was not able to detect multiple events; further simulation might provide insights into this problem.
Several studies using historical samples have suggested changes in effective size (see [38] for review). However, to our knowledge, ours is the first to use ABC on both mitochondrial and microsatellite data to better understand the timing and magnitude of population decline. While these approaches have been applied extensively to ancient genetic data (see [32] for review), our study broadly demonstrates the power of ABC analyses for recent time scales. Our results also suggest that at historical time scales, our microsatellite data may not provide enough power to investigate timing of decline. If this is true generally, it might be the reason several microsatellite datasets comparing modern and historical samples do not show clear demographic signatures.
(d). Implications
The increased population structure in modern tigers has conservation implications. First, Lorenzen et al. [60] have shown that increased population structure correlates with extinction, suggesting that the trends we observe represent a conservation ‘red flag’. Second, the broad ‘populations’ our analyses suggest (from historical and modern data) can be used to guide management. Our results will help delineate multiple protected areas falling within the same population. Finally, our results suggest that semi-arid tigers are under considerable threat from isolation, and the two remaining populations should be monitored in future conservation efforts.
Genetic variation can be crucial for species survival and adaptation to environmental change [64], and its judicious management is now required in the Convention on Biological Diversity's 2020 targets [65]. Although recent studies demonstrate that Indian tigers retain the highest proportion of the species's genetic variation [5], this study demonstrates that some components of that genetic variation have been lost, and what remains is now subdivided. Current conservation efforts focus on numbers alone (e.g. [3,66]), while others suggest the importance of maintaining connectivity [67]. Our results suggest that in order to enhance conservation efforts seeking to halt the loss of genetic diversity for this culturally valuable species, it is critical to maintain within-population variation, as well as increasing population connectivity. Current conservation initiatives discuss maintaining connectivity at a landscape level [66]. However, maintaining genetic variation will require population connectivity at much larger spatial scales (e.g. within peninsular India). Maintaining connectivity in regions with high habitat fragmentation (such as south and southeast Asia [68]) is complex. However, whether conservation initiatives at such large scales (potentially involving multiple countries) can be sustained could determine the future survival of tigers.
Acknowledgements
We thank Louise Tomsett, Richard Sabin and Paula Jenkins from the Natural History Museum, London for providing historical tiger samples. We also thank Dr Andrew Kitchener and Mr Jill Mackay for providing valuable samples from the National Museum of Scotland. Initial standardization for all the work was performed on samples from Bombay Natural History Society, and we thank Dr Rahmani for those samples. We also thank K. Garg, V. Verma and all other members of Bruford and Ramakrishnan laboratories for their help. We thank three anonymous referees for comments on an earlier version of this manuscript. S.M. was funded by the Department of Science and Technology, Government of India. The Royal Society (International Joint Project), NCBS and Cardiff University funded this work.
References
- 1.Kumar NS, Bindra PS. 2012. India refines tiger monitoring protocols. Oryx 46, 480. 10.1017/S0030605312001147 (doi:10.1017/S0030605312001147) [DOI] [Google Scholar]
- 2.Sanderson E, Forrest J, Loucks C, Ginsberg J, et al. 2006. Setting priorities for the conservation and recovery of wild tigers: 2005–2015. Washington, DC: World Wildlife Fund.
- 3.Walston J, et al. 2010. Bringing the tiger back from the brink—the six percent solution. PLoS Biol. 8, e1000485. 10.1371/journal.pbio.1000485 (doi:10.1371/journal.pbio.1000485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rangarajan M. 2006. India's wildlife history: an introduction. New York, NY: Orient Longman [Google Scholar]
- 5.Mondol S, Karanth KU, Ramakrishnan U. 2009. Why the Indian subcontinent holds the key to global tiger recovery. PLoS Genet. 5, e1000585. 10.1371/journal.pgen.1000585 (doi:10.1371/journal.pgen.1000585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranganathan J, Chan K, Karanth KU, Smith JLD. 2007. Where can tigers persist in the future? A landscape-scale, density-based population model for the Indian subcontinent. Biol. Conserv. 141, 67–77 10.1016/j.biocon.2007.09.003 (doi:10.1016/j.biocon.2007.09.003) [DOI] [Google Scholar]
- 7.Sharma R, Stuckas H, Bhaskar R, Rajput S, Khan I, Goyal SP, Tiedemann R. 2008. mtDNA indicates profound population structure in Indian tiger (Panthera tigris tigris). Conserv. Gen. 10.1007/s10592-008-8568-3 (doi:10.1007/s10592-008-8568-3) [DOI] [Google Scholar]
- 8.Luo SJ, et al. 2004. Phylogeography and genetic ancestry of tigers (Panthera tigris). PLoS Biol. 2, e442. 10.1371/journal.pbio.0020442 (doi:10.1371/journal.pbio.0020442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry P, Miquelle D, Sugimoto T, Mccullough DR, Caccone A, Russello MA. 2009. In situ population structure and ex situ representation of the endangered Amur tiger. Mol. Ecol. 18, 3173–3184 10.1111/j.1365-294X.2009.04266.x (doi:10.1111/j.1365-294X.2009.04266.x) [DOI] [PubMed] [Google Scholar]
- 10.Driscoll CA, Yamaguchi N, Bar-Gal GK, Roca AL, Luo S, Macdonald DW, O'Brien SJ, Brembs B. 2009. Mitochondrial phylogeography illuminates the origin of the extinct Caspian tiger and its relationship to the Amur tiger. PLoS ONE 4, e4125. 10.1371/journal.pone.0004125 (doi:10.1371/journal.pone.0004125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flagstad Ø, Walker CW, Vila` C, Sundqvist AK, Fernholm B, Hufthammer AK, Wiig O, Koyola I, Ellegren H. 2003. Two centuries of the Scandinavian wolf population: patterns of genetic variability and migration during an era of dramatic decline. Mol. Ecol. 12, 869–880 10.1046/j.1365-294X.2003.01784.x (doi:10.1046/j.1365-294X.2003.01784.x) [DOI] [PubMed] [Google Scholar]
- 12.Aspi J, Roininen E, Ruokonen M, Kojola I, Vila C. 2006. Genetic diversity, population structure, effective population size and demographic history of the Finnish wolf population. Mol. Ecol. 15, 1561–1576 10.1111/j.1365-294X.2006.02877.x (doi:10.1111/j.1365-294X.2006.02877.x) [DOI] [PubMed] [Google Scholar]
- 13.O'brien SJ, Wildt DE, Goldman D, Merril CR, Bush M. 1983. The cheetah is depauperate in genetic variation. Science 221, 459–462 10.1126/science.221.4609.459 (doi:10.1126/science.221.4609.459) [DOI] [PubMed] [Google Scholar]
- 14.O'Brien SJ, et al. 1985. Genetic basis for species vulnerability in the cheetah. Science 227, 1428–1434 10.1126/science.2983425 (doi:10.1126/science.2983425) [DOI] [PubMed] [Google Scholar]
- 15.Crooks KR, Sanjayan MA, Doak DF. 1998. New insights on cheetah conservation through demographic modelling. Conserv. Biol. 12, 889–895 10.1046/j.1523-1739.1998.97054.x (doi:10.1046/j.1523-1739.1998.97054.x) [DOI] [Google Scholar]
- 16.Holbrook JD, DeYoung RW, Janecka JE, Tewes ME, Honeycutt JL, Young JH. 2012. Genetic diversity, population structure, and movements of mountain lions (Puma concolor) in Texas. J. Mammal. 93, 989–1000 10.1644/11-MAMM-A-326.2 (doi:10.1644/11-MAMM-A-326.2) [DOI] [Google Scholar]
- 17.Holbrook JD, Deyoung RW, Tewes ME, Young JH. 2012. Demographic history of an elusive carnivore: using museums to inform management. Evol. Appl. 5, 619–628 10.1111/j.1752-4571.2012.00241.x (doi:10.1111/j.1752-4571.2012.00241.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menzies BR, Renfree MB, Heider T, Mayer F, Hildebrandt TB, Pask AJ, Orlando L. 2012. Limited genetic diversity preceded extinction of the Tasmanian tiger. PLoS ONE 7, e35433. 10.1371/journal.pone.0035433 (doi:10.1371/journal.pone.0035433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karanth KK, Nichols JD, Karanth KU, Hines JE, Christensen NL. 2010. The shrinking ark: patterns of mammal extinctions in India. Proc. R. Soc. B 277, 1971–1979 10.1098/rspb.2010.0171 (doi:10.1098/rspb.2010.0171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lande R. 1988. Genetics and demography in biological conservation. Science 241, 1455–1460 10.1126/science.3420403 (doi:10.1126/science.3420403) [DOI] [PubMed] [Google Scholar]
- 21.Lande R. 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 142, 911–927 10.1086/285580 (doi:10.1086/285580) [DOI] [PubMed] [Google Scholar]
- 22.Lacy RC. 1997. Importance of genetic variation to the viability of mammalian populations. J. Mammal. 78, 320–335 10.2307/1382885 (doi:10.2307/1382885) [DOI] [Google Scholar]
- 23.Hensen I, Oberprieler C. 2005. Effects of population size on genetic diversity and seed production in the rare Dictamnus albus (Rutaceae) in central Germany. Conserv. Genet. 6, 63–73 10.1007/s10592-004-7745-6 (doi:10.1007/s10592-004-7745-6) [DOI] [Google Scholar]
- 24.Frankham R. 1996. Relationship of genetic variation to population size in wildlife. Cons. Biol. 10, 1500–1508 10.1046/j.1523-1739.1996.10061500.x (doi:10.1046/j.1523-1739.1996.10061500.x) [DOI] [Google Scholar]
- 25.Frankham R, Lees K, Montgomery ME, England PR, Lowe E, Briscoe DA. 1999. Do population size bottlenecks reduce evolutionary potential? Anim. Conserv. 2, 255–260 10.1111/j.1469-1795.1999.tb00071.x (doi:10.1111/j.1469-1795.1999.tb00071.x) [DOI] [Google Scholar]
- 26.Keller LF, Waller DM. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 1, 230–241 [Google Scholar]
- 27.Storz JF, Ramakrishnan U, Alberts S. 2001. Determinants of effective population size for loci with different modes of inheritance. J. Hered. 92, 497–502 10.1093/jhered/92.6.497 (doi:10.1093/jhered/92.6.497) [DOI] [PubMed] [Google Scholar]
- 28.Chen SF, Jones G, Rossiter S. 2008. Sex biased gene flow and colonization in the Formosan lesser horseshoe bat: inference from nuclear and mitochondrial markers. J. Zool. 274, 207–215 10.1111/j.1469-7998.2007.00391.x (doi:10.1111/j.1469-7998.2007.00391.x) [DOI] [Google Scholar]
- 29.Segelbacher G, Hoglund J, Storch I. 2003. From connectivity to isolation: genetic consequences of population fragmentation in capercaillie across Europe. Mol. Ecol. 12, 1773–1780 10.1046/j.1365-294X.2003.01873.x (doi:10.1046/j.1365-294X.2003.01873.x) [DOI] [PubMed] [Google Scholar]
- 30.Keyghobadi N, Roland J, Matter SF, Strobeck C. 2005. Among- and within-patch components of genetic diversity respond at different rates to habitat fragmentation: an empirical demonstration. Proc. R. Soc. B 272, 553–560 10.1098/rspb.2004.2976 (doi:10.1098/rspb.2004.2976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heller R, Okello JBA, Siegismund H. 2010. Can small wildlife conservancies maintain genetically stable populations of large mammals? Evidence for increased genetic drift in geographically restricted populations of Cape buffalo in East Africa. Mol. Ecol. 19, 1324–1334 10.1111/j.1365-294X.2010.04589.x (doi:10.1111/j.1365-294X.2010.04589.x) [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan U, Hadly EA. 2009. Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Mol. Ecol. 18, 1310–1330 10.1111/j.1365-294X.2009.04092.x (doi:10.1111/j.1365-294X.2009.04092.x) [DOI] [PubMed] [Google Scholar]
- 33.Karanth KU. 2011. India's tiger counts: the long march to reliable science. Econ. Polit. Weekly 18, 22–25 10.1016/S0169-5347(02)02489-8 (doi:10.1016/S0169-5347(02)02489-8) [DOI] [Google Scholar]
- 34.Ministry of Environment and Forests 2011. India tiger estimate 2010. See http://moef.nic.in/downloads/public-information/tiger-brochure.pdf [Google Scholar]
- 35.Segelbacher G, et al. 2010. Applications of landscape genetics in conservation biology: concepts and challenges. Conserv. Genet. 11, 375–385 10.1007/s10592-009-0044-5 (doi:10.1007/s10592-009-0044-5) [DOI] [Google Scholar]
- 36.Mondol S, Karanth KU, Kumar NS, Gopalaswamy AM, Andheria A, Ramakrishnan U. 2009. Evaluation of noninvasive genetic sampling methods for estimating tiger population size. Biol. Conserv. 242, 2350–2360 10.1016/j.biocon.2009.05.014 (doi:10.1016/j.biocon.2009.05.014) [DOI] [Google Scholar]
- 37.Paabo S, et al. 2004. Genetic analyses from ancient DNA. Annu. Rev. Gen. 38, 645–679 10.1146/annurev.genet.37.110801.143214 (doi:10.1146/annurev.genet.37.110801.143214) [DOI] [PubMed] [Google Scholar]
- 38.Wandeler P, Hoeck PE, Keller LF. 2007. Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 10.1016/j.tree.2007.08.017 (doi:10.1016/j.tree.2007.08.017) [DOI] [PubMed] [Google Scholar]
- 39.Van Oosterhout C, Hutchinson B, Wills D, Shipley P. 2004. Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538 10.1111/j.1471-8286.2004.00684.x (doi:10.1111/j.1471-8286.2004.00684.x) [DOI] [Google Scholar]
- 40.Zhan XJ, Zheng XD, Bruford MW, Wei FW, Tao Y. 2010. A new method for quantifying genotyping errors for noninvasive genetic studies . Conserv. Genet. 11, 1567–1571 10.1007/s10592-009-9950-9 (doi:10.1007/s10592-009-9950-9) [DOI] [Google Scholar]
- 41.Kalinowski ST, Wagner AP, Taper ML. 2006. ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes 6, 576–579 10.1111/j.1471-8286.2006.01256.x (doi:10.1111/j.1471-8286.2006.01256.x) [DOI] [Google Scholar]
- 42.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mora MS, Mapelli FJ, Gaggiotti OE, Kittlein MJ, Lessa EP. 2010. Dispersal and population structure at different spatial scales in the subterranean rodent Ctenomys australis. BMC Genet. 11, 9. 10.1186/1471-2156-11-9 (doi:10.1186/1471-2156-11-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinf Online 1, 47. [PMC free article] [PubMed] [Google Scholar]
- 46.Bandelt H-J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 10.1093/oxfordjournals.molbev.a026036 (doi:10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 47.Prost S, Anderson CNK. 2011. TempNet: A method to display statistical parsimony networks for heterochronous DNA sequence data. Meth. Ecol. Evol. 2, 663–667 10.1111/j.2041-210X.2011.00129.x (doi:10.1111/j.2041-210X.2011.00129.x) [DOI] [Google Scholar]
- 48.Marjoram P, Tavare S. 2006. Modern computational approaches for analyzing molecular genetic variation data. Nat. Rev. Genet. 7, 759–770 10.1038/nrg1961 (doi:10.1038/nrg1961) [DOI] [PubMed] [Google Scholar]
- 49.Rodrigo AG, Felsenstein J. 1999. Coalescent approaches to HIV-1 population genetics. In The evolution of HIV (ed. Crandall KA.), pp. 233–272 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 50.Ramakrishnan U, Hadly EA, Mountain JL. 2005. Detecting past population bottlenecks using temporal genetic data. Mol. Ecol. 14, 2915–2922 10.1111/j.1365-294X.2005.02586.x (doi:10.1111/j.1365-294X.2005.02586.x) [DOI] [PubMed] [Google Scholar]
- 51.Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA. 2005. Serial SimCoal: a population genetic model for data from multiple populations and points in time. Bioinformatics 21, 1733–1734 10.1093/bioinformatics/bti154 (doi:10.1093/bioinformatics/bti154) [DOI] [PubMed] [Google Scholar]
- 52.Smith JLD. 1993. The role of dispersal in structuring the Chitwan tiger population. Behaviour 124, 165–195 10.1163/156853993X00560 (doi:10.1163/156853993X00560) [DOI] [Google Scholar]
- 53.Mundy NI, Unitt P, Woodruff DS. 1997. Skin from feet of museum specimens as a non-destructive source of DNA for avian genotyping. Auk 114, 126–129 10.2307/4089075 (doi:10.2307/4089075) [DOI] [Google Scholar]
- 54.Nyström V, Angerbjörn A, Dalén L. 2006. Genetic consequences of a demographic bottleneck in the Scandinavian arctic fox. Oikos 114, 84–94 10.1111/j.2006.0030-1299.14701.x (doi:10.1111/j.2006.0030-1299.14701.x) [DOI] [Google Scholar]
- 55.Tucker JM, Schwartz MK, Truex RL, Pilgrim KL, Allendorf FW. 2012. Historical and contemporary DNA indicate Fisher decline and isolation occurred prior to the European settlement of California. PLoS ONE 7, e52803. 10.1371/journal.pone.0052803 (doi:10.1371/journal.pone.0052803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pertoldi C, Hansen MM, Loeschoke V, Madsen AB, Jacobsen L, Baagoe H. 2001. Genetic consequences of population decline in the European otter (Lutra lutra): an assessment of microsatellite DNA variation in Danish otters from 1883 to 1993. Proc. R. Soc. Lond. B 268, 1775–1781 10.1098/rspb.2001.1762 (doi:10.1098/rspb.2001.1762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenbaum HC, Egan MG, Clapham PJ, Brownell JRL, Malik S, Brown MW, White BN, Walsh P, DeSalle R. 2000. Utility of North American right whale museum specimens for assessing changes in genetic diversity. Conserv. Biol. 14, 1837–1842 10.1046/j.1523-1739.2000.99310.x (doi:10.1046/j.1523-1739.2000.99310.x) [DOI] [PubMed] [Google Scholar]
- 58.Patil N, Kumar NS, Gopalaswamy AM, Karanth KU. 2011. Dispersing tiger makes a point. Oryx 45, 472–475 10.1017/S0030605311001591 (doi:10.1017/S0030605311001591) [DOI] [Google Scholar]
- 59.Valdiosera CE, et al. 2008. Surprising migration and population size dynamics in ancient Iberian brown bears (Ursus arctos). Proc. Natl Acad. Sci. USA 105, 5123–5128 10.1073/pnas.0712223105 (doi:10.1073/pnas.0712223105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364 10.1038/nature10574 (doi:10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Cruz B, Godoy JA, Negro JJ. 2007. Population fragmentation leads to spatial and temporal genetic structure in the endangered Spanish imperial eagle. Mol. Ecol. 16, 477–486 10.1111/j.1365-294X.2007.03147.x (doi:10.1111/j.1365-294X.2007.03147.x) [DOI] [PubMed] [Google Scholar]
- 62.Gadgil M, Guha R. 1992. This fissured land. Delhi, India: Oxford University Press [Google Scholar]
- 63.Kothari A. 1993. Conservation of biological diversity in India. New Delhi, India: Indian Institute of Public Administration [Google Scholar]
- 64.Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. 1998. Inbreeding and extinction in a butterfly metapopulation. Nature 392, 491–494 10.1038/33136 (doi:10.1038/33136) [DOI] [Google Scholar]
- 65.Convention on Biological Diversity. 2010. Strategic plan for biodiversity 2011–2020, including Aichi Biodiversity Targets. See www.cbd.int/sp/default.shtml .
- 66.Panthera. 2013. Tigers forever. See www.panthera.org/programs/tiger/tigers-forever .
- 67.Wikramanayake E, et al. 2011. A landscape-based conservation strategy to double the wild tiger population. Conserv. Lett. 4, 219. 10.1111/j.1755-263X.2010.00162.x (doi:10.1111/j.1755-263X.2010.00162.x) [DOI] [Google Scholar]
- 68.Crooks KR, Burdett CL, Theobald DM, Rondinini C, Boitani L. 2011. Global patterns of fragmentation and connectivity of mammalian carnivore habitat. Phil. Trans. R. Soc. B 366, 2642–2651 10.1098/rstb.2011.0120 (doi:10.1098/rstb.2011.0120) [DOI] [PMC free article] [PubMed] [Google Scholar]