Abstract
Swine populations are known to be an important source of new human strains of influenza A, including those responsible for global pandemics. Yet our knowledge of the epidemiology of influenza in swine is dismayingly poor, as highlighted by the emergence of the 2009 pandemic strain and the paucity of data describing its origins. Here, we analyse a unique dataset arising from surveillance of swine influenza at a Hong Kong abattoir from 1998 to 2010. We introduce a state–space model that estimates disease exposure histories by joint inference from multiple modes of surveillance, integrating both virological and serological data. We find that an observed decrease in virus isolation rates is not due to a reduction in the regional prevalence of influenza. Instead, a more likely explanation is increased infection of swine in production farms, creating greater immunity to disease early in life. Consistent with this, we find that the weekly risk of exposure on farms equals or exceeds the exposure risk during transport to slaughter. We discuss potential causes for these patterns, including competition between influenza strains and shifts in the Chinese pork industry, and suggest opportunities to improve knowledge and reduce prevalence of influenza in the region.
Keywords: influenza, swine, disease ecology, infectious disease surveillance, zoonosis, state–space model
1. Introduction
The 2009 pandemic highlighted the inadequate state of influenza surveillance in swine populations around the world. Mounting evidence indicates that pigs play a central role in the emergence of many pandemic influenza strains [1,2], and it is now known that influenza A (H1N1)pdm09 was created by reassortment of Eurasian avian-like (EA) and triple reassortant (TR) strains [3,4]. Phylogenetic reconstruction indicates that elements of the pandemic strain had been co-circulating undetected in swine for more than 10 years [3,4]. This finding underscores the need to study the epidemiology of influenza in swine populations at regional scales, and to understand the factors driving transmission dynamics. Achieving this goal requires long-term, systematic data from structured surveillance activities, but unfortunately almost no such datasets exist. One conspicuous exception is a dataset describing long-term virological and serological surveillance at a Hong Kong abattoir from 1998 to 2010, drawn from source farms across southern and southeastern China. These data have already yielded important insights about the co-circulation of influenza strains, strain replacement and viral reassortment in swine populations [5,6]. However, fundamental questions have yet to be addressed. What epidemiological processes give rise to the observed rates of virus isolation? Where in the swine production system, from farms to abattoirs, does most transmission of influenza occur? How are transmission dynamics affected by strain interactions and the ongoing industrialization of swine production? Answering these questions would guide efforts to control influenza in swine, and would shed light on mechanisms underpinning future pandemic risks. Yet inferring these patterns of disease transmission, a challenge in the best of circumstances, is particularly difficult here, because the available dataset was collected at a single location, just prior to slaughter, preventing the direct study of transmission at different points in the swine production system.
Here, we introduce a statistical inference framework that uses a Bayesian state–space model (BSSM) to integrate simultaneously observed data streams to infer hidden patterns of transmission [7,8]. This approach analyses virological and serological data together, informed by studies of the progression of these clinical signs of infection in individual pigs [6,9,10]. Experimental infection studies demonstrate that a naive pig infected with influenza can be expected to shed virus for 5–7 days. We define the window of recent exposure as the week prior to the sampling date; successful virus isolation indicates exposure during this period. The shedding period ends as antibodies to the infecting influenza strain increase; seropositivity reflects older exposure, which we define as infection more than one week prior to the sampling date. Virological and serological data are typically analysed separately, even when presented in the same study, but joint analysis draws additional insights from these data streams by exploiting their biological connection. Because samples are drawn simultaneously at fixed points in space and time, the differing time scales of the clinical signs provide insights into when transmission occurred, and, given knowledge of host movements, where transmission occurred.
China has long been recognized as a priority for influenza surveillance owing to the high densities of humans, swine and fowl in the region [11]. Currently, China produces and consumes almost 50 per cent of the world's pork, requiring an enormous swine population [12,13]. Over the past decade, swine production systems in China have responded to rising pork demand from a human population growing in size and affluence, and have also been impacted by rising feed costs, natural disasters and significant disease outbreaks [12–14]. Resulting changes in the farming facilities, transport systems and patterns of trade will be reflected in the long-term influenza surveillance dataset collected at the abattoir, as will any epidemiological differences among circulating strains. In the week before slaughter, pigs are exposed to increased population mixing in trucks and holding pens, as well as ‘transport stress’ that may affect immunity to infection [15,16]. During their earlier life on the farms, infection risk will be influenced by animal density, facility design and biosecurity [17]. We investigate how these risks of exposure to influenza have changed over the past decade in Chinese swine, by using our inference framework to estimate older exposures (corresponding to time spent on the farm) and recent exposures (corresponding to transport and holding just before slaughter).
2. Model and results
(a). Framework for analysis of surveillance data
The dataset consists of regular samples from active surveillance at a Hong Kong abattoir from May 1998 through January 2010. Tracheal and nasal swabs were collected fortnightly from swine at slaughter, and monthly viral isolation rates, as well as corresponding strain typing, are available for almost all of the surveillance period. Sera were collected from swine at slaughter, and monthly seroprevalence data are available for the years 2000, 2004 and 2009 (see §4 for detailed information about the abattoir, viral isolation, strain typing and serology processing methods). Unavailable data are treated as missing values and imputed using the model.
The BSSM provides a probabilistic description of hidden process dynamics and their relation to the observed virus isolation and serological data (figure 1; see also the electronic supplementary material). Following convention [7,8], we divide the state–space model into (i) a data model, which provides the connection between model parameters and data in the form of the likelihood, (ii) a process model, which describes hidden dynamics and their relation to observed data, and (iii) a parameter model. Observed data include the number of strain-specific virus isolations per month, which we denote by ni(t) for strain i in month t. Monthly counts of seropositive samples (defined as having a titre of greater than or equal to 1 : 40 to any test antigen) are designated as ns(t). Serological data were not divided into strain-specific responses owing to ambiguities in assignment. This use of serological data assumes that seropositivity reflects protection from contemporary viruses (see electronic supplementary material, figure S1; see discussion in §3b). The data model treats the virus isolation samples for each month as multinomial, 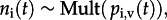 where the probability of isolating strain i, pi,v(t), is the product of the probability of virus isolation pv(t) and the probability of finding type i given virus isolation, pi|v(t). Serological data are binomially distributed,
where the probability of isolating strain i, pi,v(t), is the product of the probability of virus isolation pv(t) and the probability of finding type i given virus isolation, pi|v(t). Serological data are binomially distributed, 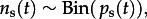 where ps(t) is the probability of being seropositive in month t.
where ps(t) is the probability of being seropositive in month t.
Figure 1.
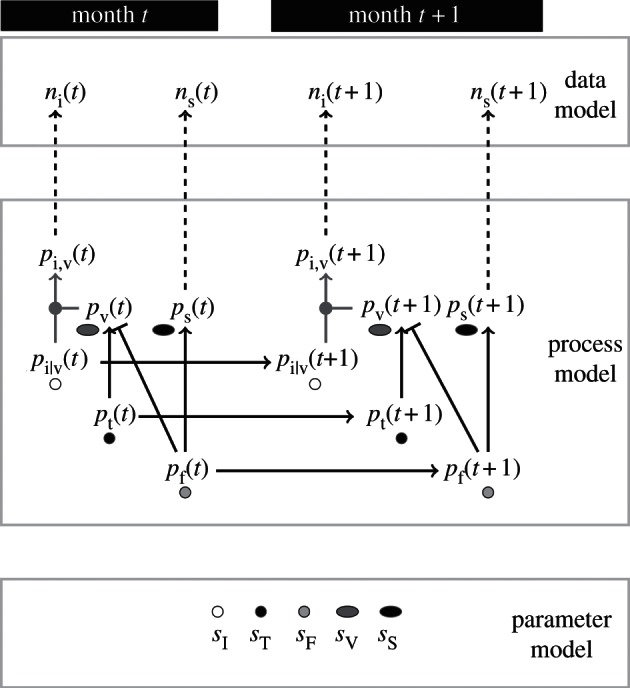
A diagram of the Bayesian state–space model consisting of data, process and parameter models (see main text for definition of all parameters). Dashed lines connect observations in the data model to relevant probabilities for each month t. Solid black lines show relations between variables that make up the process model. Arrows indicate probabilities that covary positively, whereas a bar indicates suppression of one probability by another using a factor of one minus the relevant probability. Solid grey arrows indicate a deterministic relationship as in pi,v(t) = pv(t)pi|v(t). The parameter model includes constant scale factors that represent the size of uncertainties by allowing different levels of variance in relations between parameters, as well as initial conditions pi|v(0), pt(0) and pf(0) (not shown in the figure).
The next layer of the process model connects the monthly data to the unobserved transmission dynamics described by the total probabilities of exposure to influenza on the farm or during transport to slaughter, denoted pf(t) and pt(t), respectively. We assume that previous exposure on the farm provides immunity to circulating strains of influenza, so that virus isolation is possible only when a naive animal is exposed within the week before sampling. This is reflected by the expectation 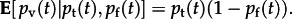 Seropositivity is directly related to the total probability of exposure on the farm,
Seropositivity is directly related to the total probability of exposure on the farm, 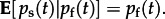 Using this model structure, both virus isolation and serostatus affect the inferred values of pf(t) and pt(t). The model includes scale parameters that control the variance in these statistical relations (see the electronic supplementary material). These parameters allow for uncertainty owing to unspecified sources such as assay accuracy, individual variation in animal response to exposure and month-to-month changes in the farms supplying the abattoir. All scale parameters are inferred along with the rest of the model parameters using Markov chain Monte Carlo (MCMC) sampling of the BSSM (see the electronic supplementary material, table S1).
Using this model structure, both virus isolation and serostatus affect the inferred values of pf(t) and pt(t). The model includes scale parameters that control the variance in these statistical relations (see the electronic supplementary material). These parameters allow for uncertainty owing to unspecified sources such as assay accuracy, individual variation in animal response to exposure and month-to-month changes in the farms supplying the abattoir. All scale parameters are inferred along with the rest of the model parameters using Markov chain Monte Carlo (MCMC) sampling of the BSSM (see the electronic supplementary material, table S1).
The final element of the state–space model describes the temporal variation of the exposure parameters pf(t), pt(t) and pi|v(t). Each of these parameters is assumed to vary as a Markov process, with expected value for the next month equal to the value in the current month. The month-to-month variance is determined by a scale parameter that is inferred from the data, as discussed earlier. This construct provides a constraint on the values of the hidden process dynamics, and potentially smooths outliers that arise from erratic sampling of a broad geographical region. It also allows us to impute missing data that make up part of our inference problem.
(b). Virus isolation and seropositivity
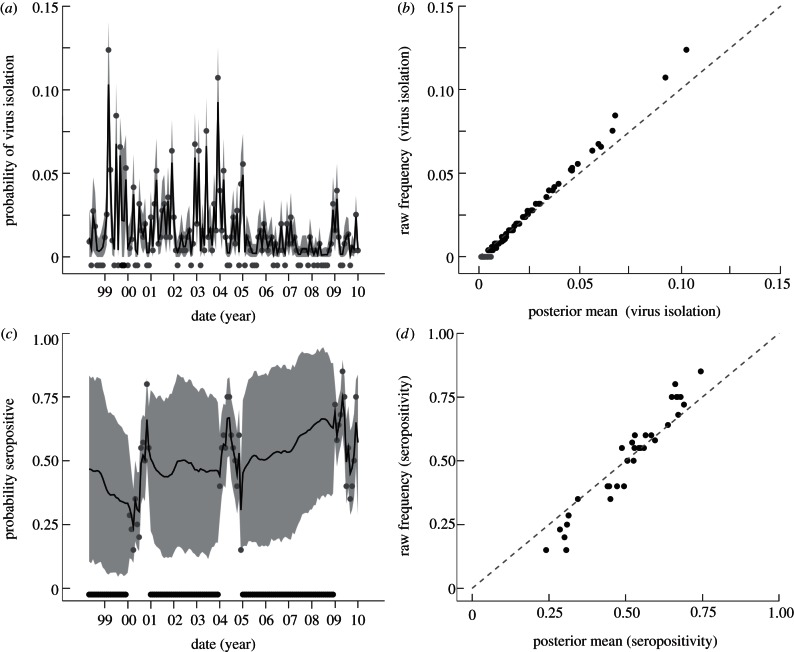
The posterior mean for the probability of virus isolation, without regard for strain type, follows the trends exhibited in the raw data (figure 2a). As noted in earlier work, virus isolation rates appear to decline over the past decade, with a protracted period of low isolation rates from 2005 to 2008 [6]. The posterior mean exhibits less variation than the raw data owing to the process constraints in our state–space model and the month-to-month variation in the raw data (figure 2b). Importantly, the raw frequencies are included in the 95 per cent high probability density (HPD) region for all months where virus was isolated (figure 2a). Only in months where there were no virus isolations, resulting in a raw frequency equal to zero, does this value fall outside the HPD. In these cases, the posterior mean for the probability of virus isolation is estimated to be in the order of 10–3. This small probability is consistent with the lack of successful virus isolation given the number of samples taken. Instances where the raw frequency falls outside of the HPD for this reason are indicated by grey symbols in figure 2b.
Figure 2.
Observed and imputed patterns of virological and serological surveillance for influenza in Chinese swine. Monthly probabilities of (a) viral isolation pv(t) and (c) seropositivity ps(t) are provided, as well as a comparison of raw frequency and posterior mean estimates for (b) viral isolation and (d) seropositivity. In (a,c), grey dots show raw frequency estimates of the monthly probabilities; estimates equal to zero are shown below the zero-line for clarity. Missing data are indicated by black dots below the zero-line. The solid black lines show the posterior means of the probabilities inferred from joint data analysis and the grey bands show the region of 95% HPD. In (b,d), the dashed line shows equality between raw frequency and posterior mean estimates; months where raw frequencies fall outside the HPD region are indicated by grey dots. With the exception of months with no virus isolation, raw frequencies fall within the HPD region, indicated by black dots.
The inferred probability of seropositivity shows a contrasting trend, rising slightly over the course of the decade (figure 2c). When there is a missing data point, the state–space model imputes the value of ps(t) using constraints imposed by the model and information from the virological data. Again, the raw frequencies for available seropositivity data are more variable than the posterior mean probabilities (figure 2d). However, the breadth of the 95 per cent HPD region includes all raw frequency estimates and reflects the uncertainty generated by the presence or absence of data through time.
(c). Inference of exposure probabilities
The monthly patterns of virus isolation and seropositivity provide a noisy and indirect picture of the unobserved transmission dynamics. Our joint analysis enables us to infer the probabilities of exposure to influenza during different phases of the pigs’ lives prior to slaughter. Estimates of exposure during transport show that this probability has been approximately constant for the decade of surveillance (figure 3a). The pattern of lifetime exposure on the swine farms is more dynamic, with the posterior mean showing an increasing trend (figure 3b). Sample sizes and missing data limit the precision with which this trend can be estimated, but the region of 95 per cent HPD does not admit a constant probability over the decade. As expected, the 2009 serology data are crucial to discerning the increase in farm exposure; if these data are removed from the BSSM fit, then the increase is not evident but is still consistent with the uncertainty in the refit model (see the electronic supplementary material, figure S2).
Figure 3.
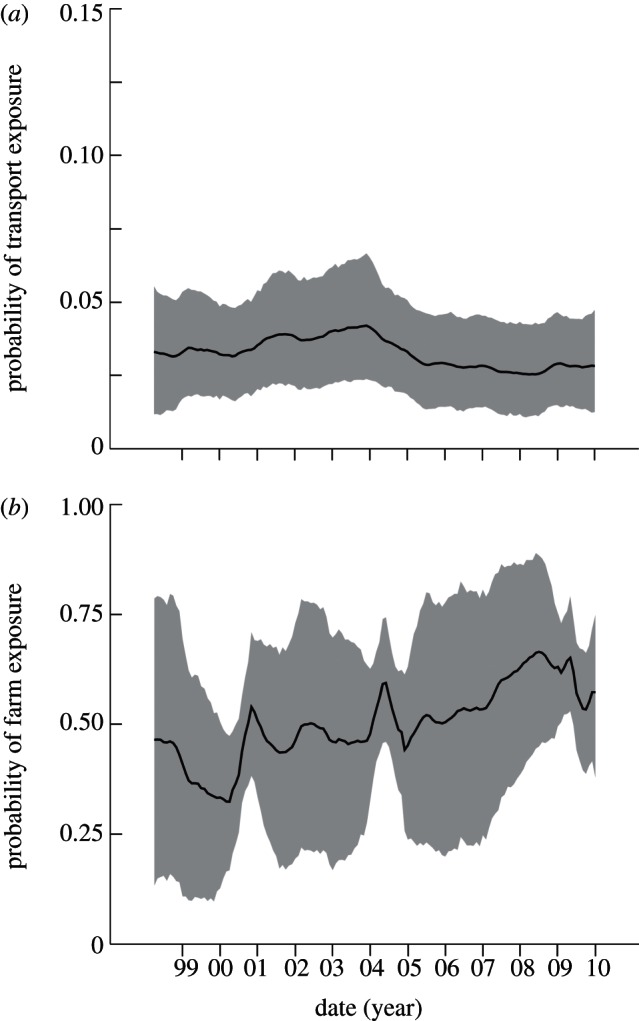
Total probabilities of exposure during (a) transport and holding in the week before slaughter (pt(t)), and (b) earlier life on the swine farm or production facility (pf(t)). Inferred values for each month are plotted. The solid black lines show the posterior means of the probabilities and the grey bands show the 95% HPD regions. No data are shown because these transmission dynamics are not directly observed.
It is conspicuous that the virus isolation rate does not mirror the probability of exposure during transport (figures 2a and 3a). Successful virus isolation at the abattoir occurs only when exposure during transport coincides with a lack of earlier exposure on the farm. A monthly estimate of this combined probability, given by 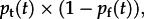 shows that the posterior mean for recent exposure of naive animals has decreased (see the electronic supplementary material, figure S3). Although this trend is not significant at the 95 per cent level, owing to the multiplication of uncertainties, it is consistent with the observed decrease in virus isolation rates over the decade. It is possible that pigs systematically spent longer in transport during periods of lower virus isolation than they did during periods of higher virus isolation. However, because of economic pressures this confounding variation in transportation time seems unlikely to have occurred. In any case, the inferred force of infection (FOI) during transport is insufficient to create this effect (i.e. the susceptible pool would not be exhausted within any reasonable period of transport, so new infections would continue to occur).
shows that the posterior mean for recent exposure of naive animals has decreased (see the electronic supplementary material, figure S3). Although this trend is not significant at the 95 per cent level, owing to the multiplication of uncertainties, it is consistent with the observed decrease in virus isolation rates over the decade. It is possible that pigs systematically spent longer in transport during periods of lower virus isolation than they did during periods of higher virus isolation. However, because of economic pressures this confounding variation in transportation time seems unlikely to have occurred. In any case, the inferred force of infection (FOI) during transport is insufficient to create this effect (i.e. the susceptible pool would not be exhausted within any reasonable period of transport, so new infections would continue to occur).
(d). Force of infection for swine influenza
The inference of the probabilities of exposure in transport and on the farm enables us to estimate the weekly FOI in each setting (figure 4). For a given exposure time Te and probability pe, the FOI can be estimated using 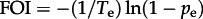 . This relation is obtained by a simple rearrangement of the more familiar form:
. This relation is obtained by a simple rearrangement of the more familiar form: 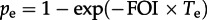 (see [18] for derivation). Because pt(t) describes exposures in the week before slaughter, the weekly FOI during transport is virtually identical to figure 3a and has remained approximately constant at roughly 0.03 per week over the decade (Te = 1 week and pe = pt(t) for each month). Estimation of the FOI for pigs on the farm depends on their age at slaughter and the duration of protection by maternally derived antibodies, which may prevent infection and development of haemagglutination inhibition (HI) titres [10]. Given typical ranges of 16–24 weeks of age at slaughter and 4–8 weeks of protection by maternal antibodies, we used possible exposure durations of 8, 16 and 24 weeks to estimate a range of possible FOIs (Te = 8, 16, 24 weeks and pe = pf(t) for each month). Vaccination of swine against influenza is not reported for Chinese farms and should not affect exposure durations in these calculations [13,19].
(see [18] for derivation). Because pt(t) describes exposures in the week before slaughter, the weekly FOI during transport is virtually identical to figure 3a and has remained approximately constant at roughly 0.03 per week over the decade (Te = 1 week and pe = pt(t) for each month). Estimation of the FOI for pigs on the farm depends on their age at slaughter and the duration of protection by maternally derived antibodies, which may prevent infection and development of haemagglutination inhibition (HI) titres [10]. Given typical ranges of 16–24 weeks of age at slaughter and 4–8 weeks of protection by maternal antibodies, we used possible exposure durations of 8, 16 and 24 weeks to estimate a range of possible FOIs (Te = 8, 16, 24 weeks and pe = pf(t) for each month). Vaccination of swine against influenza is not reported for Chinese farms and should not affect exposure durations in these calculations [13,19].
Figure 4.
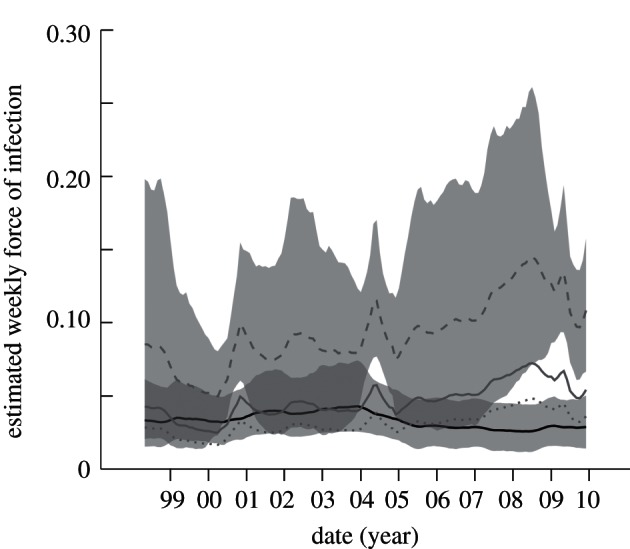
The weekly force of infection (FOI) during transport and holding before slaughter, and during earlier life on swine farms. The solid black line provides the posterior mean for transport FOI with 95% HPD indicated by the lower grey region. Three estimates of the posterior mean for FOI on farms assume a total exposure time of 8 (grey dashed line; 95% HPD shown as upper grey region), 16 (grey solid line) or 24 weeks (grey dotted line) to represent different scenarios for age at slaughter and the duration of protection by maternally derived antibodies.
Estimates of the weekly FOI on swine farms and during transport to slaughter are shown in figure 4. The 95 per cent HPD region is provided for the transport FOI and one farm FOI to give an indication of the scale of uncertainties; HPD regions for the farm curves are larger due to the greater uncertainty in seropositivity (figure 2c). For most assumptions about exposure duration on the farm, the weekly FOI is statistically indistinguishable between farm and transport phases over the decade. However, when the period of exposure on farms is assumed to last just eight weeks, the weekly FOI is significantly higher on farms than in transport from 2007 onwards. The robust pattern across all scenarios is that, despite the stress and increased mixing associated with transport and holding before slaughter, the weekly risk of exposure to influenza appears to be equal or higher on the production farms.
(e). Strain replacement
During the decade of surveillance, one of the major features observed was an replacement of the classic swine H1N1 (CS) with an EA strain as the dominant viral strain in southern China (previously described by Vijaykrishna et al. [6]). Experimental infection studies demonstrated a competitive advantage of EA over CS strains, as well as limited immunity in the population, suggesting a biological mechanism for this replacement [6]. The state–space model provides insights into the dynamics of strain replacement by inferring monthly, strain-specific viral isolation rates (see the electronic supplementary material, figures S4 and S5).
The emergence of a fitter influenza strain could account for the rise in on-farm exposure revealed by our analysis. However, the EA strain appeared in 2000–2001 and became dominant in 2003–2005, whereas the rise in exposure on farms occurred mostly from 2004 to 2008 (figure 3b). This suggests that the strain replacement was not the immediate cause of the changing epidemiological pattern. Instead, we propose that large-scale changes in the swine industry are the more likely explanation, and indeed may have played an important role in the strain dynamics via introduction of novel strains and emergence of reassortant strains. The geographical sourcing of swine for the sampled abattoir shifted significantly over the decade [6]. From 2000 to 2007, 15–20 per cent of the pigs were farmed near Hong Kong, with the remainder imported from several provinces in China; by 2008, the local proportion had fallen to 5 per cent. The percentage of swine sourced from nearby Guangdong province increased from 31.1 in 2003 to 51.8 in 2010. These shifts in industry-wide patterns, along with accompanying changes in conditions at production facilities, should be considered as potential causes of the observed shifts in epidemiology and strain frequencies, alongside the biological mechanisms already proposed.
3. Discussion
The long-term, active surveillance data considered here provide a rare glimpse into the transmission dynamics of influenza in swine populations over the decade leading up to the 2009 pandemic. While the pandemic did not arise in China, the co-circulation of multiple lineages and genetic reassortment events observed in the course of this surveillance provide insight into likely virus transmission dynamics leading up to pandemic emergence in the Americas [5,6]. The joint analysis of virological and serological data allows us to infer time-varying risks of influenza exposure across production stages, despite the fact that samples were taken at a single location. To untangle this information, we used knowledge gained from experimental infection studies of influenza in swine to define recent and older time frames for infection, indicated by the potential for virus shedding and detection of antibodies, respectively. Using this connection, we separated exposure risks during transportation and holding the week before sampling from exposures occurring during earlier life on swine farms in southern and southeastern China.
The model presented here provides a statistical framework to link experimental infection studies with surveillance data streams to quantify epidemiological processes, both observed and hidden. We can infer that influenza prevalence in the region has not decreased despite an apparent reduction in virus isolation rates (see figure 2; electronic supplementary material, figure S1). Instead, we find that the FOI during transport in the week before slaughter has remained remarkably constant at approximately 3 per cent per week over the decade. By contrast, the cumulative probability of exposure on the farm has increased over this period. We conclude that the observed reduction in virus isolation rates reflects the rise in exposure early in life, which results in immunity that prevents infection in the week before sampling. This finding has two important implications: (i) a perceived reduction in influenza prevalence at slaughter is not due to improved biosecurity or other preventative measures; and (ii) the overall prevalence of influenza in the swine population has not decreased, and hence the risk of spillover to susceptible human and bird populations is constant or growing. Our findings show that the risk, as measured by the FOI, is at least as high on swine farms as it is in transport and holding settings. Further, influenza on the farms appears to have increased over the decade, although the statistical significance of this trend would be strengthened with additional data.
(a). Epidemiology of swine influenza
The increasing scale of the pork industry in China provides the backdrop for the findings reported here. Over the past decade, a series of market-driven and natural perturbations (including rising feed costs, the 2008 Sichuan earthquake and outbreaks of non-influenza disease) have led to major disruptions in the Chinese swine population [12–14,19,20]. The resulting economic impacts motivated a push towards production in larger, more regulated facilities in an effort to stabilize supply, limit price increases and minimize disease outbreaks [13,19]. The growing size of swine farming facilities has increased the potential for sustained influenza transmission, particularly given the apparent lack of vaccination against influenza. The rise in on-farm exposure reported here could be attributed to these factors, and related changes linked to industrialization [21].
Our study reveals long-term patterns of influenza circulation under these changing conditions, inferred from data collected from a random subset of animals passing through the abattoir in Hong Kong. We estimate the rate of virus isolation to range from 0.1 to 10 per cent, with a mean of 1.6 per cent over the decade (see figure 2; electronic supplementary material, figure S3), whereas seropositivity ranges from 24 to 74 per cent, with a mean of 50 per cent (figure 2). Many of the high rates of isolation occur in the early 2000s, with relatively low seroprevalence; this pattern shifts to lower isolation rates and higher seropositivity late in the decade. Here, we place our observations in the context of shorter-term epidemiological studies conducted elsewhere in the world.
Samples collected in 1997–1998, during fortnightly surveillance of swine at an abattoir in the United States, showed a mean virus isolation rate of 2.2 per cent, with monthly peaks as high as 16 per cent [22] (see also [23]). The mean seroprevalence, using abattoir samples as well as sera sent to the Wisconsin Animal Health Laboratory for pseudorabies virus testing, was 27.7 per cent for a strain of CS H1 influenza, with monthly values ranging from 10 to 60 per cent. These observations are consistent with our data from Hong Kong, both on average and with regard to the wide variation in monthly rates; intriguingly, the quantitative match is closer during the 1998–2004 period, when our data were also dominated by the CS strain.
A survey of Spanish swine farms during 2008–2009 found a seroprevalence of 75.4 per cent to at least one of three strains of influenza (H1N1, H1N2 and H3N2) [17]. These results were broken down by age, with seropositivity among younger fattening pigs (11–20 weeks of age, a closer match to animals included in our study) measured to be 53.1 per cent, while that among older cows was 89.9 per cent. Similar values for sows of varying ages were obtained for pig-dense European countries during a 2002–2003 survey with 85.2 per cent seroprevalence in Germany and 94 per cent in Belgium [24]. A recent survey of influenza prevalence in other Asian countries showed mean levels of virus isolation and seropositivity similar to our results for Hong Kong [21,25]. We conclude that the epidemiology of swine influenza in southern China is broadly consistent with available data from other industrialized countries worldwide.
Our findings show that efforts to limit the prevalence of influenza A in Chinese swine populations could usefully be targeted to improving practices in production facilities. Such control efforts could be informed by recently identified risk factors for influenza on Spanish swine farms: (i) high replacement rates that introduce new susceptibles or new subclinically infected animals; (ii) lack of solid separations to prevent transmission between pens; and (iii) uncontrolled entrance to farms [17]. In contrast to the situation on farms, we found that the probability of influenza exposure during the week before slaughter has not risen or fallen as a result of industry growth over the past decade. This suggests that there are unrealized opportunities to reduce spread during transport and limit potential for mixing of geographically separated strains. For example, transmission between pens holding pigs from different farms has been observed at a quarantine point in Shenzhen and provides an opportunity for improvement (see §4). However, the validity of comparisons between this system and Spanish farms remains unclear, and the economics of swine production raise challenges in motivating changes in practice for influenza control.
(b). Impact of model assumptions
The results of our analysis arise from the probabilistic relations that make up the BSSM. The strongest assumption we made is that antibodies above a certain titre are protective against challenge by any contemporary influenza strain. In our analysis, exposure probabilities are inferred by considering virus isolation and serostatus data independent of the strain type or detailed serological profile. Patterns of serological cross-reactivity and immunity in influenza are complicated, even when the test antigen is known and experimental infection is done under laboratory conditions [6,9,26]. As a result it is not possible to definitively connect a given serological profile to prior infection by one or more known strains. Under field conditions, this problem is amplified by the fact that the observed serological profiles might be caused by one or more viral strains that are not used as n strains in HI panels, and have not been characterized by experimental infection studies.
A significant proportion of swine sampled in our dataset had titres greater than or equal to 1 : 40 to many, or all, strains tested (see the electronic supplementary material, figure S1). This pattern could arise from cross-reactivity among the strains or from multiple infections by different strains. However, the age of pigs at the time of slaughter ranges from four to six months, and it is usually assumed that only a single infection is possible during this time (though this assumption may not hold in all cases [27,28]). As a result, we argue that the assumption of immunity to contemporary strains is appropriate, and that strain-specific analyses of our data are not possible given current knowledge of influenza serology. If the assumption is too strong, and cross-immunity among these strains is not complete, then our results would overestimate the probability of recent exposure (because we would have underestimated the true proportion of swine that are susceptible to infection). To explore this possibility, we re-ran the BSSM assuming 50 per cent protection for seropositive animals. This model, which could account for second infections by different strains or for limited protection from previous exposure, reduces the posterior mean probability of recent exposure to 2.4–2.5 per cent (from approx. 3%) over the decade. The difference in these estimates, given the related uncertainties, does not substantially change the conclusions presented earlier. Further progress on this problem will require experimental infection studies, including multiple exposures and longitudinal sampling of serum from individual animals, as well as development of methods that elucidate the connections between immunity and profiles derived from field serological data [9,29].
The model includes constant scale parameters that determine the variances in association between elements of the process model (see figure 1; electronic supplementary material). In table S1 of the electronic supplementary material, we show the inferred values, which vary inversely with the variance, along with regions of 95 per cent HPD. The scales for virus isolation and serostatus are low, and allow for considerable variance in the relation to the hidden probabilities of exposure. This uncertainty can be attributed to month-to-month variation in source farms, assay sensitivity and individual variation, as well as other unknown factors. The estimated scale parameters for hidden process dynamics are larger, allowing for less variation and resulting in smoothed dynamics, hence limiting inordinate weight to data from any single month.
(c). Broader applications of multi-stream surveillance analysis
We have introduced a mechanistic state–space model to infer unobserved transmission histories from joint analysis of virological and serological surveillance data. Our model provides robust inference of epidemiological parameters, smoothing anomalous data and imputing missing data based on mechanistic assumptions. Multiple modes of surveillance are commonly conducted in parallel, and other studies could benefit from similar joint analyses of these data streams [25,30–32]. In practice, sampling design is very important when applying this idea, because all data streams need to be sampled at the same time points, from the same population. Ideally, this sampling would occur at regular intervals from randomly selected animals, providing an unbiased picture of disease dynamics in the focal population.
This approach can be adapted to any disease for which the time course of multiple clinical outcomes, including diagnostic measures or signs of disease, have been characterized by experimental infection studies or close observation of natural infections [33]. Given different time scales associated with different clinical outcomes, such analyses would open broader opportunities to reconstruct the histories of exposure prior to sampling. Further quantitative precision could be obtained by expanding experimental infection studies to characterize individual-specific variation, and estimate the sensitivity and specificity of different assays. In principle, the kinetics of some assays could be characterized so that quantitative results could be used, instead of binary outcomes, to derive more specific information about the time since infection. Further theoretical and statistical tools to integrate these classes of data will also need to be developed. Combined with a carefully designed surveillance plan, these approaches could enable the inference of disease transmission histories in systems where host individuals can only be sampled once, an all-too-common limitation in settings ranging from livestock inspections to wildlife studies.
4. Material and methods
(a). Surveillance data
Systematic influenza surveillance was conducted between May 1998 and January 2010 at an abattoir in Hong Kong where roughly 4500 pigs per day are processed (current data are available online at http://www.fehd.gov.hk/english/sh/index.html). Tracheal or nasal swabs (109–512 per month) and serum samples (20–50 per month) were collected from a random sample of slaughtered swine [6] (see the electronic supplementary material, figure S1 for raw data). Sampled swine were between the ages of four and six months, and were sourced from provinces throughout southern and southeastern China [6].
Information that connects source farm, transport time, transport method and disease status of individual animals is unavailable. We have no information about the size of farms that provide pigs to this abattoir, but McOrist et al. [19] provide data on the size of pig farms in China and their growth between 2008 and 2011. Pigs are transported in trains and lorries, with the transport period varying depending on the starting farm location (see supplementary information in [6] for information on source farms). The pigs are held at a border quarantine station in Shenzhen, just across the border from Hong Kong, to check health status before transport to Hong Kong. Pigs from each source farm are held in separate pens, but they may be next to a pen with animals from a different farm and there is evidence of transmission among pens. After the quarantine station, each consignment is shipped separately to the abattoir in Hong Kong.
(b). Virus isolation, strain typing and seroprevalence
Swab materials were inoculated into 9- to 10-day-old embryonated chicken eggs and Madin Darby canine kidney cells; virus isolates were identified and subtyped by HI assays as previously described [6]. In addition, strains included in the study were fully sequenced and typed based on phylogenetic relationships of all gene segments [4]. Antibody prevalence against the major swine influenza lineages was characterized using the HI assays with six representative viruses—A/Sw/HK/4167/1999 (CS), A/Sw/HK/1110/2006 (TR), A/CA/4/2009 (Pand), A/SW/HK/1304/2003 (CS), A/Sw/HK/NS29/2009 (EA) and A/SW/HK/1559/2008 (EA)—starting at a dilution of 1 : 10. Titres greater than or equal to 1 : 40 were taken as seropositive [6].
(c). Bayesian state-space model and Markov chain Monte Carlo sampling
The BSSM is a hierarchical model with time-dependent associations as developed in earlier studies [7,8]. Complete details of the statistical framework are provided in the electronic supplementary material. Computational elements of the MCMC inference process use the Metropolis–Hastings algorithm with tuning during burn-in to sample from the posterior distribution, as implemented by the PyMC Python package [34].
Acknowledgements
This work was funded by a grant from the National Center for Foreign Animal and Zoonotic Disease Defense, a Department of Homeland Security Science and Technology Center of Excellence. Data collection was supported by the National Institute of Allergy and Infectious Diseases (contract no. HHSN266200700005C). J.O.L.-S. and S.R. are supported by the RAPIDD programme of the Science and Technology Directorate of the Department of Homeland Security and NIH Fogarty International Center; J.O.L.-S. is grateful for support from the De Logi Chair in Biological Sciences. We thank the Institute for Digital Research and Education at UCLA for use of the hoffman2 cluster.
References
- 1.Thacker E, Janke B. 2008. Swine influenza virus: zoonotic potential and vaccination strategies for the control of avian and swine influenzas. J. Infect. Dis. 197, S19–S24 10.1086/524988 (doi:10.1086/524988) [DOI] [PubMed] [Google Scholar]
- 2.Smith GJD, Bahl J, Vijaykrishna D, Zhang J, Poon LLM, Chen H, Webster RG, Peiris JSM, Guan Y. 2009. Dating the emergence of pandemic influenza viruses. Proc. Natl Acad. Sci. USA 106, 11 709–11 712 10.1073/pnas.0904991106 (doi:10.1073/pnas.0904991106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garten RJ, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325, 197–201 10.1126/science.1176225 (doi:10.1126/science.1176225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith GJD, et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125 10.1038/nature08182 (doi:10.1038/nature08182) [DOI] [PubMed] [Google Scholar]
- 5.Peiris JSM, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. 2001. Cocirculation of avian H9N2 and contemporary ‘human’ H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75, 9679–9686 10.1128/JVI.75.20.9679-9686.2001 (doi:10.1128/JVI.75.20.9679-9686.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijaykrishna D, et al. 2011. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473, 519–522 10.1038/nature10004 (doi:10.1038/nature10004) [DOI] [PubMed] [Google Scholar]
- 7.West M, Harrison PJ, Migon HS. 1985. Dynamic generalized linear models and Bayesian forecasting. J. Am. Stat. Assoc. 80, 73–83 10.1080/01621459.1985.10477131 (doi:10.1080/01621459.1985.10477131) [DOI] [Google Scholar]
- 8.Clark JS, Bjornstad ON. 2004. Population time series: process variability, observation errors, missing values, lags, and hidden states. Ecology 85, 3140–3150 10.1890/03-0520 (doi:10.1890/03-0520) [DOI] [Google Scholar]
- 9.Van Reeth K, Labarque G, Pensaert M. 2006. Serological profiles after consecutive experimental infections of pigs with European H1N1, H3N2, and H1N2 swine influenza viruses. Viral Immunol. 19, 373–382 10.1089/vim.2006.19.373 (doi:10.1089/vim.2006.19.373) [DOI] [PubMed] [Google Scholar]
- 10.Markowska-Daniel I, Pomorska-Mól M, Pejsak Z. 2011. The influence of age and maternal antibodies on the postvaccinal response against swine influenza viruses in pigs. Vet. Immunol. Immunopathol. 142, 81–86 10.1016/j.vetimm.2011.03.019 (doi:10.1016/j.vetimm.2011.03.019) [DOI] [PubMed] [Google Scholar]
- 11.Shortridge KF, Stuart-Harris CH. 1982. An influenza epicentre? Lancet 320, 812–813 10.1016/S0140-6736(82)92693-9 (doi:10.1016/S0140-6736(82)92693-9) [DOI] [PubMed] [Google Scholar]
- 12.Wang R. 2006. China: pork powerhouse of the world. Adv. Pork Prod. 17, 33–46 [Google Scholar]
- 13.Gale F, Marti D, Hu D. 2012. China's volatile pork industry. USDA outlook report no. LDPM-21101. See http://www.ers.usda.gov
- 14.Zhang J, Beckman C. 2008. People's Republic of China, agricultural situation: livestock and products. USDA GAIN report no. CH8078. See http://gain.fas.usda.gov
- 15.Hurd HS, McKean JD, Griffith RW, Wesley IV, Rostagno MH. 2002. Salmonella enterica infections in market swine with and without transport and holding. Appl. Environ. Microbiol. 68, 2376–2381 10.1128/AEM.68.5.2376-2381.2002 (doi:10.1128/AEM.68.5.2376-2381.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams JL, Minton JE, Patterson JA, Marchant Forde J, Eicher SD. 2008. Lairage during transport of eighteen-kilogram pigs has an impact on innate immunity and commensal bacteria diversity in the intestines. J. Anim. Sci. 86, 1232–1244 10.2527/jas.2007-0592 (doi:10.2527/jas.2007-0592) [DOI] [PubMed] [Google Scholar]
- 17.Simon-Grifé M, Martín-Valls GE, Vilar MJ, García-Bocanegra I, Mora M, Martín M, Mateu E, Casal J. 2011. Seroprevalence and risk factors of swine influenza in Spain. Vet. Microbiol. 149, 56–63 10.1016/j.vetmic.2010.10.015 (doi:10.1016/j.vetmic.2010.10.015) [DOI] [PubMed] [Google Scholar]
- 18.Anderson RM, May RM. 1991. Infectious diseases in humans. Oxford, UK: Oxford University Press [Google Scholar]
- 19.McOrist S, Khampee K, Guo A. 2011. Modern pig farming in the People's Republic of China: growth and veterinary challenges. Rev. Sci. Tech. 30, 961–968 [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. 2007. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the mid-eastern region of China. Vet. J. 174, 577–584 10.1016/j.tvjl.2007.07.032 (doi:10.1016/j.tvjl.2007.07.032) [DOI] [PubMed] [Google Scholar]
- 21.Trevennec K, Cowling BJ, Peyre M, Baudon E, Martineau GP, Roger F. 2011. Swine influenza surveillance in east and southeast Asia: a systematic review. Anim. Health Res. Rev. 12, 213–223 10.1017/S1466252311000181 (doi:10.1017/S1466252311000181) [DOI] [PubMed] [Google Scholar]
- 22.Olsen CW, Carey S, Hinshaw L, Karasin AI. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145, 1399–1419 10.1007/s007050070098 (doi:10.1007/s007050070098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webby R. 2012. H5N1: How to track a flu virus. Nature 483, 535–536 10.1038/483535a (doi:10.1038/483535a) [DOI] [PubMed] [Google Scholar]
- 24.Van Reeth K, et al. 2008. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002–2003. Influenza Other Resp. Viruses 2, 99–105 10.1111/j.1750-2659.2008.00043.x (doi:10.1111/j.1750-2659.2008.00043.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W, Wei M-T, Tong Y, Tang F, Zhang L, Fang L, Yang H, Cao W-C. 2011. Seroprevalence and genetic characteristics of five subtypes of influenza A viruses in the Chinese pig population: a pooled data analysis. Vet. J. 187, 200–206 10.1016/j.tvjl.2009.10.026 (doi:10.1016/j.tvjl.2009.10.026) [DOI] [PubMed] [Google Scholar]
- 26.Park AW, Daly JM, Lewis NS, Smith DJ, Wood JLN, Grenfell BT. 2009. Quantifying the impact of immune escape on transmission dynamics of influenza. Science 326, 726–728 10.1126/science.1175980 (doi:10.1126/science.1175980) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeffen WLA, Heinen PP, Bianchi ATJ, Hunneman WA, Verheijden JHM. 2003. Effect of maternally derived antibodies on the clinical signs and immune response in pigs after primary and secondary infection with an influenza H1N1 virus. Vet. Immunol. Immunopathol. 92, 23–35 10.1016/S0165-2427(03)00019-9 (doi:10.1016/S0165-2427(03)00019-9) [DOI] [PubMed] [Google Scholar]
- 28.Van Reeth K, Brown I, Essen S, Pensaert M. 2004. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 103, 115–124 10.1016/j.virusres.2004.02.023 (doi:10.1016/j.virusres.2004.02.023) [DOI] [PubMed] [Google Scholar]
- 29.Lorusso A, et al. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J. Gen. Virol. 92, 919–930 10.1099/vir.0.027557-0 (doi:10.1099/vir.0.027557-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown IH. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74, 29–46 10.1016/S0378-1135(00)00164-4 (doi:10.1016/S0378-1135(00)00164-4) [DOI] [PubMed] [Google Scholar]
- 31.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 103, 67–73 10.1016/j.virusres.2004.02.015 (doi:10.1016/j.virusres.2004.02.015) [DOI] [PubMed] [Google Scholar]
- 32.Brockwell-Staats C, Webster RG, Webby RJ. 2009. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza Other Resp. Viruses 3, 207–213 10.1111/j.1750-2659.2009.00096.x (doi:10.1111/j.1750-2659.2009.00096.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charleston B, et al. 2011. Relationship between clinical signs and transmission of an infectious disease and the implications for control. Science 332, 726–729 10.1126/science.1199884 (doi:10.1126/science.1199884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patil A, Huard D, Fonnesbeck CJ. 2010. PyMC: Bayesian stochastic modelling in Python. J. Stat. Softw. 35, 1–81 [PMC free article] [PubMed] [Google Scholar]