Abstract
In nature, biological systems gradually evolve through complex, algorithmic processes involving mutation and differential selection. Evolution has optimized biological macromolecules for a variety of functions to provide a comparative advantage. However, nature does not optimize molecules for use in human-made devices, as it would gain no survival advantage in such cooperation. Recent advancements in genetic engineering, most notably directed evolution, have allowed for the stepwise manipulation of the properties of living organisms, promoting the expansion of protein-based devices in nanotechnology. In this review, we highlight the use of directed evolution to optimize photoactive proteins, with an emphasis on bacteriorhodopsin (BR), for device applications. BR, a highly stable light-activated proton pump, has shown great promise in three-dimensional optical memories, real-time holographic processors and artificial retinas.
Keywords: bionanotechnology, bacteriorhodopsin, photoactive proteins, retinal proteins, directed evolution, protein-based devices
1. Introduction
Scientists and engineers are continuously seeking new ways to harness the resources of the natural world to improve and advance the fields of nanotechnology and nanobiotechnology. The desire of humans to seek help from nature follows naturally from a recognition that nature has been operating at the nanoscale for billions of years. By using multi-disciplinary approaches, researchers are starting to gain control over the intrinsic properties of naturally occurring materials, significantly changing the way that scientists approach and solve complex problems [1]. The integration and unique interplay between protein engineering and biology can transform a number of scientific disciplines, as well as biologically inspired technologies.
Developments in nanoscience are characterized by the manipulation of biological and non-biological matter at the nanoscale regime. Starting with the early proposals of Richard Feynman in 1979 to the more recent innovations in materials science, the goals of nanoscience remain the same: to develop useful architectures with at least one dimension less than 100 nm [2]. Nanoelectronics, originally at the forefront of nanotechnology, centred on electrical, mechanical and materials engineering, and began with the continual minimization of the transistor feature sizes on integrated circuits, a trend described by Moore's Law [3]. Today, the field of nanoelectronics encompasses the processing, transmission, storage and retrieval of information at the molecular level. As a consequence of the increasing demand for miniaturization, scientists are forced to search for alternatives to the more conventional and fundamental methods of research to meet the demands of modern technology [4,5].
Nanotechnological paradigms can be divided into two contrasting efforts: a top-down approach and a bottom-up approach [6–8]. Although commonly applied to information processing and knowledge ordering in software and computer development, these approaches can also be used in any scientific field. In a top-down approach, nanoscale objects are made through the processing of larger objects, such as silicon crystals or graphite [6]. The bottom-up approach focuses on the measurement, prediction and construction of nanostructures from smaller building blocks, such as atoms and molecules. A classic and sophisticated analogy of nanotechnology can be found in nature, where biological systems have carried out the direct assembly of the structures necessary to sustain life [9,10]. Biologists and engineers are working to imitate this natural capability to produce small clusters of atoms that can then self-assemble into more complex structures. Using a bottom-up approach, scientists can generate smaller structures than is possible via a top-down approach and can do so with greater efficiency and less material waste [10–12].
As the emphasis of nanotechnology switches from mechanical and materials engineering to a more biologically influenced field, researchers have started to redefine the definition of nanotechnology to stress the development of novel chemicals, as well as new materials and devices to study biological systems. Using a scale set up by nature, one made up of cells, organelles, proteins and molecules, researchers have started to broaden the scope of traditional nanotechnology to include the natural sciences and biology, a term loosely defined as nanobiotechnology [13,14]. Nanobiotechnology refers to the application of nanotechnology to target problems directly relevant to biology [15]. The closely related term, bionanotechnology, is often used interchangeably with nanobiotechnology; however, when distinction is needed, bionanotechnology is used to refer to the general overlap of biology and nanotechnology, and exploits natural biomimetic systems, such as DNA nanotechnology and self-assembly systems. Bionanotechnology also encompasses the study of how nanotechnology can be directed through a better understanding of biology and the integration of biological ideas and motifs into improving nanotechnologies [16]. Conversely, nanobiotechnology highlights the advances in and applications of nanotechnology to biological problems [15]. Some of the target applications of these technologies include drug delivery [17–19], gene therapy [20] and regenerative medicine [21,22].
This review explores some of the strategies for tailoring the electronic and photochemical properties of proteins for performance in nanotechnological devices. In particular, we will focus on directed evolution, as a method to genetically engineer microbial rhodopsins and photoactive proteins for application in non-native environments. Bacteriorhodopsin (BR), a photoactive protein found in the salt marsh archaeon, Halobacterium salinarum, is a leading candidate for application in devices, including three-dimensional optical memories, real-time holographic media, photovoltaic cells and artificial retinas [23–28].
2. Directed evolution
Directed evolution has emerged as a powerful tool for optimizing biological macromolecules to function outside of the native organism. By using directed evolution, researchers have been able to create novel proteins and biomolecules with properties not normally found in nature [29–32]. Combined with high-throughput screening methods, directed evolution has been successful in generating proteins to function as protein-based tools and devices, including biosensors, therapeutics and biocatalysts [33–38].
Evolution yields proteins that have been optimized to function in a particular biological system. Unfortunately, nature finds no comparative advantage in optimizing proteins for devices, and thus applied technologies using proteins and other biological molecules have only found limited success. The most that one can hope for is that nature has optimized a protein for a biological function that corresponds in some fashion to the intended application of the researcher. In nature, the slow accumulation of genetic mutations and adaptations are manifested in changes in the structure and function of biological macromolecules. The accumulation of genetic mutations can be positive, negative or have no effect on the organism. Beneficial mutations often enable the host organism to overcome a selective environmental pressure, while lethal mutations lead to cell death. Most protein structures are so complex that predicting how to alter the structure and function of the protein for a specific function is impossible [39].
Directed evolution (otherwise referred to as laboratory evolution, molecular evolution, evolution in vitro or evolution in a test tube) can be used to optimize the inherent properties of biological macromolecules via iterative rounds of diversification and differential selection [40]. Classically, investigators have used directed evolution to enhance and alter specific traits, such as thermal and chemical stability, which are not apparent in the parent molecule [41]. Directed evolution employs both chemical biology and combinatorial chemistry via multiple rounds of mutagenesis to improve or alter the efficiency and specificity of proteins, with the ultimate goal of generating new molecules with useful properties for applied technologies.
In nature, environmental pressures, such as temperature and availability of sunlight, often affect the overall fitness of the organism. Applying selective environmental pressures in the laboratory can alter the behaviour or fitness of an organism, thereby leading to atypical cell phenotypes or proteins, which thrive outside of the context of biology. However, in the research laboratory this type of directed evolution often requires the development of new technologies, which have the potential to improve the design process, such as novel screening systems and computer programs [33]. With rapid advancements in synthetic and computational tools, the functional space of protein engineering is only limited by the ingenuity of the protein scientist. Innovations in nanobiotechnology are providing new opportunities for the directed evolution of proteins.
Three main steps characterize directed evolution: diversification, selection and amplification. The first step involves the diversification of a mutant library and is accomplished using mutagenesis to create a large library of gene variants. High-throughput screening and various other selection methods are then used to test for the presence of mutants with a particular property, trait or characteristic. Finally, amplification of the mutant library is used to allow for further characterization of the mutant DNA. Using a combinatorial library, a diverse pool of mutants are generated randomly in a test tube, and then screened for a particular property or phenotype through expansive screening and selection methods [38].
With the emergence of biotechnology and nanotechnology, investigators are looking to optimize photoactive proteins to function in optoelectronic, photovoltaic and biomedical devices. This review will explore some of the advancements in genetic engineering and describe some of the advantages of combining biology and nanotechnology. More specifically, we will describe the application of directed evolution and other genetic engineering strategies on the optimization of photoactive proteins, using BR as the model system.
3. Bacteriorhodopsin
BR is one of the most studied proteins for use in bioelectronic and biomimetic devices [42,43]. Located in the outer membrane of the salt marsh archaeon, H. salinarum, this 27-kDa protein functions as a light-transducing proton pump. When environmental oxygen becomes scarce and drops below a level necessary to sustain oxidative phosphorylation, H. salinarum expresses BR to generate energy via photosynthesis [44]. The protein, comprised of seven transmembrane alpha helices, is arranged in a two-dimensional hexagonal lattice of trimers in the lipid bilayer, often referred to as the purple membrane [45–47]. The inherent stability of the semicrystalline matrix allows the protein to survive and remain functionally active in the most extreme environments [48–51].
The primary light-absorbing moiety of BR is an all-trans retinal chromophore that is bound to the protein via a protonated Schiff base linkage to lysine-216 [43]. When the retinal chromophore absorbs a photon, a photocycle is initiated (figure 1). Through a complex process involving a series of spectrally discrete intermediates, BR translocates a proton from the intracellular to extracellular milieu. This photocycle is used to generate a proton gradient that is used to drive membrane-bound ATPase to synthesize adenosine triphosphate (ATP) under anaerobic conditions [55–57]. After decades of research, the photocyclic mechanism of proton pumping of BR is well characterized and several models have been generated to help describe the complex nature of the BR photocycle [58–63].
Figure 1.
The photocycle of light-adapted BR (bR) (top) and the three chromophore configurations represented in the main and branched photocycles. The main photocycle pumps a proton with Asp-96 (D96) and Asp-85 (D85) serving as the source and destination of the Schiff base proton, respectively [52]. The Schiff base linkage is protonated in all intermediates of the main photocycle except the M1 and M2 states. The branched photocycle is accessed via excitation of the O state (all-trans, 15-anti) to form the P state (9-cis, 15-anti) [53]. The P state spontaneously forms the Q state, which contains a hydrolysed 9-cis retinal captured within the binding site [54].
The main photocycle of BR is made up of a series of transient photochemical and conformational intermediates labelled the bR, K, L, M, N and O photostates (figure 1) [64]. Each BR photocycle lasts approximately 10–15 ms and results in the net translocation of a single proton across the membrane [65]. The primary photochemical event, which initiates the photocycle, operates with a quantum efficiency of 0.65 [66,67]. This high quantum efficiency is identical to that of the visual photopigment, rhodopsin, which is found in the photoreceptor cells of the retina [68,69].
One of the unique features of the BR photocycle is the branched photocycle [25,70–72]. During the classical photocycle of BR, the last and most red-shifted intermediate, designated as the O state, contains an all-trans configuration of the retinal chromophore (figure 1). Unless the O state is illuminated with a second red photon, it will thermally decay back to the resting state (bR). However, if the retinal chromophore absorbs a photon of red light during the O state, it will photoisomerize to form a 9-cis photoproduct that is unstable in the binding pocket [70]. This 9-cis chromophore is characteristic of the branched photocycle and is comprised of the P and Q states (figure 1). The P state contains a bound 9-cis chromophore, and was originally defined as having an absorption maximum (λmax) of approximately 490 nm [73]; however, further research shows that the P state is actually a mixture of two photostates, described as P1(λmax = 525 nm) and P2 (λmax = 445 nm). These photostates undergo a dynamic equilibrium after the absorption of a photon [54], and because 9-cis retinal is unstable in the binding pocket, the chromophore thermally decays to form the Q state (λmax = 380 nm) [54,70,74,75]. The Q photoproduct contains a hydrolysed 9-cis retinal chromophore trapped inside the BR binding pocket (figure 1) [70]. With an activation barrier of approximately 190 kJ mol−1 for the spontaneous formation of the bR resting state, the Q state is the only thermally stable photoproduct of the protein with a lifetime of approximately 7–12 years at ambient temperature [25]. Reversion of the Q state to bR is possible via absorption of a blue photon (360–400 nm) by the 9-cis chromophore [75].
The biological origin of the branched photocycle remains uncertain, although it is hypothesized that this photochemistry is an unwanted photochemical artefact minimized by years of evolution [54]. Additionally the observation that a significant fraction of random mutants have a lower efficiency of formation suggests that nature has adjusted the efficiency of the formation of the branched photocycle rather than eliminate the formation of this state [41]. Possible explanations for this branching reaction are that the P and Q states serve as a way to minimize DNA photodamage [54]. Additionally, it is possible that at high light intensity, the protein converts part of the photoactive protein to the Q state to avoid pH damage owing to the generation of too large of a pH gradient [76]. Recall that formation of the branched photocycle and the Q photoproduct prevents the protein from carrying out the biological function of proton pumping. Although the origin of the Q state remains unclear, the presence of a branched photocycle provides an excellent template for biomimetic and biophotonic device applications [77].
The absorption of a photon by the retinal chromophore of BR is one of the most efficient and stable photochemical reactions found in nature [78,79]. The photochemical stability of BR, commonly referred to as the cyclicity, is a measure of the number of times that the protein can be converted between two states before 37 per cent (1/e) of the material denatures. At ambient temperatures, the cyclicity of BR has been found to exceed 106 [25]. The thermal stability of BR, which is commonly cited to exceed 80°C [49–51], has also made BR a remarkable candidate for device applications. With excellent photochemical efficiency and stability, coupled with the ability to withstand high levels of light flux and chemical stress from a self-induced pH gradient, BR has been a leading candidate for application in numerous biophotonic, biomimetic and biomedical applications.
4. Biophotonic device applications of bacteriorhodopsin
Shortly following the discovery of BR by Oesterhelt and Stoeckenius in 1971 [55], Soviet scientists first recognized the potential of using this photoactive material in protein-based computing and processing applications. At the time, interest in pursuing bioelectronic devices was tied to military development and international competition towards designing new nanobiotechnologies, which had first elicited a holographic processor based on BR thin films through the project, Biochrome [80,81]. In the subsequent decades of research, BR has emerged as one of the most rigorously investigated biomaterial for use in electronic applications, culminating in the development of numerous prototypes of devices that include Fourier-transform holographic associative processors [25,42,82–84], three-dimensional optical memories [25,82,85,86], biosensors [87,88], photovoltaic cells [89–91] and protein-based retinal prostheses [28,92]. Recent advances in genetic engineering and directed evolution have further invigorated the pursuit of optimizing these protein-based devices, thereby increasing the potential of introducing a commercial BR-based technology in the near future [38,41,76]. Here, we briefly highlight select device applications of BR, with a focus on those that incorporate the Q state photoproduct into the device architectures.
4.1. Holographic associative processors
Associative memories and processors differ significantly from the serial memories that dominate modern computing technologies. These memories mimic the function of the human brain, in which associative recall is used to search through the entire memory bank simultaneously to match an input data block [93,94]. In application, the memory would return the corresponding data block that matches the input criteria, or would return the closest match if no exact copy is identified. Because of this functional paradigm, associative processors are often considered a target design for the development of true artificial intelligence [95]. Fourier-transform holographic optical loops, which were first proposed by Paek and Psaltis, have long been considered an optimal architecture necessary to realize the implementation of this memory class [84,96–98]. Bunkin et al. [80] had first proposed thin films of BR as a viable medium for the technology owing to the inherent holographic and photochromic properties of the photoactive protein. During the photocycle of BR, the biomaterial undergoes a series of spectrally distinct photointermediates, which are coupled with a significant transient modulation of the refractive index [64,99]. There are two options for the design of BR-based holographic memories: (i) a real-time processor based on the transient M state intermediate and (ii) a long-term memory based on the Q state photoproduct. We describe both schemes below.
Of particular interest for real-time holography is the formation and reversion of the M state, which is the photointermediate that displays the largest hypsochromic shift in λmax from the bR state (570 nm → 410 nm) and contains a reasonable time constant (approx. 10 ms) for real-time holographic processing [100]. Recall that the primary photochemical event of BR is followed by a series of secondary dark reactions, corresponding to the formation and decay of the K, L, M, N and O states (figure 1) [64]. In addition to this thermal decay pathway, the bR state is capable of being regenerated directly from the M state by the absorption of light with a wavelength of approximately 410 nm [101]. This ability describes the behaviour of a photochromic pair, in which photochromism is defined as the reversible conversion between two chemical forms with different absorption maxima via photonic stimuli. Both photochemical reactions (i.e., bR → M and M → bR) exhibit unprecedented quantum efficiencies (approx. 0.65) among alternative media options for holographic associative processors [66,67]. In application, the exploitation of these photochromic properties circumvents the inherent limitation of depending on a finite photochemical intermediate. The lifetime of the M state, however, is prohibitively short in the native protein. Through the use of genetic engineering and chemical modification, Hampp [26] and Hampp et al. [102,103] have enhanced the diffraction efficiency of protein thin films and increased the M state lifetime up to the order of seconds, thereby enabling success in designing a real-time holographic interferometer. Target residues for site-directed mutagenesis involve those that are directly involved with proton translocation during the BR photocycle [26,102]. Using the directed evolution methodology outlined below, researchers have been able to identify BR mutants that further enhance M state characteristics, thus improving the prospect of optimizing the protein for this application [104].
Despite past successes in developing a Fourier-transform holographic associative processor based on BR, the transient nature of the M state precludes the design of a long-term data storage device based on this scheme. The branched photocycle and the Q state have been considered for use in holographic processing architectures because of a similarly blue-shifted λmax (approx. 380 nm) and a temporal stability that nullifies the transient photochemistry of BR [25,75]. A recent review describes the incorporation of the Q state into holographic memory architectures by using chemical and mutational modification to generate the blue membrane form of the protein [105]. The photochromic pair that involves the Q state differs from the bR/M state system in two ways. First, the Q state is stable for months or years in contrast to the transient BR photocycle [25]. Secondly, the quantum efficiencies of the forward and reverse reactions to the branched photocycle are extremely small [71,106]. This feature, however, is easily circumvented by modern high-energy diode lasers and can even be advantageous for long-term storage applications. In the following section, we describe an alternate device architecture, which implements a volumetric approach to harnessing the sequential two-photon stimulation requirement of Q state formation in the purple membrane.
4.2. Three-dimensional optical memories
Within an increasingly technology-driven society, consumer demand for faster computing speeds and smaller device interfaces have led to a reevaluation of the efficacy of modern computers. One promising architectural motif, among a diverse collection of forthcoming molecular and biomolecular device structures, involves a three-dimensional (or volumetric) optical memory design [25,85]. BR is capable of serving as an efficient data storage medium when the purple membrane is fixed in a polymer-based suspension [25]. In combination with the bR resting state, the Q state photoproduct allows for the assignment of binary bit 0 and binary bit 1, respectively, within the volumetric matrix [25,85,86]. Because the branched photocycle is accessed using a sequential two-photon pulse sequence, the BR medium is placed within the beam path of orthogonally positioned diode lasers to selectively drive data blocks within the volume into the Q state. Unique volumetric patterns within the medium can then be used to store and retrieve information in a computing system based on differential absorptivity. The photochemistry of BR, the architectural scheme, and the nanoscale feature size facilitate a unique potential to write, read and erase data with high speed and a high storage capacity. In the following paragraphs, we briefly outline the specific routines to write, read and erase data; however, a more thorough review of this technology can be found in [25,82,85,86,107].
Both the writing and the reading processes begin by using a green paging laser (approx. 570 nm), which initiates the photocycle within a thin region of the BR medium. To write data within this page, a second laser pulse (approx. 640 nm) must be directed orthogonally to the page within the window of time determined by the BR photocycle lifetime. Ideally, the conversion to the branched photocycle via the second pulse should be accomplished once the O state reaches a maximum concentration. Information is spatially encoded in the incident red writing laser using a spatial light modulator (SLM), which converts the protein into the Q state in the desired volumetric pattern. The writing function is an example of an optical AND gate, in which data are written if and only if the two required photochemical conditions are satisfied.
The reading operation begins with an identical first condition, in which the green paging laser activates the BR photocycle (figure 1) within a select region of the medium. In this case, the page being probed contains some volume that is already converted into the Q state and transparent to the incident laser pulses. The page is then exposed to red light during the O state, however, the intensity of the red writing laser is much lower and is not modulated using a SLM to encode information. A charge-coupled detector (CCD) array is then capable of reading the presence and absence of the Q state. In other words, the CCDs measure the pattern of binary bit 0 and binary bit 1 within the page.
To erase data written within the BR medium in the described architecture, the Q state must be reverted back to the bR resting state by using blue light (approx. 380 nm). The Q → bR reversion can be achieved using two methods. First, a blue laser of coherent light can be used to selectively convert individual volumes within the BR medium. Second, the entire BR medium can be erased simultaneously by globally exposing the medium to blue incoherent light. Both options are available in the design of these volumetric optical memories. In general, the hardware and optics necessary to realize this architecture are widely available, and a number of prototypes have been developed that successfully implement the design [25,82]. Methods in directed evolution have now allowed Q state formation and reversion efficiencies to catch up to levels imposed by the hardware capabilities [38,41,76,104].
4.3. Protein-based retinal implants
BR has remained at the forefront of protein-based device development for the past several decades, however, this trend has only recently permeated into the field of biomedical devices and prosthetics [28,92]. More specifically, BR is currently being investigated as the photoactive element in a protein-based retinal prosthesis for patients suffering from retinal degenerative diseases such as age-related macular degeneration and retinitis pigmentosa. The proposed retinal implant mimics the light-absorbing capabilities of the homologous native visual pigments and generates a unidirectional proton gradient that is sufficient to stimulate the remaining neural network of the damaged retina [92]. The implant consists of a multilayered BR thin film that is anchored between two ion permeable and biologically inert membrane surfaces. The multiple layers of BR are adsorbed onto the membrane surface using an alternating layer-by-layer approach, with a polycation used to electrostatically interact with each applied layer of BR [108]. The oriented, multilayer thin film is necessary to absorb sufficient incident light and drive the proton-pumping mechanism that is required to generate a neural response [92]. In application, the Q state photoproduct can be used to further manipulate the active surface of the thin film. Retinal degenerative diseases are inherently difficult to treat with prosthetics because of extreme variability of pathologies, which are unique to the individual patient. Driving select areas of the film to the Q state can turn down or turn off extraneous pixels, which allows for the selective tuning of the active surface area of the implant that is required to generate meaningful vision.
5. Optimization strategies
The optimization of BR for a synthetic environment requires enhancing properties of the protein, which are either irrelevant or non-essential for biological function. Although researchers are actively studying the structural motifs and intramolecular interactions responsible for the photophysical properties of BR, many of these interactions remain unknown. Despite access to rigorous theoretical models and fast computers, neither software nor hardware can adequately predict which mutations will provide the desired properties. More expansive experimental methods and high-throughput screening techniques must be developed in order to make protein-based devices commercially viable. Consequently, devices, which are based on the Q state of BR, have been met with limited success because the native protein does not efficiently access the branched photocycle.
Advancements in genetic engineering techniques have changed the way scientists approach the field of nanotechnology. The redesign of proteins for use in synthetic environments has gained momentum owing to improvements in site-directed mutagenesis, site-specific saturation mutagenesis, semi-random mutagenesis, random mutagenesis and now directed evolution. Perhaps the most versatile method for improving protein performance is through the use of site-directed mutagenesis [109,110]. Site-directed mutagenesis is a targeted technique, which has been used to characterize structure–function relationships proteins from a variety of organisms. This technique is used to introduce a specific mutation at a point of interest on the primary sequence of the protein. Mutations are introduced using oligonucleotides that are complementary to a part of single-stranded DNA, but contain an internal mismatch, which directs the mutation. Site-directed mutagenesis is a powerful method when one knows what sites on the primary protein sequence to target. However, this technique is often inefficient when large protein sequences must be probed. Saturation mutagenesis is similar to site-directed mutagenesis, but has a higher efficiency of generating more mutations. With saturation mutagenesis, many mutations are generated via the saturation of a key residue or residues at a target locus on the gene of interest. In the case of site-specific saturation mutagenesis, doped oligonucleotides are used to introduce all 20 amino acids at the site of interest [111].
As a result of the limited efficiency of site-directed and saturation mutagenesis, many researchers have turned to more global techniques, implementing high-throughput screening and selection methods to optimize the genetic landscape of biological molecules. Semi-random mutagenesis and random mutagenesis have the capability to produce a large number of indiscriminate variants via chemical manipulation, ultraviolet light, error prone PCR, or the use of doped oligonucleotides [111]. These methods offer a selective advantage over classical mutagenesis strategies because they allow for a greater mutational landscape of the protein to be explored. In the case of mutagenesis for device applications, semi-random and random mutagenesis permit greater genetic diversification of the protein, allowing the researcher to narrow in on the diverse interactions, which contribute to a desired phenotype. An efficient and effective screening method must be available in order to make the generation and screening of such a large library of mutants time and cost effective.
For decades, researchers have been using semi-random or saturation mutagenesis to identify particular residues that play an integral role in protein function. Mutagenesis of this type can best be envisioned in terms of a mutational landscape, where the optimization of a single characteristic, such as the photophysical properties of the protein, is represented by fairly localized, irregular changes (figure 2b) [38]. More fluid properties, such as thermal stability, involve larger portions of the protein and a variety of complementary mutations, thus leading to a mutational landscape that is continuous and slowly varying (figure 2a). Unfortunately, for scientists trying to optimize proteins for function outside of the biological context of the organism, the challenge lies in predicting a priori which amino acid will generate the desired properties for the intended application.
Figure 2.
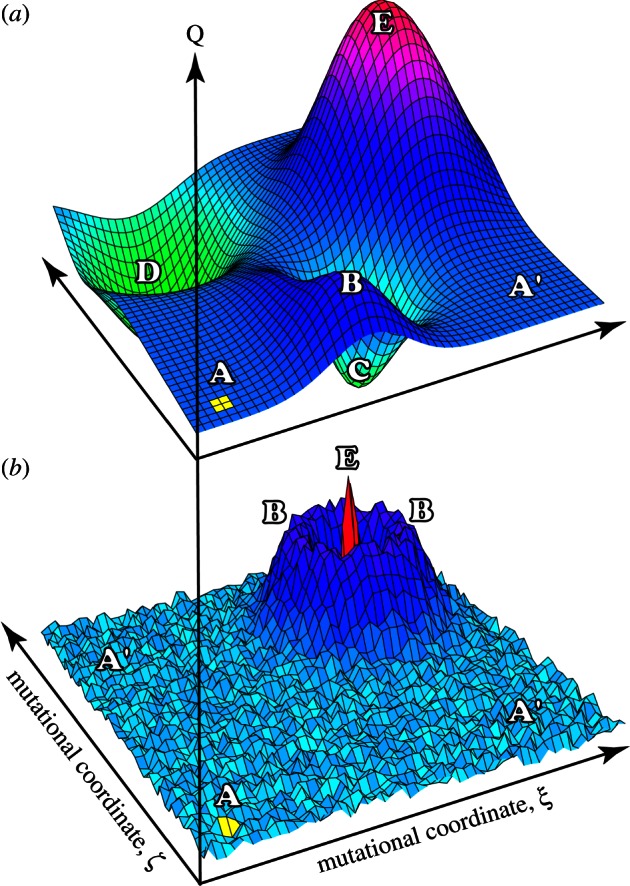
(a) Hypothetical mutational landscapes for fluid traits and (b) photochemical optimizations. The native protein is located at the squares located under A, the mutational coordinates ξ and ζ are arbitrary, and the vertical axis measures an arbitrary Q (quality) factor. Beginning at A, the goal of directed evolution is to discover the optimal protein, represented by peak E. The A’ and A’ regions signify mutations that have little impact on the Q factor, the B regions are local maxima in Q and the optimal mutation is at the peak labelled E. Cyclical optimization will typically find E in the fluid case, but is unlikely to find E in the photochemical optimization without good modelling or operator intuition. For fluid characteristics such as thermal stability, less variation is expected (a). The landscape shown in (b) depicts more complex characteristics such as photochemical properties, in which most of the performance is localized in specific regions of the protein; after fig. 3 in [38].
Characteristic targets for the mutagenesis of photoactive proteins are typically a group or series of residues that contribute to the photochemistry of the molecule. In the case of BR, a majority of mutagenesis has focused on optimizing the residues that contribute to the formation of the M and O photointermediates and the Q photoproduct because these states offer the greatest potential for use in device applications. In addition to enhancing the lifetime of different photocycle intermediates of BR, mutagenesis has been employed to enhance the innate dipole moment, gold-binding capabilities and specificity of ion pumping of BR. For a number of device platforms and architectures, the ability of BR to bind to gold is critical because gold is a chemically inert and electrically conductive [112–114]. The native protein contains no cysteine residues and thus the strategic addition of cysteine residues in the loop regions of BR via site-directed mutagenesis allows BR to covalently bind to gold. Enhancing the dipole moment of BR is critical for the application of the protein in photovoltaic devices [23,89,115]. Altering the intrinsic dipole moment through charge substitution in the helical loop regions of the protein allows the protein to pack more densely, while enhancing the photovoltaic output or signal from BR.
Genetic optimization of BR has led to the commercial development of a variety of applied technologies. In the following sections, we describe the use of type I directed evolution for the systematic optimization of BR for applied technologies. We explore the unique challenges faced in the optimization of photoactive proteins, particularly the issue of how to optimize a complex photochemical reaction, which gains stability from a two-dimensional lattice during protein expression in the native organism. Furthermore, we describe the importance of pH screening in the optimization of photoactive proteins. Enhancement of pH is a critical part of the directed evolution process and cannot be adequately accomplished through traditional cell-based techniques because the organism will either buffer the pH or die because it cannot mediate the pH fluctuations. In particular, we will focus on the optimization of the Q photoproduct of BR, a photochemical state that is rarely found in nature.
5.1. Optimization of the branched photocycle of bacteriorhodopsin by using directed evolution
In order to optimize proteins for applied technologies, a number of criteria must be satisfied. First, mutational vectors and methods must exist in order to introduce genetic variation into the coding sequence of the protein of interest. Next, an expression method must be implemented in order to obtain sufficient quantities of the macromolecule under investigation for testing. Finally, an assessment paradigm must be available to identify, screen and characterize the mutations generated. The ability to generate successful mutants relies heavily on the ability to identify the mutations of interest quickly, efficiently and inexpensively. High-throughput screening methods are necessary to detect improvements in the phenotype of interest. The methods and procedures presented below provide only the fundamental features of BR optimization, but provide a template for type I directed evolution of any protein for which expression methods, mutational vectors and screening methods are available.
The optimization of BR for devices that implement the branched photocycle requires the simultaneous modification of five variables: minimization of the formation lifetime of the O state, optimization of the decay of the O state, enhancement of the quantum efficiency of the O to P photochemical reaction, enhancement of the efficiency of the P to Q hydrolysis and an increase in the lifetime of the O state [38]. Prediction of all of the impactful mutations and interactions that would simultaneously lead to the optimization of each of these photochemical properties is nearly impossible. Although experimental data exist that would allow scientists to begin targeting residues of BR for specific photochemical enhancement, the time and cost associated with mutagenesis is prohibitive. Thus, directed evolution is an excellent method for the optimization of BR for a variety of applications.
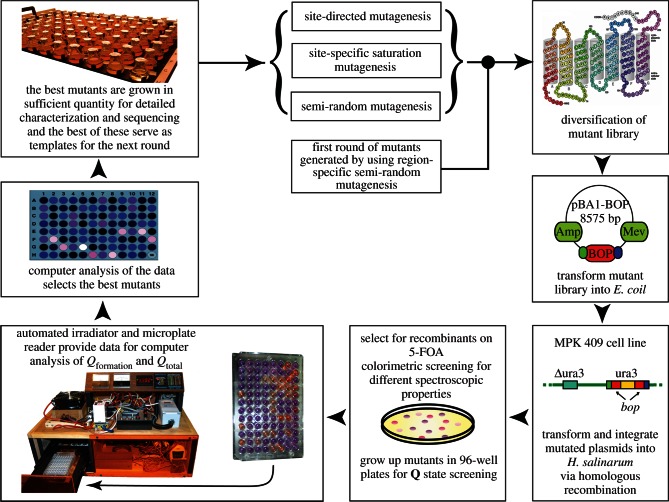
Type I directed evolution, which implements automated screening methods and microgram protein characterization, is used to generate new BR mutants through a combination of region-specific semi-random mutagenesis, site-directed mutagenesis and saturation mutagenesis. These mutants are screened and evaluated with respect to the formation and reversion efficiencies of the branched photocycle, particularly the Q state. After selection and identification of the best Q state mutants, the most efficient Q state mutants serve as the parents to the next generation of progeny. This process is reiterative and gradually improves the efficiency of the Q state photochemistry at each stage of directed evolution. In order to maximize efficiency, only the best mutants from each round of automated testing need to be sequenced, and thus the procedures are cost and time efficient.
In type I directed evolution of BR, a diverse molecular library of BR mutants is generated using region-specific semi-random mutagenesis (figure 3). First, the bacterio-opsin (bop) sequence is divided into 17 regions, approximately 15–20 amino acids in length (figure 4, inset). Next, the sequence overlap extension method [116] is employed and doped oligonucleotides are designed to overlap each of the 17 regions of interest in the bop coding sequence. The resulting diversified mutant library is then transformed into Escherichia coli for amplification using the pBA1 expression vector. Following amplification, the resulting genetically distinct colonies are pooled and allowed to grow overnight. Next, mutant DNA is extracted and transformed into the native organism, H. salinarum, via the MPK 409 cell line. Transformants are selected using antibiotic markers (mevinolin) and recombinants are screened using 5-fluoroorotic acid (5-FOA). Colonies isolated from the 5-FOA plates are then replicated in 96-well plates for sequencing, and are expressed in rich media supplemented with uracil for protein synthesis. Following purification via multiple rounds of differential ultracentrifugation, the protein is isolated and placed in 96-well plates for screening.
Figure 3.
Overview of the multistaged type I directed evolution process implemented in this study. Most of the routine tasks were automated for efficiency and reproducibility. Region-specific semi-random mutagenesis [38] was carried out via the strand overlap extension method [116] in order to amplify a 15–20 amino acid region of the bacterio-opsin (bop) gene. The diversified genetic library was transformed into Escherichia coli using the pBA1 expression vector. The resulting genetically distinct colonies were pooled and DNA was extracted. Mutant DNA was transformed into the native organism, Halobacterium salinarum, via the MPK 409 cell line. Transformants were selected using mevinolin resistance and recombinants were selected using 5-fluoroorotic acid (5-FOA) [117]. Individual colonies were picked from the 5-FOA plates and replicated in 96-well plates. An automated irradiator was used to provide data for computer analysis of Qformation and Qreversion and the best mutants were selected based on a computer algorithm. The result is a visual colorimetric display of the best Q state formers with lighter colours representing high Q state formation and darker colours signifying low or no Q state formation. Mutants that have a high Qtotal are selected to serve as the parents to the next generation of mutants.
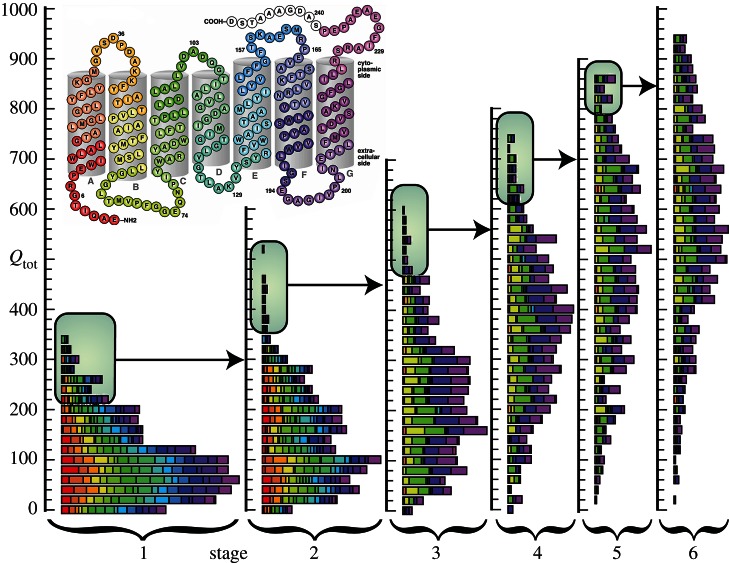
Figure 4.
Type I directed evolution of BR to enhance formation of the Q state. Progression of Qtotal efficiency through six stages of Type I directed evolution is shown. The shading corresponds to the parent mutation, as shown in the figure insert at upper left. Only those mutants with high Qtotal were sequenced so that these mutations could serve as a parent protein in the subsequent stage of evolution.
Screening of BR involves the characterization of select photophysical properties of the protein. An in-house automated irradiator is used to provide data for computer analysis of Q formation (bR → Q) and Q reversion (Q → bR) (figure 3). The automated irradiator system uses a set of 12 640 nm Luxeon III Lambertian LEDs, each driven at 850 mA to uniformly irradiate the 96-well plates containing purified BR mutants. The 640 nm light is important for coupling the absorption bands of the bR resting state and the O state. Each plate is then irradiated for 3 h under constant temperature (35 ± 0.1°C), and then scanned using a microplate reader to measure the absorption spectrum of the protein in each of the wells. The best mutants are selected based on a computer algorithm, which analyses the amount of Q state formed for each mutant. Following Q state formation, the plate is returned to the irradiator for further illumination via a set of 28 510 mcd LEDs at 395 nm for 2 h. The plate is then scanned using the microplate reader and the absorption spectrum is recorded for evaluation of the efficiency of reversion from (Q → bR). Computer analysis of the resulting absorption spectra is used to calculate the formation and reversion efficiencies of the Q state. The output of the computer analysis is a visual colorimetric display of the best Q state mutants, where lighter colours represent a greater formation of the Q state and darker colours signify low or no formation of the Q state.
The computer algorithm functions by assigning a quality to each variable, Q, which should not be confused with the Q state. The greater the Qtotal, the more efficient the formation and reversion of the Q state. Qformation represents the multistep process of forming the Q state (bR → Q), while Qreversion represents the process of reverting the protein back to the resting state (Q → bR):
 |
The ξ multipliers are arbitrary multipliers, which define the scaling. The formation and reversion efficiencies are critical to the application of the Q state of BR in read–write devices. The key is to use fully characterized proteins as internal standards to assign both Qformation and Qreversion.
At the conclusion of each stage of mutagenesis, the top 10–20 mutants with the highest Qtotal values are sequenced. New mutants are then constructed using the best Q state mutants from the previous round to serve as the parents to the next generation of progeny. The goal of this iterative process is to only pass on useful mutations and to avoid carrying silent mutations forward.
An important characteristic to consider when optimizing photoactive proteins is the inclusion of pH as a variable. The pH of a solution has a significant role in the protonation states of the binding site residues. In stages 4–6 of the directed evolution of BR, pH was included as a screening parameter (figure 4). In order to implement this screening method, the best mutants at pH 7 were grown up at larger volumes and scanned from pH = 6 to 10.5 in increments of 0.5, and the Qtotal value for each mutant was registered at the pH value that yielded the best result. Buffers below a pH of 6 were excluded to avoid formation of the blue membrane [71,118,119].
After six generations of optimization via directed evolution, involving over 10 000 mutants, a number of mutants have been discovered with excellent Q state formation and reversion efficiency. The results of each of the six stages of mutagenesis to optimize Qtotal can be seen in figure 4. Important to note is the fact that many of the mutations to BR had little or no impact on protein function (figure 5). Additionally, the mutations that yielded a high Qtotal were mutations outside of the binding site (figure 6), which was an unexpected result. Moreover, without the inclusion of pH as a variable, the best mutants would have not been discovered. The best mutant, V49A/I119T/T121S/A126T, was discovered after six stages of directed evolution and has a Qtotal of 977, which is approximately 70 times greater than wild-type BR (figure 7). The best single mutant, V49A, has a Qtotal of 924, nearly 62 times greater than wild-type BR. Both of these mutants offer significant advantages in the formation and reversion of the Q state, and have allowed for improved commercialization of BR in devices.
Figure 5.
Number of mutations at each residue following six stages of directed evolution on BR using the methods and procedures outlined in this paper. Note that a majority of the mutations had no significant impact, and only a handful yielded significant improvement.
Figure 6.
The improvement in the variable Qtotal as a function of mutational position following six stages of directed evolution is shown in (a). The number of mutational hits (N) at each site during these stages is shown in (b). Note that only mutations that result in the formation of photochemically active (purple) membrane are included, and that hits at a given site may involve identical site mutations or are combined with other sites to yield a unique mutant. The residues in white were either not included in the search (238–248 on C-terminus end) or, when mutated, yielded inactive proteins or blue membranes.
Figure 7.
Location of the residues modified to form the high Q mutant V49A/I119T/T121S/A126T, with the relevant residues marked with red lettering. Other residues that are important to the photochemical properties or proton translocation process are also shown. The proton channel is shaded. The contours of the electrostatic difference map associated with the quadruple mutation are shown in the background. The key electrostatic impact of the mutation is to increase negative charge in the Asp-85 (D85) and Asp-212 (D212) region, which stabilizes the protonation of D85 and prolongs the yield and lifetime of the O state. The V49A component of this mutation also preferentially stabilizes the 9-cis conformation. Contour lines are drawn at the following energies (J/mol): ±1, ±6, ±21, ±49, ±96, ±170, ±260, ±390, ±560, ±760, ±1000, ±1300, ±1700, ±2100, ±2600, ±3100, ±3800, ±4500, ±5200, ±6100.
Understanding the key mechanisms of Q state enhancement in V49A and V49A/I119T/T121S/A126T is important to further enhance BR through directed evolution. We predict that the mechanism of Q state enhancement in V49A is associated with stabilizing the hydrolysed 9-cis chromophore in the binding site. By providing a binding site that preferentially accommodates a 9-cis chromophore, V49A appears to not only enhance the stability of the P state, but also appears to lower the activation barrier to the formation of the P state. The V49A/I119T/T121S/A126T mutant adds hydroxyl groups, which alter the electrostatics of the binding site (figure 7) and lower the pH for optimal performance. The latter is important for making polymeric matrices, which are more stable at near-neutral pH values.
5.2. Optimization of protein stability
Any protein that is to be used as the photoactive element of a device must be resistant to both thermal and photochemical stress [25,101,120]. Type I directed evolution of BR includes cyclicity as a variable, and mutants with poor photochemical reversion will therefore have a low Qtotal and will not be selected for further investigation (see discussion above). However, the above methods are not designed to optimize the thermal stability of the protein, and thus a mutant with outstanding Qtotal may also diminish the structural integrity of BR. Moreover, the reversion efficiency optimized here does not translate directly into high photochemical cyclicity.
Photochemical cyclicity measures how many times the protein can undergo a photocycle before 1/e (approx. 37%) of the protein ensemble has denatured [120]. A proper study of photochemical cyclicity is an arduous process, particularly when the protein has a high cyclicity [120]. We have found that the vast majority of BR mutants with a λmax between 540–570 nm have excellent photochemical cyclicity, comparable to the native protein [120]. The V49A and V49A/I119T/T121S/A126T mutants are both being studied in more detail to establish a formal cyclicity, but preliminary results suggest these mutants are within approximately 20 per cent of the native protein. The photochemical cyclicity required for a given device depends on the application and varies significantly. Volumetric memories can operate with a cyclicity as low as 103, whereas most Fourier-transform optical associative processors require a cyclicity above 105 [25,101,105,120]. This observation parallels the range observed in living organisms. Halobacterium salinarum requires a native protein with a high photochemical cyclicity (approx. 106), whereas the deep sea bacteria that uses blue proteorhodopsin experiences a low enough light flux to require a protein with a much lower photochemical cyclicity (approx. 104) [120]. Organisms have a significant advantage in that they can express additional protein if the existing population is compromised. As a result, a device will place a higher burden on photochemical cyclicity than an organism. Hence, the remarkable photochemical cyclicity of BR is an important attribute that provides a comparative advantage over the majority of photochemical proteins and organic molecules.
Structural stability is the second important attribute of a photochromic material, particularly for devices that operate above ambient temperatures. Stability is often evaluated by measuring thermally induced conformational changes in the protein structure via differential scanning calorimetry. These transitions are often reported in terms of a melting temperature (TM), or the temperature at which the transition exhibits the greatest change in heat. Two such transitions are observed for BR in an unbuffered solution (figure 8): a reversible relaxation of the rigid protein structure at approximately 80°C and an irreversible denaturation at approximately 98°C [49]. This stability is sensitive to several factors that include pH [121,122], manipulation of the lipid environment [123,124] and chemical effects [125–127]. Unsurprisingly, BR exhibits the greatest stability in the native membrane. Hence, the TM of BR mutants is usually measured in the purified native membrane and suspended in deionized water. In general, most BR mutants yield data similar to the native protein, with TM values at approximately 80°C and 90–100°C. Some mutants (e.g. V49F, R82G) exhibit complex profiles with a skewed baseline, which indicate that the protein is unstable (figure 8). These mutants are removed from the pool of candidate proteins.
Figure 8.

Differential scanning calorimetry traces of mutant BR relative to native wild-type (WT) BR. The relative heat capacity of each mutant is used as a comparative measure to highlight observed differences in melting temperature (TM) and the general nature of the reversible and irreversible thermal transitions of the protein. Some mutants have stabilities roughly equivalent to WT (V49A, R82K and E204Q), while others can be considered less stable (I119T/T121S/A126T and T90A) or prohibitively unstable (V49F, R82G and L206P) for use in device applications. Note that V49A exhibits a TM equivalent, if not superior, to WT, whereas I119T/T121S/A126T is slightly less stable. The stability of the V49A/I119T/T121S/A126T mutant is currently uncharacterized, however, the data suggest that the combination of V49A and the triple mutation will lead to a protein with sufficient stability for use in biophotonic devices.
BR is a kinetically stabilized protein, meaning that the irreversible TM of BR is sensitive to the rate of heating. Measuring the TM of mutants is therefore only the first step of screening the stability of candidates. Mutants with native-like TM values are subjected to kinetic denaturation experiments that estimate the amount of total thermal energy required to denature the protein. Data from three to five traces, collected at various scanning rates, are fit to an Arrhenius model:
where v is the scan rate (K min−1), TM is the melting temperature from differential scanning calorimetry data (K), EAPP is the apparent energy of thermal denaturation (kJ mol−1) and R is the gas constant (8.314 J mol−1 K−1) [128]. In water, native BR exhibits a stability of 900 kJ mol−1 and the V49A mutant, for example, demonstrates a similar stability of 850 kJ mol−1. Suspension in buffered solutions at slightly alkaline pH (e.g. pH 8.5), where Qtotal is more efficient, roughly doubles this stability. This method consumes a considerable amount of protein and is only done for the top performing mutants.
6. Conclusions
Advances in genetic engineering and, more significantly, directed evolution have enhanced the potential of developing commercially viable protein-based devices. The approach presented in this review serves as a general template for researchers interested in creating a mutant library that would ultimately lead to a desired phenotype for application outside of the biological context of the organism. While the properties relevant to Qtotal are capable of being measured by using absorption spectroscopy on BR mutants, we suggest that other spectroscopic or analytical methods could be exploited to monitor a similar series of stages of directed evolution when in pursuit of a non-native physical property. Using a combination of mutagenesis methods in conjunction with high-throughput screening techniques, the overall yield, lifetime and formation and reversion efficiencies of the Q state of BR have been enhanced for applications in bioelectronics. After six stages of directed evolution, nearly 10 000 BR mutants were generated, with a majority of those mutants offering improvements in Qtotal. Two of the best mutants, V49A and V49A/I119T/T121S/A126T, have made it possible to use low intensity write lasers in three-dimensional optical memories. Efforts are being carried out to study these mutants in more detail and to further investigate the potential of these mutants as the photoactive element in photonic devices.
Acknowledgements
This research was supported in parts by grants from the National Science Foundation (EMT-0829916, EIA-0129731), the National Institutes of Health (GM-34548), DARPA (HR0011-05-1-0027), the Army Research Office (MURI, DAAD 19-99-1-0198) and the Harold S. Schwenk Sr. Distinguished Chair in Chemistry.
References
- 1.Heath JR, Kuekes PJ, Snider GS, Williams SR. 1998. A defect-tolerant computer architecture: opportunities for nanotechnology. Science 280, 1716–1721 10.1126/science.280.5370.1716 (doi:10.1126/science.280.5370.1716) [DOI] [Google Scholar]
- 2.Whitesides GM. 2003. The ‘right’ size in nanobiotechnology. Nat. Biotechnol. 21, 1161–1165 10.1038/nbt872 (doi:10.1038/nbt872) [DOI] [PubMed] [Google Scholar]
- 3.Moore G. 1965. Cramming more components onto integrated circuits. Electronics 38, 114–117 [Google Scholar]
- 4.Agarwal R, Lieber CM. 2006. Semiconductor nanowires: optics and optoelectronics. Appl. Phys. A-Mater. 85, 209–215 10.1007/s00339-006-3720-z (doi:10.1007/s00339-006-3720-z) [DOI] [Google Scholar]
- 5.Lu W, Lieber CM. 2007. Nanoelectronics from the bottom up. Nat. Mater. 6, 841–850 10.1038/nmat2028 (doi:10.1038/nmat2028) [DOI] [PubMed] [Google Scholar]
- 6.Mansoori GA, Fauzi Soelaiman TA. 2005. Nanotechnology—an introduction for the standards community. J. ASTM Int. 2, 1–21 10.1520/JAI12938 (doi:10.1520/JAI12938) [DOI] [Google Scholar]
- 7.Haberland H. 2013. Looking from both sides. Nature 494, E1–E2 10.1038/nature11886 (doi:10.1038/nature11886) [DOI] [PubMed] [Google Scholar]
- 8.Konstas K, Langford SJ, Latter MJ. 2010. Advances toward synthetic machines at the molecular and nanoscale level. Int. J. Mol. Sci. 11, 2453–2472 10.3390/ijms11062453 (doi:10.3390/ijms11062453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu RS, Tirrell M. 2003. Bottom-up design of biomimetic assemblies. Adv. Drug Del. Rev. 56, 1537–1563 [DOI] [PubMed] [Google Scholar]
- 10.Balzani V. 2008. Nanoscience and nanotechnology: the bottom-up construction of molecular devices and machines. Pure Appl. Chem 80, 1631–1650 10.1351/pac200880081631 (doi:10.1351/pac200880081631) [DOI] [Google Scholar]
- 11.Boncheva M, Whitesides GM. 2004. Biomimetic approaches to the design of functional self-assembling systems. In Dekkar encyclopedia of nanoscience and nanotechnology (eds JA Schwarz, CI Contescu, K Putyera), pp. 287–294 New York, NY: Marcel Dekker [Google Scholar]
- 12.Bausch AR, Kroy K. 2006. A bottom-up approach to cell mechanics. J. ASTM Int. 2, 231–238 [Google Scholar]
- 13.Nguyen P, Meyyappan M, Yiu SC. 2010. Applications in nanobiotechnology in ophthalmology. Ophthal. Res. 44, 1–14 10.1159/000279436 (doi:10.1159/000279436) [DOI] [PubMed] [Google Scholar]
- 14.Whitesides GM. 2004. Nanoscience, nanotechnology, and chemistry. Small 1, 172–179 10.1002/smll.200400130 (doi:10.1002/smll.200400130) [DOI] [PubMed] [Google Scholar]
- 15.Bruchez MP. 2007. Commercialization and future developments in bionanotechnology. In Frontiers of engineering: reports on leading-edge engineering from the 2006 symposium. Washington, DC: The National Academies Press [Google Scholar]
- 16.Nikolov L, Slavchev S, Stoychev S. 2008. The convergence of biotechnology and nanobiotechnology as an accelerator of the development of biofilm technologies. Ecol. Eng. Environ. Prot. 7, 36–47 [Google Scholar]
- 17.Caraglia M, Rosa G, Salzano G, Santini D, Lamberti M, Sperlongano P, Lombardi A, Abbruzzese A, Addeo R. 2012. Nanotech revolution for the anti-cancer drug delivery through blood–brain-barrier. Curr. Cancer Drug Tar. 12, 186–196 10.2174/156800912799277421 (doi:10.2174/156800912799277421) [DOI] [PubMed] [Google Scholar]
- 18.Cario E. 2012. Nanotechnology-based drug delivery in mucosal immune diseases: hype or hope? Mucosal Immunol. 5, 1–3 10.1038/mi.2011.59 (doi:10.1038/mi.2011.59) [DOI] [Google Scholar]
- 19.Mazzola L. 2003. Commercializing nanotechnology. Nat. Biotechnol. 21, 1137–1143 10.1038/nbt1003-1137 (doi:10.1038/nbt1003-1137) [DOI] [PubMed] [Google Scholar]
- 20.Cronin T, Bennett J. 2011. Switching on the lights: the use of optogenetics to advance retinal gene therapy. Mol. Ther. 19, 1190–1192 10.1038/mt.2011.115 (doi:10.1038/mt.2011.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora P, Sindhu A, RajaKumar G, Dilbaghi N, Rahuman AA, Chaudhury A. 2012. Nano-regenerative medicine towards clinical outcome of stem cell and tissue engineering in humans. J. Cell. Mol. Med 16, 1991–2000 10.1111/j.1582-4934.2012.01534.x (doi:10.1111/j.1582-4934.2012.01534.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mata A, Palmer L, Tejeda-Montes E, Stupp SI. 2012. Design of biomolecules for nanoengineered biomaterials for regenerative medicine. In Nanotechnology in regerenative medicine: methods and protocols, methods in molecular biology (eds Navarro M, Planell JA.), pp. 39–47 Berlin, Germany: Springer; [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Bhattacharya P, Varo G. 2004. Monolithically integrated bacteriorhodopsin/semiconductor opto-electronic integrated circuit for a bio-photoreceiver. Biosen. Bioelectron. 19, 885–892 10.1016/j.bios.2003.08.018 (doi:10.1016/j.bios.2003.08.018) [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Stuart JA, Birge RR, Xu J, Stickrath A, Bhattacharya P. 2004. Photoelectric response of polarization sensitive bacteriorhodopsin films. Biosens. Bioelectron. 15, 869–874 10.1016/j.bios.2003.08.017 (doi:10.1016/j.bios.2003.08.017) [DOI] [PubMed] [Google Scholar]
- 25.Birge RR, et al. 1999. Biomolecular electronics: protein-based associative processors and volumetric memories. J. Phys. Chem. B 103, 10 746–10 766 10.1021/jp991883n (doi:10.1021/jp991883n) [DOI] [Google Scholar]
- 26.Hampp N. 2000. Bacteriorhodopsin: mutating a biomaterial into an optoelectronic material. Appl. Microbiol. Biotechnol. 53, 633–639 10.1007/s002539900311 (doi:10.1007/s002539900311) [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Govender D, Gross R, Birge R. 1995. Advances in protein-based three-dimensional optical memories. BioSystems 35, 145–151 10.1016/0303-2647(94)01503-Y (doi:10.1016/0303-2647(94)01503-Y) [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Birge RR. 1993. Protein based artificial retinas. Trends Biotech. 11, 292–300 10.1016/0167-7799(93)90017-4 (doi:10.1016/0167-7799(93)90017-4) [DOI] [PubMed] [Google Scholar]
- 29.Umeno D, Tobias AV, Arnold FH. 2005. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 69, 51–58 10.1128/MMBR.69.1.51-78.2005 (doi:10.1128/MMBR.69.1.51-78.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäckel C, Kast P, Hilvert D. 2008. Protein design by directed evolution. Annu. Rev. Biophys. 37, 153–173 10.1146/annurev.biophys.37.032807.125832 (doi:10.1146/annurev.biophys.37.032807.125832) [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki K, Arnold FH. 1999. Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J. Mol. Evol. 49, 716–720 10.1007/PL00006593 (doi:10.1007/PL00006593) [DOI] [PubMed] [Google Scholar]
- 32.Bornscheuer UT, Bessler C, Srinivas R, Krishna SH. 2002. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol. 20, 433–437 10.1016/S0167-7799(02)02046-2 (doi:10.1016/S0167-7799(02)02046-2) [DOI] [PubMed] [Google Scholar]
- 33.Dougherty MJ, Arnold FH. 2009. Directed evolution: new parts and optimized function. Curr. Opin. Biotech. 20, 1–6 10.1016/j.copbio.2009.04.001 (doi:10.1016/j.copbio.2009.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasserot AP, Dickinson CD, Tang Y, Huse WD, Manchester KS, Watkins JD. 2003. Optimization of protein therapeutics by directed evolution. Drug Discov. Today 8, 118–126 10.1016/S1359-6446(02)02590-4 (doi:10.1016/S1359-6446(02)02590-4) [DOI] [PubMed] [Google Scholar]
- 35.Hibbert EG, Baganz F, Hailes HC, Ward JM, Lye GJ, Woodley JM, Dalby PA. 2005. Directed evolution of biocatalytic processes. Biomol. Eng. 22, 11–19 10.1016/j.bioeng.2004.09.003 (doi:10.1016/j.bioeng.2004.09.003) [DOI] [PubMed] [Google Scholar]
- 36.Choi EJ, Jacak R, Kuhlman B. 2013. A structural bioinformatics approach for identifying proteins predisposed to bind linear epitopes on pre-selected target proteins. Prot. Eng. Des. Sel 26, 283–289 10.1093/protein/gzs108 (doi:10.1093/protein/gzs108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A, Singh S. 2012. Directed evolution: tailoring biocatalysts for industrial applications. Crit. Rev. Biotechnol., 1–14 10.3109/07388551.2012.716810 (doi:10.3109/07388551.2012.716810) [DOI] [PubMed] [Google Scholar]
- 38.Wise KJ, Gillespie NB, Stuart JA, Krebs MP, Birge RR. 2002. Optimization of bacteriorhodopsin for bioelectronic devices. Trends Biotechnol. 20, 387–394 10.1016/S0167-7799(02)02023-1 (doi:10.1016/S0167-7799(02)02023-1) [DOI] [PubMed] [Google Scholar]
- 39.Liang J, Luo Y, Zhao H. 2011. Synthetic biology: putting synthesis into biology. Rev. Sys. Biol. Med 3, 7–20 10.1002/wsbm.104 (doi:10.1002/wsbm.104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold F. 1998. Design by directed evolution. Acc. Chem. Res. 31, 125–131 10.1021/ar960017f (doi:10.1021/ar960017f) [DOI] [Google Scholar]
- 41.Hillebrecht JR, Wise KJ, Birge RR. 2004. Directed evolution of bacteriorhodopsin for device applications. Method Enzymol. 388, 333–347 10.1016/S0076-6879(04)88026-1 (doi:10.1016/S0076-6879(04)88026-1) [DOI] [PubMed] [Google Scholar]
- 42.Oesterhelt D, Brauchle C, Hampp N. 1991. Bacteriorhodopsin: a biological material for information processing. Quart. Rev. Biophys. 24, 425–478 10.1017/S0033583500003863 (doi:10.1017/S0033583500003863) [DOI] [PubMed] [Google Scholar]
- 43.Birge RR. 1990. Photophysics and molecular electronic applications of the rhodopsins. Annu. Rev. Phys. Chem. 41, 683–733 10.1146/annurev.pc.41.100190.003343 (doi:10.1146/annurev.pc.41.100190.003343) [DOI] [PubMed] [Google Scholar]
- 44.Lanyi JK. 2004. Bacteriorhodopsin. Annu. Rev. Physiol. 66, 665–688 10.1146/annurev.physiol.66.032102.150049 (doi:10.1146/annurev.physiol.66.032102.150049) [DOI] [PubMed] [Google Scholar]
- 45.Blaurock AE, Stoeckenius W. 1971. Structure of the purple membrane. Nat. New Biol. 233, 152–155 10.1038/233152a0 (doi:10.1038/233152a0) [DOI] [PubMed] [Google Scholar]
- 46.Blaurock AE. 1975. Bacteriorhodopsin: a trans-membrane pump containing α-helix. J. Mol. Biol. 93, 139–142 10.1016/0022-2836(75)90124-2 (doi:10.1016/0022-2836(75)90124-2) [DOI] [PubMed] [Google Scholar]
- 47.Henderson R. 1975. The structure of the purple membrane from Halobacterium halobium. Analysis of the X-ray diffraction pattern. J. Mol. Biol. 93, 123–138 10.1016/0022-2836(75)90123-0 (doi:10.1016/0022-2836(75)90123-0) [DOI] [PubMed] [Google Scholar]
- 48.Krebs M, Isenbarger T. 2000. Structural determinants of purple membrane assembly. Biochim. Biophys. Acta 1460, 15–26 10.1016/S0005-2728(00)00126-2 (doi:10.1016/S0005-2728(00)00126-2) [DOI] [PubMed] [Google Scholar]
- 49.Jackson MB, Sturtevant JM. 1978. Phase transitions of the purple membranes of Halobacterium halobium. Biochemistry 17, 911–915 10.1021/bi00598a026 (doi:10.1021/bi00598a026) [DOI] [PubMed] [Google Scholar]
- 50.Shen Y, Safinya CR, Liang KS, Ruppert AF, Rothschild KJ. 1993. Stabilization of the membrane protein bacteriorhodopsin to 140°C in two-dimensional films. Nature 366, 48–50 10.1038/366048a0 (doi:10.1038/366048a0) [DOI] [Google Scholar]
- 51.Lukashev EP, Robertson B. 1995. Bacteriorhodopsin retains its light-induced proton-pumping function after being heated to 140°C. Bioelectrochem. Bioenerg. 37, 157–160 10.1016/0302-4598(94)05017-O (doi:10.1016/0302-4598(94)05017-O) [DOI] [Google Scholar]
- 52.Gerwert K, Hess B, Engelhard M. 1990. Proline residues undergo structural changes during proton pumping in bacteriorhodopsin. FEBS Lett. 261, 449–454 10.1016/0014-5793(90)80613-N (doi:10.1016/0014-5793(90)80613-N) [DOI] [Google Scholar]
- 53.Gillespie NB, Ren L, Ramos L, Daniell H, Dews D, Utzat KA, Stuart JA, Buck CH, Birge RR. 2005. Characterization and photochemistry of 13-desmethyl bacteriorhodopsin. J. Phys. Chem. B 109, 16 142–16 152 10.1021/jp052124+ (doi:10.1021/jp052124+) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillespie NB, et al. 2002. Characterization of the branched-photocycle intermediates P and Q of bacteriorhodopsin. J. Phys. Chem. B 106, 13 352–13 361 10.1021/jp021221p (doi:10.1021/jp021221p) [DOI] [Google Scholar]
- 55.Oesterhelt D, Stoeckenius W. 1971. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature 233, 149–152 [DOI] [PubMed] [Google Scholar]
- 56.Luecke H, Richter HT, Lanyi JK. 1998. Proton transfer pathways in bacteriorhodopsin at 2.3Å resolution. Science 280, 1934–1937 10.1126/science.280.5371.1934 (doi:10.1126/science.280.5371.1934) [DOI] [PubMed] [Google Scholar]
- 57.Spudich JL, McCain DA, Nakanishi K, Okabe M, Shimizu N, Rodman H, Honig B, Bogomolni RA. 1986. Chromophore/protein interaction in bacterial sensory rhodopsin and bacteriorhodopsin. Biophys. J. 49, 479–483 10.1016/S0006-3495(86)83657-8 (doi:10.1016/S0006-3495(86)83657-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haupts U, Tittor J, Bamberg E, Oesterhelt D. 1997. General concept for ion translocation by halobacterial retinal proteins: the iosmerization/switch/transfer (IST) model. Biochemistry 36, 2–7 10.1021/bi962014g (doi:10.1021/bi962014g) [DOI] [PubMed] [Google Scholar]
- 59.Lanyi JK. 1998. Understanding structure and function in the light-driven proton pump bacteriorhodopsin. J. Struct. Biol. 124, 164–178 10.1006/jsbi.1998.4044 (doi:10.1006/jsbi.1998.4044) [DOI] [PubMed] [Google Scholar]
- 60.Lanyi JK. 1998. The local-access mechanism of proton transport by bacteriorhodopsin. Biochim. Biophys. Acta 1365, 17–22 10.1016/S0005-2728(98)00036-X (doi:10.1016/S0005-2728(98)00036-X) [DOI] [Google Scholar]
- 61.Brown L, Dioumaev A, Needleman R, Lanyi J. 1998. Local-access model for proton transfer in bacteriorhodopsin. Biochemistry 37, 3982–3993 10.1021/bi9728396 (doi:10.1021/bi9728396) [DOI] [PubMed] [Google Scholar]
- 62.Tittor J, Haupts U, Haupts C, Oesterhelt D, Becker A, Bamberg E. 1997. Chloride and proton transport in bacteriorhodopsin mutant D85T: different modes of ion translocation in a retinal protein. J. Mol. Biol. 271, 405–416 10.1006/jmbi.1997.1204 (doi:10.1006/jmbi.1997.1204) [DOI] [PubMed] [Google Scholar]
- 63.Kandori H. 2004. Hydration switch model for the protonation transfer in the Schiff base region of bacteriorhodopsin. Biochim. Biophys. Acta 1658, 72–79 10.1016/j.bbabio.2004.03.015 (doi:10.1016/j.bbabio.2004.03.015) [DOI] [PubMed] [Google Scholar]
- 64.Lozier RH, Bogomolni RA, Stoeckenius W. 1975. Bacteriorhodopsin: a light-driven proton pump in Halobacterium halobium. Biophys. J. 15, 955–962 10.1016/S0006-3495(75)85875-9 (doi:10.1016/S0006-3495(75)85875-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birge RR. 1994. Bacteriorhodopsin, a nonlinear proton pump. Nature 371, 659–660 10.1038/371659a0 (doi:10.1038/371659a0) [DOI] [Google Scholar]
- 66.Govindjee R, Ebrey TG, Crofts AR. 1980. The quantum efficiency of proton pumping by the purple membrane of Halobacterium halobium. Biophys. J. 30, 231–240 10.1016/S0006-3495(80)85091-0 (doi:10.1016/S0006-3495(80)85091-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Birge RR, Cooper TM, Lawrence AF, Masthay MB, Vasilakis C, Zhang CF, Zidovetzki R. 1989. A spectroscopic, photocalorimetric and theoretical investigation of the quantum efficiency of the primary event in bacteriorhodopsin. J. Am. Chem. Soc. 111, 4063–4074 10.1021/ja00193a044 (doi:10.1021/ja00193a044) [DOI] [Google Scholar]
- 68.Stuart JA, Birge RR. 1996. Characterization of the primary photochemical events in bacteriorhodopsin and rhodopsin. In Biomembranes (ed. Lee AG.), pp. 33–140 London, UK: JAI Press [Google Scholar]
- 69.Jang G-F, McBee JK, Alekseev AM, Haeseleer F, Palczewski K. 2000. Stereoisomeric specificity of the retinoid cycle in the vertebrate retina. J. Biol. Chem 275, 28 128–28 138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popp A, Wolperdinger M, Hampp N, Bräuchle C, Oesterhelt D. 1993. Photochemical conversion of the O-intermediate to 9-cis-retinal-containing products in bacteriorhodopsin films. Biophys. J. 65, 1449–1459 10.1016/S0006-3495(93)81214-1 (doi:10.1016/S0006-3495(93)81214-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tallent JR, Stuart JA, Song QW, Schmidt EJ, Martin CH, Birge RR. 1998. Photochemistry in dried polymer films incorporating the deionized blue membrane form of bacteriorhodopsin. Biophys. J. 75, 1619–1634 10.1016/S0006-3495(98)77605-2 (doi:10.1016/S0006-3495(98)77605-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Masthay MB, Sammeth DM, Helvenston MC, Buckman CB, Li W, Cde-Baca MJ, Kofron JT. 2002. The laser-induced blue state of bacteriorhodopsin: mechanistic and color regulatory roles of protein–protein interactions, protein–lipid interactions, and metal ions. J. Amer. Chem. Soc. 124, 3418–3430 10.1021/ja010116a (doi:10.1021/ja010116a) [DOI] [PubMed] [Google Scholar]
- 73.Chang C-H, Liu SY, Jonas R, Govindjee R. 1987. The pink membrane: the stable photoproduct of deionized purple membrane. Biophys. J. 52, 617–623 10.1016/S0006-3495(87)83252-6 (doi:10.1016/S0006-3495(87)83252-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dancshazy Z, Tokaji Z, Der A. 1999. Bleaching of bacteriorhodopsin by continuous light. FEBS Lett. 450, 154–157 10.1016/S0014-5793(99)00487-1 (doi:10.1016/S0014-5793(99)00487-1) [DOI] [PubMed] [Google Scholar]
- 75.Dancshazy Z, Tokaji Z. 2000. Blue light regeneration of bacteriorhodopsin bleached by continuous light. FEBS Lett. 476, 171–173 10.1016/S0014-5793(00)01714-2 (doi:10.1016/S0014-5793(00)01714-2) [DOI] [PubMed] [Google Scholar]
- 76.Hillebrecht JR, Koscielecki JF, Wise KJ, Krebs MP, Stuart JA, Birge RR. 2008. Optimizing photoactive proteins for optoelectronic environments by using directed evolution. In NanobioTechnology: bioinspired devices and materials of the future (eds Shoseyov O, Levy I.), pp. 121–140 Totowa, NJ: Humana Press Inc [Google Scholar]
- 77.Ranaghan MJ, Wagner NL, Sandberg MN, Birge RR. 2012. Optical applications of biomolecules. In Optical biomimetics: materials and applications (ed. Large M.). Cambridge, UK: Woodhead Publishing [Google Scholar]
- 78.Birge RR. 1990. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim. Biophys. Acta 1016, 293–327 10.1016/0005-2728(90)90163-X (doi:10.1016/0005-2728(90)90163-X) [DOI] [PubMed] [Google Scholar]
- 79.Birge RR. 1981. Photophysics of light transduction in rhodopsin and bacteriorhodopsin. Annu. Rev. Biophys. Bioeng. 10, 315–354 10.1146/annurev.bb.10.060181.001531 (doi:10.1146/annurev.bb.10.060181.001531) [DOI] [PubMed] [Google Scholar]
- 80.Bunkin FV, Vsevolodov NN, Druzhko AB, Mitsner BI, Prokhorov AM, Savranskii VV, Tkachenko NW, Shevchenko TB. 1981. Diffraction efficiency of bacteriorhodopsin and its analogs. Sov. Tech. Phys. Lett. 7, 630–631 [Google Scholar]
- 81.Vsevolodov NN, Poltoratskii VA. 1985. Holograms in biochrome, a biological photochromic material. Sov. Phys. Tech. Phys. 30, 1235 [Google Scholar]
- 82.Greco JA, Wagner NL, Ranaghan MJ, Rajasekaran S, Birge RR. 2012. Biomolecular electronics and protein-based optical computing. In Biomolecular information processing: from logic systems to smart sensors and actuators (ed. Katz E.), pp. 33–59 Weinheim, Germany: Wiley-VCH [Google Scholar]
- 83.Bräuchle C, Hampp N, Oesterhelt D. 1991. Optical applications of bacteriorhodopsin and its mutated variants. Adv. Mater. 3, 420–428 10.1002/adma.19910030903 (doi:10.1002/adma.19910030903) [DOI] [Google Scholar]
- 84.Birge RR, Fleitz PA, Gross RB, Izgi JC, Lawrence AF, Stuart JA, Tallent JR. 1990. Spatial light modulators and optical associative memories based on bacteriorhodopsin. Proc. IEEE EMBS 12, 1788–1789 [Google Scholar]
- 85.Stuart JA, Tallent JR, Tan EHL, Birge RR. 1996. Protein-based volumetric memory. Proc. IEEE Nonvol. Mem. Tech. (INVMTC) 6, 45–51 10.1109/NVMT.1996.534668 (doi:10.1109/NVMT.1996.534668) [DOI] [Google Scholar]
- 86.Stuart JA, Marcy DL, Wise KJ, Birge RR. 2002. Volumetric optical memory based on bacteriorhodopsin. Synth. Met. 127, 3–15 10.1016/S0379-6779(01)00586-0 (doi:10.1016/S0379-6779(01)00586-0) [DOI] [Google Scholar]
- 87.Boucher F, Taneva S, Elouatik S, Déry M, Messaoudi S, Harvey-Girard E, Beaudoin N. 1996. Reversible inhibition of proton release activity and the anesthetic-induced acid–base equilibrium between the 480 and 570 nm forms of bacteriorhodopsin. Biophys. J. 70, 948–961 10.1016/S0006-3495(96)79638-8 (doi:10.1016/S0006-3495(96)79638-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lanyi J, Luecke H. 2001. Bacteriorhodopsin. Curr. Opin. Struct. Biol. 11, 415–419 10.1016/S0959-440X(00)00226-8 (doi:10.1016/S0959-440X(00)00226-8) [DOI] [PubMed] [Google Scholar]
- 89.Hong FT. 1994. Retinal proteins in photovoltaic devices. Adv. Chem. 240, 527–560 10.1021/ba-1994-0240.ch022 (doi:10.1021/ba-1994-0240.ch022) [DOI] [Google Scholar]
- 90.Hong FT. 1999. Interfacial photochemistry of retinal proteins. Prog. Surf. Sci. 62, 1–237 10.1016/S0079-6816(99)00014-3 (doi:10.1016/S0079-6816(99)00014-3) [DOI] [Google Scholar]
- 91.Xu J, Stickrath AB, Bhattacharya P, Nees J, Varo G, Hillebrecht JR, Ren L, Birge RR. 2003. Direct measurement of the photoelectric response time of bacteriorhodopsin via electro-optic sampling. Biophys. J. 85, 1128–1134 10.1016/S0006-3495(03)74549-4 (doi:10.1016/S0006-3495(03)74549-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wagner NL, Greco JA, Birge RR. 2013. Visual restoration using microbial rhodopsins. In Bionanotechnology: biological self-assembly and its applications (ed. Rehm BHA.), pp. 205–240 Norfolk, UK: Caister Academic Press [Google Scholar]
- 93.Cutsuridis V, Wennekers T. 2009. Hippocampus, microcircuits and associative memory. Neural Networks 22, 1120–1128 10.1016/j.neunet.2009.07.009 (doi:10.1016/j.neunet.2009.07.009) [DOI] [PubMed] [Google Scholar]
- 94.Reijmers LG, Perkins BL, Matsuo N, Mayford M. 2007. Localization of a stable neural correlate of associative memory. Science 317, 1230–1233 10.1126/science.1143839 (doi:10.1126/science.1143839) [DOI] [PubMed] [Google Scholar]
- 95.Abu-Mostafa YS, Psaltis D. 1987. Optical neural computers. Sci. Am. 256, 88–95 10.1038/scientificamerican0387-88 (doi:10.1038/scientificamerican0387-88) [DOI] [Google Scholar]
- 96.Paek EG, Psaltis D. 1987. Optical associative memory using Fourier transform holograms. Opt. Eng. 26, 428–433 [Google Scholar]
- 97.Gross RB, Izgi KC, Birge RR. 1992. Holographic thin films, spatial light modulators and optical associative memories based on bacteriorhodopsin. Proc. SPIE 1662, 186–196 10.1117/12.58502 (doi:10.1117/12.58502) [DOI] [Google Scholar]
- 98.Soffer BH, Dunning G, Owechko Y, Marom E. 1986. Associative holographic memory with feedback using phase-conjugate mirrors. Opt. Lett. 11, 118–120 10.1364/OL.11.000118 (doi:10.1364/OL.11.000118) [DOI] [PubMed] [Google Scholar]
- 99.Zhang C, Song QW, Ku CY, Gross RB, Birge RR. 1994. Determination of the refractive index of a bacteriorhodopsin film. Opt. Lett. 19, 1406–1411 10.1364/OL.19.001406 (doi:10.1364/OL.19.001406) [DOI] [PubMed] [Google Scholar]
- 100.Subramaniam S, Greenhalgh DA, Khorana HG. 1992. Aspartic acid 85 in bacteriorhodopsin functions both as proton acceptor and negative counterion to the Schiff base. J. Biol. Chem. 267, 25 730–25 733 [PubMed] [Google Scholar]
- 101.Hampp N. 2000. Bacteriorhodopsin as a photochromic retinal protein for optical memories. Chem. Rev. 100, 1755–1776 10.1021/cr980072x (doi:10.1021/cr980072x) [DOI] [PubMed] [Google Scholar]
- 102.Hampp N, Popp A, Bräuchle C, Oesterhelt D. 1992. Diffraction efficiency of bacteriorhodopsin films for holography containing bacteriorhodopsin wildtype BRWT and its variants BRD85E and BRD96N. J. Phys. Chem. 96, 4679–4685 10.1021/j100190a098 (doi:10.1021/j100190a098) [DOI] [Google Scholar]
- 103.Hampp N, Thoma R, Zeisel D, Bräuchle C, Oesterhelt D. 1994. Bacteriorhodopsin variants for holographic pattern recognition. Adv. Chem. 240, 511–526 10.1021/ba-1994-0240.ch021 (doi:10.1021/ba-1994-0240.ch021) [DOI] [Google Scholar]
- 104.Hillebrecht JR, et al. 2005. Optimization of protein-based volumetric optical memories and associative processors by using directed evolution. NanoBiotechnology 1, 141–152 10.1385/NBT:1:2:141 (doi:10.1385/NBT:1:2:141) [DOI] [Google Scholar]
- 105.Greco JA, Wagner NL, Birge RR. 2012. Fourier transform holographic associative processors based on bacteriorhodopsin. Int. J. Unconventional Comput. 8, 433–457 [Google Scholar]
- 106.Liu SY, Ebrey TG. 1987. The quantum efficiency for the interphotoconversion of the blue and pink forms of purple membrane. Photochem. Photobiol. 46, 263–267 10.1111/j.1751-1097.1987.tb04765.x (doi:10.1111/j.1751-1097.1987.tb04765.x) [DOI] [Google Scholar]
- 107.Birge RR, Parsons B, Song QW, Tallent JR. 1997. Protein-based three-dimensional memories and associative processors. In Molecular electronics (eds Ratner MA, Jortner J.), pp. 439–471 Oxford, UK: Blackwell Science Ltd [Google Scholar]
- 108.He J-A, Samuelson L, Li L, Kumar J, Tripathy SK. 1998. Photoelectric properties of oriented bacteriorhodopsin/polycation multilayers by electrostatic layer by layer assembly. J. Phys. Chem. B 102, 7067–7072 10.1021/jp981612x (doi:10.1021/jp981612x) [DOI] [Google Scholar]
- 109.Carrigan PE, Ballar P, Tuzmen S. 2011. Site-directed mutagenesis. Methods Mol. Biol. 700, 107–124 10.1007/978-1-61737-954-3_8 (doi:10.1007/978-1-61737-954-3_8) [DOI] [PubMed] [Google Scholar]
- 110.Greener A, Callahan M, Jerpseth B. 1996. An efficient random mutagenesis technique using an E. coli mutator strain. Methods Mol. Biol. 57, 375–385 [DOI] [PubMed] [Google Scholar]
- 111.Nannemann DP, Birmingham W, Scism RA, Bachmann BO. 2001. Assessing directed evolution methods for the generation of biosynthetic enzymes with potential in drug biosynthesis. Future Med. Chem. 3, 809–819 10.4155/fmc.11.48 (doi:10.4155/fmc.11.48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Berthoumieu O, Patil A, Wang X, Aslimovska L, Davis J, Watts A. 2012. Molecular scale conductance photoswitching in engineered bacteriorhodopsin. Nano Lett. 12, 899–903 10.1021/nl203965w (doi:10.1021/nl203965w) [DOI] [PubMed] [Google Scholar]
- 113.Patil A, Premaruban T, Berthoumieu O, Watts A, Davis J. 2012. Enhanced photocurrent in engineered bacteriorhodopsin monolayer films. J. Phys. Chem. B 116, 683–689 10.1021/jp210520k (doi:10.1021/jp210520k) [DOI] [PubMed] [Google Scholar]
- 114.Crameri A, Raillart S, Bermudez E, Stemmer WPC. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 10.1038/34663 (doi:10.1038/34663) [DOI] [PubMed] [Google Scholar]
- 115.Hong FT. 1997. Molecular sensors based on the photoelectric effect of bacteriorhodopsin. FEBS Lett. 109, 189–193 [Google Scholar]
- 116.Georgescu R, Bandara G, Sun L. 2003. Saturation mutagenesis. Methods Mol. Biol. 231, 75–83 [DOI] [PubMed] [Google Scholar]
- 117.Peck RF, DasSarma S, Krebs MP. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35, 667–676 10.1046/j.1365-2958.2000.01739.x (doi:10.1046/j.1365-2958.2000.01739.x) [DOI] [PubMed] [Google Scholar]
- 118.Fischer UC, Towner P, Oesterheldt D. 1981. Light induced isomerization, at acidic pH, initiates hydrolysis of bacteriorhodopsin to bacterio-opsin and 9-cis-retinal. Photochem. Photobiol. 33, 529–537 10.1111/j.1751-1097.1981.tb05456.x (doi:10.1111/j.1751-1097.1981.tb05456.x) [DOI] [Google Scholar]
- 119.Turner GJ, Miercke LJW, Thorgeirsson TE, Kliger DS, Betlach MC, Stroud RM. 1993. Bacteriorhodopsin D85N: three spectroscopic species in equilibrium. Biochemistry 32, 1332–1337 10.1021/bi00056a019 (doi:10.1021/bi00056a019) [DOI] [PubMed] [Google Scholar]
- 120.Ranaghan MJ, Shima S, Ramos L, Poulin DS, Whited G, Rajasekaran S, Stuart JA, Albert AD, Birge RR. 2010. Photochemical and thermal stability of green and blue proteorhodopsins: implications for protein-based bioelectronic devices. J. Phys. Chem. B 114, 14 064–14 070 10.1021/jp106633w (doi:10.1021/jp106633w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kresheck GC, Lin CT, Williamson LN, Mason WR, Jang D-J, El-Sayed MA. 1990. The thermal stability of native, delipidated, deionized and regenerated bacteriorhodopsin. J. Photochem. Photobiol. B 7, 289–302 [Google Scholar]
- 122.Brouillette C, Muccio D, Finney T. 1987. pH dependence of bacteriorhodpsin thermal unfolding. Biochemistry 26, 7431–7438 10.1021/bi00397a035 (doi:10.1021/bi00397a035) [DOI] [PubMed] [Google Scholar]
- 123.Alonso A, Restall CJ, Turner M, Gomez-Fernandez JC, Goñi FM, Chapman D. 1982. Protein–lipid interactions and differential scanning calorimetric studies of bacteriorhodopsin reconstituted lipid–water systems. Biochim. Biophys. Acta Biomembr. 689, 283–289 10.1016/0005-2736(82)90261-9 (doi:10.1016/0005-2736(82)90261-9) [DOI] [Google Scholar]
- 124.Brouillette CG, McMichens RB, Stern LJ, Khorana HG. 1989. Structure and thermal stability of monomeric bacteriorhodopsin in mixed pospholipid/detergent micelles. Proteins: Struct. Funct. Gen. 5, 38–46 10.1002/prot.340050106 (doi:10.1002/prot.340050106) [DOI] [PubMed] [Google Scholar]
- 125.Gao MM, Boucher F. 1998. The uncoupling of bacteriorhodopsin by high temperature and anaesthetics. Toxicol. Lett. 100–101, 393–396 10.1016/S0378-4274(98)00212-4 (doi:10.1016/S0378-4274(98)00212-4) [DOI] [PubMed] [Google Scholar]
- 126.Cladera J, Galisteo ML, Duñach M, Mateo PL, Padrós E. 1988. Thermal denaturation of deionized and native purple membranes. Biochim. Biophys. Acta 943, 148–156 10.1016/0005-2736(88)90546-9 (doi:10.1016/0005-2736(88)90546-9) [DOI] [Google Scholar]
- 127.Cladera J, Galisteo ML, Sabés M, Mateo PL, Padrós E. 1992. The role of retinal in the thermal stability of the purple membrane. Eur. J. Biochem. 207, 581–585 10.1111/j.1432-1033.1992.tb17084.x (doi:10.1111/j.1432-1033.1992.tb17084.x) [DOI] [PubMed] [Google Scholar]
- 128.Galisteo M, Sanchez-Ruiz K. 1993. Kinetic study into the irreversible thermal denaturation of bacteriorhodopsin. Eur. Biophys. J. 22, 25–30 [Google Scholar]