Abstract
Motivation: It has been known for more than 2 decades that after RNA polymerase II (RNAPII) initiates transcription, it can enter into a paused or stalled state immediately downstream of the transcription start site before productive elongation. Recent advances in high-throughput genomic technologies facilitated the discovery that RNAPII pausing at promoters is a widespread physiologically regulated phenomenon. The molecular underpinnings of pausing are incompletely understood. The CCCTC-factor (CTCF) is a ubiquitous nuclear factor that has diverse regulatory functions, including a recently discovered role in promoting RNAPII pausing at splice sites.
Results: In this study, we analyzed CTCF binding sites and nascent transcriptomic data from three different cell types, and found that promoter-proximal CTCF binding is significantly associated with RNAPII pausing.
Contact: praveen_sethupathy@med.unc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Promoter-proximal pausing of RNA polymerase II (RNAPII) has emerged as a widespread and important step in the regulation of transcription (Core et al., 2008; Gilchrist and Adelman, 2012; Li and Gilmour, 2011; Seila et al., 2009). The role of pausing is not completely understood, but initial evidence suggests that it allows for precise and rapid transcriptional response to stimuli such as developmental cues or cellular stressors (Espinosa, 2010; Li and Gilmour, 2011). The underlying mechanisms of RNAPII pausing are thought to be complex (Landick, 2009). Recent publications have reported a few key factors that regulate polymerase pause site entry and release, including GAGA (Lee et al., 2008), c-MYC (Rahl et al., 2010), ELL (Byun et al., 2012) and XRN2 (Brannan et al., 2012); however, it is not clear whether and how many other complexes are also involved. As such, this area merits further investigation.
The CCCTC-factor (CTCF) is a multifaceted gene regulator, with demonstrated roles in transcriptional activation/repression, insulation, imprinting and chromatin remodeling (Ohlsson et al., 2010; Phillips and Corces, 2009). In 2011, Shukla et al. published a seminal article, which reported for the first time that CTCF can facilitate exon inclusion by promoting RNAPII pausing at splice sites (Shukla et al., 2011). Another recent study postulated that CTCF may regulate vascular endothelial growth factor transcription in part by mediating RNAPII pausing (Lu and Tang, 2012). Given these reports, and recent observations that at least 20–25% of CTCF-binding events are proximal to promoters (Jothi et al., 2008; Kim et al., 2007), we hypothesized that CTCF binding may be a cofactor in mediating promoter-proximal RNAPII pausing. To investigate this possibility, we analyzed genome-wide nascent RNA data (global nuclear run-on followed by sequencing; GRO-seq) and empirically determined CTCF-binding sites from the only three cell types for which both datasets are publicly available: human cell lines IMR90 (lung fibroblast) and MCF-7 (breast cancer) and mouse embryonic stem cells (mES).
2 METHODS
2.1 Datasets
Empirically determined CTCF-binding sites were downloaded from http://licr-renlab.ucsd.edu/download.html (CTCF chromatin immunoprecipitation followed by microarray analysis [ChIP-chip] in IMR90 cells), http://genome.ucsc.edu/ENCODE/downloads.html (CTCF chromatin immunoprecipitation followed by high-throughput sequencing [ChIP-seq] in MCF-7 cells) and Chen et al., 2008 (mES ChIP-seq). All coordinates were mapped to either the mm9 (mES) or the hg18 (IMR90, MCF-7) genome builds using the command line liftOver program with the –minMatch parameter set to 0.95. CTCF-binding sites were also computationally predicted by scanning for matches to the canonical motif (Kim et al., 2007) using the PWM-scan algorithm (Levy and Hannenhalli, 2002). Global nuclear run-on followed by high-throughput sequencing (GRO-seq) data were downloaded from Core et al., 2008 (IMR90), Hah et al., 2011 (MCF-7) and Min et al., 2011 (mES).
2.2 Analysis
Promoter-proximal pausing indices were calculated in the following manner: first, for each annotated RefSeq transcript longer than 3 kb (n = 27 863 for human, n = 22 547 for mouse), we set the transcription start site (TSS) as position 0, and defined 0–500 nt downstream as the promoter-proximal region and 500-gene end as the gene body. Second, we computed the density of RNAPII in both promoter-proximal regions and gene bodies by calculating GRO-seq reads/nt/mapability using the same mapability data as in Melgar et al., 2011. Third, for all genes that had at least one read mapping to the gene body (n = 24 988 for IMR90, n = 24 613 for MCF-7, n = 20 512 for mES), we computed pausing index as the ratio of RNAPII density in the promoter-proximal region to RNAPII density in the gene body, similar to previous studies (Core et al., 2008; Min et al., 2011).
Promoters were divided into two categories: those with and those without a CTCF-binding site overlapping X to Y nt downstream of the TSS, where X and Y were set in independent experiments to 0 and 100, 101 and 200, 201 and 300, 301 and 500, 501 and 1000 or 1001 and 2000. To compare pausing indices between categories, we computed the Jaccard distance in median pausing index as follows: (x − y)/(x + y), where x is the median pausing index at CTCF-bound promoters and y is the median pausing index at promoters where CTCF is not bound. The significance of the difference in pausing indices between the two promoter categories was assessed by one-sided permutation and non-parametric Mann–Whitney–Wilcoxon (MWW) tests. This analysis was repeated for the same distances upstream of the TSS.
3 RESULTS
For each cell type, we divided promoters according to whether CTCF was bound upstream/downstream of the TSS (Section 2). The number of promoter-proximal CTCF-binding sites was highly variable across cell types (Supplementary Material 1).
GRO-seq data provide a density map of transcriptionally engaged RNAPII across the genome by purifying, sequencing and mapping nascent RNAs (Core et al., 2008). Using recently published GRO-seq data, we calculated RNAPII pausing indices at RefSeq-annotated promoters in three different cell types, and assessed whether promoter-proximal CTCF binding is associated with elevated RNAPII pausing indices (Section 2). We made two novel observations:
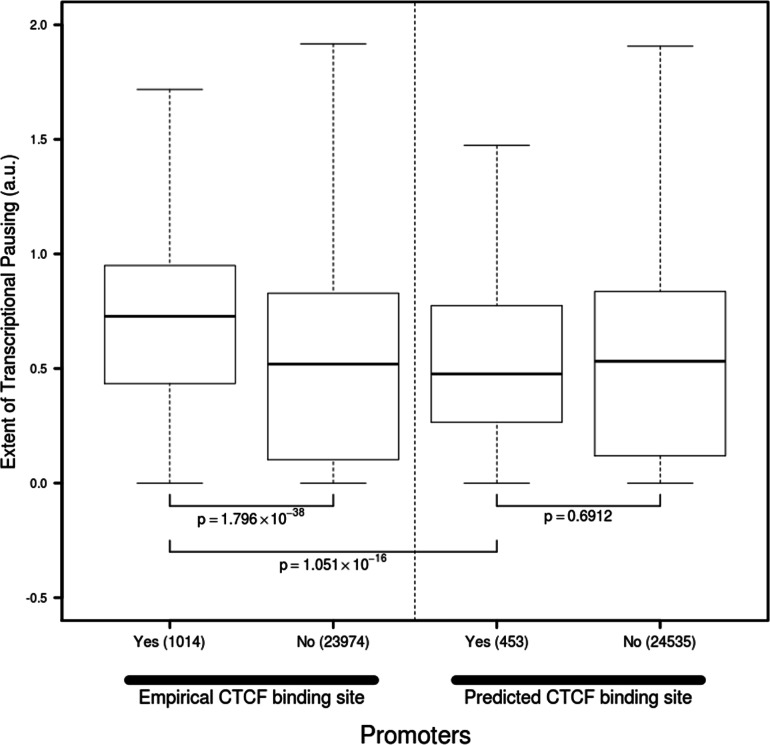
(1) Genes that exhibit promoter-proximal CTCF binding (defined as within 100 nt downstream of the TSS) have a significantly greater RNAPII pausing index than genes that do not (Supplementary Material 1). For example, in IMR90 cells, the median pausing index at promoters with proximal CTCF-binding sites is ∼2-fold greater (permutation test P < 0.0001, Mann–Whitney–Wilcoxon test P = 1.8 × 10 − 38) than at promoters without proximal CTCF-binding sites (Fig. 1). The highly overlapping range of pausing indices between the two categories of promoters indicates that while CTCF binding may be associated with pausing, it is by no means necessary or sufficient.
Fig. 1.
The association between CTCF binding and pausing index is shown. Each IMR90 promoter was classified based on the presence (Yes) or absence (No) of an empirically determined (IMR90 ChIP-chip), or computationally predicted CTCF-binding site in the first 100 nt downstream of its TSS (x-axis). The pausing index at each promoter was calculated (Section 2) and transformed using the formula log10(pausing index + 1) for visualization purposes (y-axis). The boxes in the plot cover percentiles 25–75, and the bar within each box represents the median. The whiskers extend to the most extreme data point that is no more than 1.5 times the interquartile range from the box. Mann–Whitney–Wilcoxon non-parametric test P-values are indicated for the relevant comparisons
Regions of CTCF binding are enriched for motifs with elevated cytosine (C) content (Jothi et al., 2008). As such, the observed association between CTCF-binding events and increased pausing index could be explained by nucleotide composition (i.e. cytosine enriched regions may be responsible for the association with pausing, independent of CTCF binding). To test this, we repeated the analysis in each cell type with computationally predicted CTCF-binding sites (Section 2), based on the known C-rich motif (Kim et al., 2007). We did not observe any association with pausing (Fig. 1; Supplementary Material 1), which strongly suggests that sequence composition alone is not sufficient to explain the association signal, but rather, the physical binding of CTCF is required.
(2) The association between CTCF binding and the promoter-proximal pausing index is position-dependent—that is, the further upstream/downstream from the TSS CTCF is bound, the less of an association there is with RNAPII pausing (Fig. 2; Supplementary Material 1). For example, in mES cells, CTCF binding within 101–200 nt downstream of TSSs is highly significantly associated with pausing (permutation test P < 0.0001); however, this association is dramatically weakened for CTCF binding within 1001–2000 nt upstream/downstream of TSSs (permutation test P > 0.5).
Fig. 2.

The position-specific effect on the association of CTCF binding with promoter-proximal RNAPII pausing is shown. The x-axis represents the distance (nt) from the TSS (vertical dashed line) of annotated (RefSeq) genes longer than 3 kb. The y-axis depicts a normalized difference in median pausing index between promoters with a CTCF-binding event (ChIP-seq/ChIP-chip peak) and promoters without a CTCF-binding event (no ChIP-seq/ChIP-chip peak). As such, the value 0.0 (horizontal dashed line) represents ‘no difference’ in median pausing index between the two groups, a positive value represents increased pausing in promoters with a CTCF-binding event and a negative value represents decreased pausing in promoters with a CTCF-binding event. Results are shown for three cell lines (two human: IMR90, MCF-7; one mouse: mES cells)
4 DISCUSSION
A recent study identified a novel role for CTCF in polymerase pausing at splice sites to facilitate exon inclusion (Shukla et al., 2011). Our findings provide the first indication that CTCF is associated with promoter-proximal pausing. It is important to note that the study does not demonstrate that CTCF binding is necessary or sufficient for promoter-proximal pausing. Substantial additional experimental work is required to uncover the molecular underpinnings of the observed association between CTCF binding and RNAPII dynamics.
The greatest degree of association with pausing is observed for CTCF-binding sites that are immediately downstream of the TSS. Therefore, an appealing mechanism for the observed association is steric hindrance—that is, stably bound CTCF may serve to hinder RNAPII processivity. If this is the case, it is likely that many other transcription factors with binding sites immediately downstream of TSSs also associate with RNAPII pausing. It will be possible to test this hypothesis as more transcription factor (TF) ChIP-seq and GRO-seq data are generated from the same cell type. We note, however, that because even sites that are upstream of the TSS are somewhat associated with pausing, steric hindrance is not likely to be the only mechanism involved.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Karen Adelman (National Institute of Environmental Health and Safety, National Institutes of Health) for valuable discussions, and Dr Michael L. Stitzel (National Human Genome Research Institute, National Institutes of Health) and members of the Sethupathy laboratory for helpful suggestions.
Funding: This work was supported in part by a National Institutes of Health R00 award (DK091318-02 to P.S).
Conflict of Interest: none declared.
REFERENCES
- Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol. Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun JS, et al. ELL facilitates RNA polymerase II pause site entry and release. Nat. Commun. 2012;3:633. doi: 10.1038/ncomms1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM. The meaning of pausing. Mol. Cell. 2010;40:507–508. doi: 10.1016/j.molcel.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Adelman K. Coupling polymerase pausing and chromatin landscapes for precise regulation of transcription. Biochim. Biophys. Acta. 2012;1819:700–706. doi: 10.1016/j.bbagrm.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, et al. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. Transcriptional pausing without backtracking. Proc. Natl Acad. Sci. USA. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, et al. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Hannenhalli S. Identification of transcription factor binding sites in the human genome sequence. Mamm. Genome. 2002;13:510–514. doi: 10.1007/s00335-002-2175-6. [DOI] [PubMed] [Google Scholar]
- Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 2011;21:231–235. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tang M. CTCF-dependent chromatin insulator as a built-in attenuator of angiogenesis. Transcription. 2012;3:73–77. doi: 10.4161/trns.19634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melgar MF, et al. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12:R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, et al. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, et al. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.