Abstract
Motivation: Extensive DNA sequencing of tumor and matched normal samples using exome and whole-genome sequencing technologies has enabled the discovery of recurrent genetic alterations in cancer cells, but variability in stromal contamination and subclonal heterogeneity still present a severe challenge to available detection algorithms.
Results: Here, we describe publicly available software, Shimmer, which accurately detects somatic single-nucleotide variants using statistical hypothesis testing with multiple testing correction. This program produces somatic single-nucleotide variant predictions with significantly higher sensitivity and accuracy than other available software when run on highly contaminated or heterogeneous samples, and it gives comparable sensitivity and accuracy when run on samples of high purity.
Availability: http://www.github.com/nhansen/Shimmer
Contact: nhansen@mail.nih.gov
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The development of next-generation DNA sequencing methods has led to a rapid increase in the availability of high-quality sequence data from large numbers of tumor samples, some with matched samples from the same individuals’ blood or unaffected tissue (Berger et al., 2011; Ding et al., 2012b; International Cancer Genome Consortium, 2010; Pleasance et al., 2010; Shah et al., 2009). These data have uncovered genes and pathways that are mutated in various forms of cancer (Gui et al., 2011; Lee et al., 2010; Stark et al., 2012), and the characterization of these alterations has led to a greater understanding of oncogenesis, as well as the potential for earlier diagnosis of cancers and more directed treatments (Yang et al., 2011; Zhang et al., 2007).
However, sensitive and accurate detection of somatic single-nucleotide variants (sSNVs) from next-generation sequencing data is still a challenging informatic problem because of differing levels of purity and numbers of subclonal populations represented in tumor samples. The depth of read coverage necessary to detect a mutation accurately is dependent on the prevalence of the mutation in that sample, which is in turn dependent on levels of copy number variation, levels of stromal contamination and prevalence of mutation among subclones within the sample (Carter et al., 2012; Koboldt et al., 2012). Available software for detecting differences between samples often provides little guidance with regard to how deep coverage needs to be to attain the user’s required sensitivity and accuracy.
A large number of algorithms have been developed to identify small mutations in aligned sequencing reads from pairs of samples. For sSNVs, several studies (Pleasance et al., 2010; Stark et al., 2012, Wei et al., 2011) have used a naïve ‘subtraction’ method in which variants are first called separately from both the tumor and normal samples’ reads, and then sites that have a confident call of a variant in the tumor, as well as a confident call of no variant in the normal, are assumed to be sSNVs. However, analyzing samples separately can lead to false positives when a germline variant is present in a low percentage of reads from the normal sample, as well as false negatives when mutation levels in the tumor are too low to allow an algorithm to confidently distinguish variant reads from sequencing error. Analyzing reads from the two samples simultaneously enables better discrimination of germline alleles from sequencing error.
Other algorithms do this simultaneous analysis with hypothesis testing. VarScan2 (Koboldt et al., 2012) is a comprehensive sequence analysis tool that includes sSNV detection and analyzes pairs of samples simultaneously. It predicts sSNVs using a combination of heuristic filtering and a Fisher’s exact test, but the tool reports P-values without any correction for multiple testing, and fails to report the number of tests performed or the expected underlying distribution of P-values, making it impossible to perform these corrections after running the program. Similarly, deepSNV (Gerstung et al., 2012) performs a likelihood ratio test on each site in a sequenced region, along with rigorous multiple testing correction, to report sites for which the frequency of a variant allele is significantly higher than that of modeled sequencing errors, but it gives the user only rough control over which sites are tested.
JointSNVMix2 (Roth et al., 2012) uses Bayesian probability models to infer the genotypes of both tumor and normal samples simultaneously, allowing users to train the model on the data and then classify mutations as somatic or germline with a given posterior probability. The software also now includes the ability to filter predictions using parameters learned from the data (Ding et al. 2012a). However, as it is based on a diploid model, this method is prone to errors in regions where the model does not reflect reality, for example, copy number altered regions and misaligned repetitive sequences (Roth et al., 2012). Similarly, SomaticSniper (Larson et al., 2012) uses a Bayesian model to calculate posterior probabilities of somatic mutations, using a prior that incorporates the dependence of tumor and normal genotypes from a single individual and provides software for filtering using other indicators for potential false positives. Others have used Bayesian approaches as well (Cibulskis et al., 2013; Li, 2011; Saunders et al., 2012).
In this study, we describe a new simpler approach based on hypothesis testing with correction for multiple testing. Implemented in a publicly available software tool, Shimmer, the algorithm yields highly accurate and sensitive calls on matched tumor and normal sequence, even in the presence of large amounts of stromal contamination, heterogeneity and copy number alteration, and even without any post-filtering of sSNV calls.
2 METHODS
2.1 Single-nucleotide detection algorithm
Shimmer takes as input aligned sequence reads from a tumor and its matched normal tissue in BAM format (Li et al., 2009). In a manner similar to other sSNV discovery programs, Shimmer examines the base counts for each possible allele at every genomic position covered by sequence data in both the tumor and the normal sample. Shimmer’s program options allow filtering of bases based on base quality score or reads based on read mapping quality. If the total number of reads displaying a non-reference allele in the two samples is greater than a minimum threshold nvar, a Fisher’s exact test is performed to test the null hypothesis that variant alleles are distributed randomly between the two samples (as they would be if the non-reference bases were sequencing errors or evidence of a germline variant that would also be present in the normal tissue). The optimal choice of nvar is the smallest value that will yield a near-uniform P-value distribution (Supplementary Methods), ensuring adequate power to reject the null hypothesis even when performing a large number of tests.
As thousands to millions of sites are tested for a single BAM file comparison, Shimmer performs a multiple testing correction (Benjamini and Hochberg, 1995; Noble, 2009) on the Fisher exact P-values to report only a set of results with false discovery rate (FDR) below a desired maximum q. This FDR provides a conservative estimate of the proportion of predicted variants that are not true somatic variants, but are instead the consequence of random variation in allele frequencies. In addition, after testing, only the variants for which the normal sample has a predicted genotype of homozygous reference (Teer et al., 2010) are reported as somatic mutations in variant call format (VCF) (Danacek et al., 2011) or VarSifter format (Teer et al., 2012). Shimmer will also format variants for the annotation program ANNOVAR (Wang et al., 2010) and annotate the variant file if ANNOVAR is installed separately and the annotate option has been specified. A more detailed description of the methods is given in the Supplementary Methods.
2.2 Performance on simulated datasets
To measure the sensitivity and accuracy of various sSNV calling algorithms against a known truth dataset, we introduced single base changes corresponding to known somatic mutations into next-generation sequencing data from one of two whole-exome libraries from a single-HapMap sample NA18506 (The International HapMap Consortium, 2003). The simulated sSNVs were created using Pysam (http://code.google.com/p/pysam/). For each simulated tumor/normal comparison, in the ‘tumor’ BAM file, at each position from the list of randomly selected mutations, any read spanning that position was changed at the mutated position with a probability equal to half of the desired tumor purity (e.g. for a sample that is 20% tumor, ∼10% of the reads at the indicated position are mutated, in an effort to simulate a heterozygous mutation in a diploid region of a sample with the desired tumor purity). The base qualities of the mutated bases were not altered, as they reflect real sequencing accuracy. As the two datasets originated from libraries created from the same sample, a somatic caller comparing the original, unaltered BAM files should find no sSNVs, and variants discovered that are not part of the set of introduced mutations can be considered false positives. The mutations were selected randomly from a set of 111 521 mutations downloaded from the Catalogue of Somatic Mutations in Cancer (COSMIC) (Forbes et al., 2010), all of which lie within targeted regions of the capture kit used to create the whole-exome libraries of NA1506, and simulations were performed with numbers of mutations per exome corresponding to a high (10 mutations per megabase) or low (two mutations per megabase) rate of mutation.
These simulations were repeated for high and low mutation rate, and at three levels corresponding to a heterozygous mutation with three different levels of tumor purity: 100% tumor, in which reads were mutated with probability 50%, 60% tumor, in which reads were mutated with probability 30%, and 20% tumor, in which reads were mutated with probability 10%. Multiple replicates of these simulated BAM files were compared with the corresponding normal NA18506 BAM file from the separate run using Shimmer, JointSNVMix2 (using the train and classify commands), SomaticSniper (with recommended post-filtering), VarScan2 with the ‘somaticFilter’ option and deepSNV. All predicted sSNVs present in dbSNP build 134 were filtered before evaluation of sensitivity and accuracy. Further details regarding alteration of the BAM files and versions, options and post-filtering of the programs are available in the Supplementary Methods.
2.3 Performance on sequence data from COLO-829
To assess Shimmer’s sensitivity and accuracy on a real dataset, we obtained the whole-genome sequence data of the COLO-829 melanoma cell line previously sequenced (Pleasance et al., 2010), from the European GenomePhenome Archive (EGAS00000000052) and ran Shimmer, VarScan2 and SomaticSniper with recommended parameters and filtering (Supplementary Methods). We then calculated the sensitivity as the percentage of the 497 sites previously validated (Pleasance et al., 2010) that was called by each program. Furthermore, to determine calling accuracy, we validated all protein-altering mutations predicted by the three programs by amplifying the affected region in both the melanoma and the matching normal cell line (COLO-829/COLO-829BL) using polymerase chain reaction (PCR), and then performing Sanger sequencing to classify each predicted site as confirmed somatic, germ line or wild-type.
2.4 Implementation and availability
Shimmer is distributed as a Perl script, with computationally intensive portions of the algorithms implemented in C and R. The source code is available on github at http://github.com/nhansen/Shimmer.
3 RESULTS
We compared the variants predicted by Shimmer with those predicted by VarScan2, SomaticSniper, deepSNV and JointSNVMix2 on the simulated datasets described in the previous section. In addition, we ran Shimmer, VarScan2 and SomaticSniper on the whole-genome COLO-829 data. For each dataset, we defined sensitivity as the percentage of known true variants that are predicted correctly by each program, and assessed accuracy by tallying the number of false positive variants a program predicted.
3.1 Results from simulated datasets
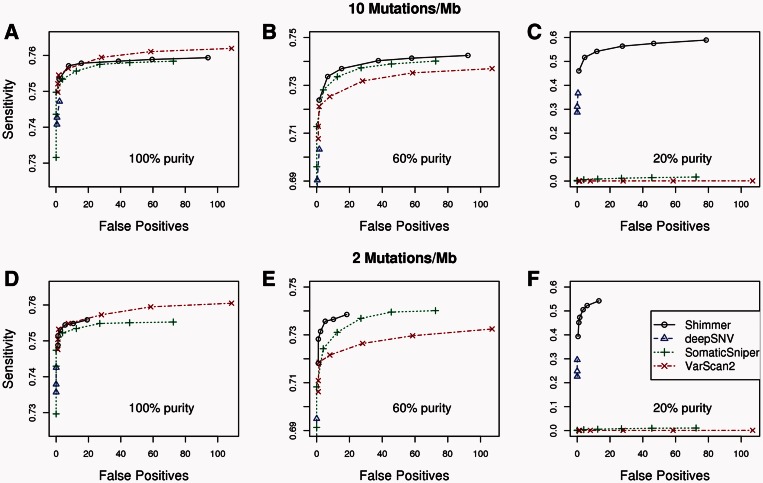
The six sets of simulated BAM files with 619 or 124 introduced sSNVs each (corresponding to mutation rates of 10 and 2 per megabase of targeted sequence), and at each of three simulated levels of purity, 20, 60 and 100%, were compared with an unaltered set of reads from a different library of NA18506 using Shimmer, JointSNVMix2 (Roth et al., 2012), SomaticSniper (Larson et al., 2012), VarScan2 (Koboldt et al., 2012) and deepSNV (Gerstung et al., 2012). The results are shown in Figure 1. JointSNVMix2 results are not shown in these plots because their mean false positive count (which is >500 for all six simulation types at the program’s highest stringency cut-off) is not easily displayed, but the mean values and errors for all five programs are available in Supplementary Table S1 (Supplementary Data). For high-purity tumors (100 and 60% purity), all five programs easily achieved sensitivities of >70%, but none achieved sensitivities beyond ∼76%. This is because the sensitivity was limited by the sequencing depth of coverage across the targeted regions of the capture, and ∼20% of the targeted regions have little or no sequencing coverage. Note that for highly pure tumors, Shimmer provides comparable sensitivity and accuracy with the other programs. In addition, at very low purity (20%), other programs either miss large numbers of variants (SomaticSniper, VarScan2 and deepSNV) or predict large numbers of false positives (JointSNVMix2), whereas Shimmer maintains high accuracy with optimal sensitivity.
Fig. 1.
Sensitivity and number of false positives for somatic variant detection at two different simulated mutation rates and three different simulated levels of tumor purity. (A–C) The mean sensitivity and number of false positives for whole exomes with a mutation rate of 10 mutations per megabase of targeted genomic regions for tumor purity levels of 100, 60 and 20%, respectively. Likewise, (D–F) Same results are observed when there are two mutations per megabase of targeted sequence. Estimated error of the mean for each point’s sensitivity is <0.005, indicating that the differences between the three programs may not be statistically significant for tumor purity levels of 60 and 100%. Cut-offs used to generate the points for each program were as follows: Shimmer: max_q = 0.01, 0.05, 0.1, 0.2, 0.3 and 0.5. VarScan2: α = 0.01, 0.005, 0.002, 0.0005, 0.0002, 0.00005 and 0.00002. SomaticSniper: somatic_score = 40, 45, 50, 60, 70, 90, 110 and 130. DeepSNV: max_fdr = 0.1, 0.2 and 0.9999
When run on the two unmutated BAM files derived from NA18506, Shimmer and deepSNV, both report zero mutations, as one would expect, as there are no true mutations in the data, and any false positive prediction would lead to a FDR of 1.0, above the level of control provided by the Benjamini–Hochberg procedure. On the other hand, SomaticSniper, VarScan2 and JointSNVMix2 all predict similar numbers of false positives when run on the unmutated BAM files, as they do on the files with simulated mutations used to generate the results in Figure 1.
3.2 Results from COLO-829 dataset
When run on EGA sequence data from the melanoma-matched tumor and normal cell lines COLO-829, Shimmer, SomaticSniper and VarScan2 with α = 0.05 all detected 96–97% of 497 previously confirmed variant sites. When VarScan2 was run with α = 0.001, however, its sensitivity dropped to 92.4%. Variants predicted by Shimmer and SomaticSniper show a markedly increased percentage of ultraviolet (UV)-induced C→T/G→A mutations (70.2% for Shimmer with max_q = 0.05, and 70.8% for SomaticSniper with recommended filtering) when compared with filtered VarScan2 predictions (67.5% with α = 0.001). Details regarding the mutation spectrum are provided in Supplementary Figure S1 (Supplementary Data).
To assess the accuracy of predicted mutations from these three programs, we examined the set of all non-synonymous, splice-site, stop-gain and stop-loss mutations predicted by Shimmer, SomaticSniper and/or VarScan2. Sixty-four of these mutations (Supplementary Data and Supplementary Table S2) had been validated previously (Pleasance et al., 2010). Of the remaining predicted mutations, we were able to design PCR primers and amplify regions surrounding 218, and the combined results of the previous and new validation experiments are shown in Table 1. When all protein-altering predictions are considered, 98.5% of mutations called by Shimmer with FDR of 0.05 were validated as true, compared with 92.7% of SomaticSniper calls and 92.6% of VarScan2 calls with α = 0.001. When we include all VarScan2 calls obtained with α = 0.05, a level that gives comparable sensitivity with the other two programs, only 56.7% of predicted somatic variants are confirmed by Sanger sequencing. Supplementary Table S2 in the Supplementary Data gives the details regarding each sSNV prediction we attempted to validate.
Table 1.
Sanger confirmation results for predicted COLO-829 somatic variants
| Program(s) | Number of 497 previously validated variants detected | Program sensitivity (%) | Total predicted variants assessed by Sanger sequencing | Number of variants confirmed by Sanger sequencing | Program accuracy (%) |
|---|---|---|---|---|---|
| Shimmer | 476 | 95.8 | 134 | 132 | 98.5 |
| SomaticSniper | 480 | 96.6 | 164 | 152 | 92.7 |
| VarScan2 | 459 | 92.4 | 148 | 137 | 92.6 |
| All three programs | 442 | 88.9 | 125 | 124 | 99.2 |
Note: ‘Total variants’ is the number of predicted somatic variants in each set that were successfully amplified and sequenced to test for validity. Shimmer was run with a maximum FDR of 0.05, and VarScan2 was run with α = 0.001.
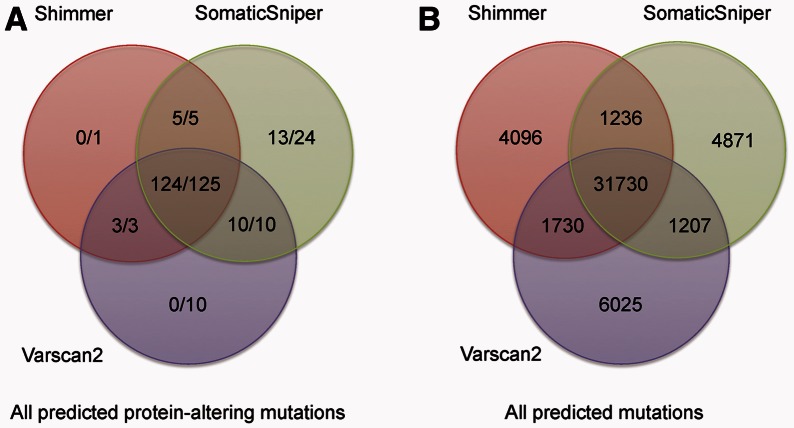
Shimmer, VarScan2 and SomaticSniper each predict variants not present in the others’ sets of predictions. Figure 2 shows the concordance of predicted variants from the three programs, both for protein-altering variants and for total sets of variants across the genome. Figure 2A also shows the number of the protein-altering variants that were confirmed by Sanger sequencing. In all, 99.3% (142 of 143) of variants predicted by at least two of the programs were shown to be true somatic variants.
Fig. 2.
Concordance among predictions of Shimmer, SomaticSniper and VarScan2. These Venn diagrams show the distribution of predicted variants among the three different programs for (A) protein altering variants and (B) across the genome. In (A), the counts are reported as ‘#TP/#Pred’, where ‘#TP’ is the number of coding variants verified by Sanger sequencing, and ‘#Pred’ is the total number of protein-altering variants predicted by the program that were amplified and subjected to Sanger sequencing. In (B), only the total number of variants predicted is shown
3.3 Analysis of errors
The generation of Sanger sequence data allows us to classify false positives in somatic variant calls as to whether they represent germ line variants that were not sequenced thoroughly in the normal sample, or wild-type sequence for which sequencing errors were mistaken for variant alleles in the tumor. All three programs made prediction errors of both of these types. For the false positives discovered in the COLO-829 validation sequencing, the breakdown was as follows: SomaticSniper incorrectly predicted that five germ line variants and seven wild-type sites were somatic, VarScan2 predicted that eight germ line variants and three wild-type sites were somatic, and Shimmer predicted that one germ line variant and one wild-type site were somatic.
Hypothesis testing methods like Shimmer are also prone to making errors in regions with very high-sequencing depth, where small differences in variant allele frequency can become statistically significant because of the large counts being tested. These regions, which are often repetitive and prone to alignment errors, can also suffer from inaccurate removal of PCR duplicates. When this happens, the assumption of independent sampling on which hypothesis testing is based is flawed, and results can be inaccurate. Shimmer’s filtering of sites for which the normal sample has a genotype that is not homozygous represents a first effort to remove these artifacts from its somatic variant predictions.
3.4 Analysis of power to detect mutations
Although most somatic variant detection methods provide little guidance about the depth of sequence coverage needed to detect all somatic mutations, the use of hypothesis testing enables us to confidently estimate the number of independent reads that will yield a given power to confidently reject the null hypothesis of equal distribution of alternate alleles between the two samples.
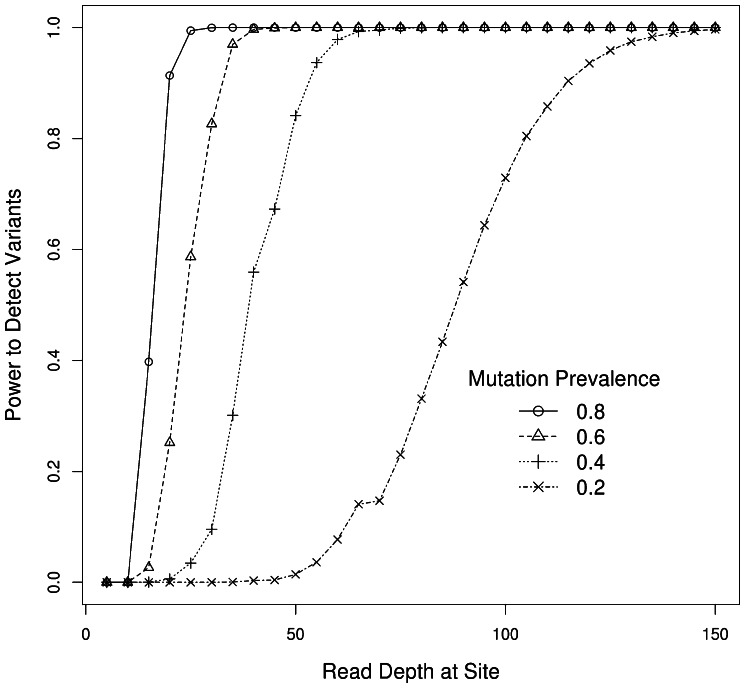
Figure 3 shows the coverage required to achieve any desired sensitivity to detect somatic mutations at different tumor variant allele frequencies. The ability to calculate expected power to detect mutations is critical to planning experiments, especially for larger projects. From the plot, which assumes 10 000 tests, a typical number of tests performed on a whole-exome sequence dataset from a tumor/normal pair, it is estimated that ∼50× coverage is needed to obtain 90% sensitivity when 40% of reads can be expected to display the alternate allele in the tumor, whereas well over 100× coverage is required to obtain the same sensitivity when only 20% of reads are expected to show the alternate allele.
Fig. 3.
Power to detect somatic variants with increasing read depth of coverage. Different curves represent different expected mean fraction of alternate allele to be observed among the sequencing reads
3.5 Program performance
Analysis of next-generation sequence data often requires impressive amounts of computational infrastructure; therefore, the performance of software for the analysis of somatic variants can be of great importance. To assess CPU usage and memory requirements of the programs we tested, we noted their running time and memory usage when they were run on our high-performance computing cluster, which consists of 64 bit Linux nodes. Shimmer, SomaticSniper, VarScan2 and deepSNV all make use of the samtools software package (Li et al., 2009), and for indexed BAM files, can be run in parallel by dividing the genome into smaller regions (e.g. chromosomes). Total CPU usage, including extraction of read counts from the BAM files, was comparable for these four programs, all on the order of 10 h for analysis of a single tumor/normal pair’s whole-exome data. In addition, all four of these programs could be run on whole exomes with significantly <4 Gb of physical memory. It should be noted that deepSNV has physical memory requirements which grow prohibitively with the sizes of the regions analyzed. For this reason, it was necessary to analyze only targeted capture regions for whole exome, instead of all regions covered by sequencing reads. For this reason, deepSNV did not scale well enough to be run on whole genome datasets.
JointSNVMix2 analysis of a whole-exome dataset required considerably more CPU time, on the order of 12–24 h for the training step and up to several days for the classify step. In addition, the training step, when run on a pair of whole exomes, required ∼6.5 Gb of memory. This program, as well as deepSNV, could not be run on the COLO-829 whole-genome data in a realistic period; therefore, they were omitted from the second part of our analysis.
4 DISCUSSION
Here, we present results from both real and simulated datasets supporting the use of simple statistical methods, without heuristics or filtering, for the determination of genetic differences between tumors and matched normal samples. It should be no surprise that the use of hypothesis testing with correction for multiple testing provides highly accurate determination of true positive variants while effectively controlling the FDR, as these statistical methods have already been successfully applied in so many fields of scientific research.
We also show that these statistical methods, as implemented in the program Shimmer, perform comparably with other currently available software, such as SomaticSniper, at typical depths of sequencing for whole exome and whole-genome sequencing experiments. Although there does not seem to be a significant advantage to using Shimmer when analyzing samples with high purity, it can be expected to give far superior results on samples that are highly contaminated or contain subclonal heterogeneity.
Furthermore, although Shimmer’s algorithm can be less sensitive at lower depths of sequencing, well-established statistical results allow the user to estimate the depth of coverage necessary to confidently detect somatic mutations present in any level in a tumor. These estimates can then guide decision-making regarding how much sequencing is cost effective in large tumor/normal sequencing studies. Barring difficulties in obtaining independent sampling of molecules because of sparse samples or low library complexity, obtaining more reads is a viable strategy for increasing power to detect mutations present in small quantities in a sample. Regardless of the depth of sequencing performed, the Shimmer algorithm allows the user to decide what their maximum acceptable level of false discoveries will be.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Matthieu LeGallo, Andrea O’Hara, Daphne Bell and Narisu Narisu for testing and giving useful feedback about the Shimmer program and to Peter Chines, Larry N. Singh and our reviewers for critical reading and valuable suggestions for this manuscript. They are also thankful to the NISC Comparative Sequencing Program for providing the NA18506 libraries and sequence data.
Funding: Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Conflict of Interest: none declared.
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Soc. Stat. B. 1995;57:289–300. [Google Scholar]
- Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danacek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, et al. Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinformatics. 2012a;28:167–175. doi: 10.1093/bioinformatics/btr629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012b;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M, et al. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat. Commun. 2012;3:811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
- Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. VarScan2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- Li H, et al. The Sequence/Alignment Map format and Samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble WS. How does multiple testing correction work? Nat. Biotechnol. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance E, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, et al. JoinSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics. 2012;28:907–913. doi: 10.1093/bioinformatics/bts053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat. Genet. 2012;44:165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, et al. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Wang K, et al. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, et al. Antitumor activity of BRAF inhibitor Vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res. 2011;72:779–789. doi: 10.1158/0008-5472.CAN-11-2941. [DOI] [PubMed] [Google Scholar]
- Zhang H, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J. Clin. Invest. 2007;117:2051–2058. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.