Abstract
Summary: Mendel is one of the few statistical genetics packages that provide a full spectrum of gene mapping methods, ranging from parametric linkage in large pedigrees to genome-wide association with rare variants. Our latest additions to Mendel anticipate and respond to the needs of the genetics community. Compared with earlier versions, Mendel is faster and easier to use and has a wider range of applications. Supported platforms include Linux, MacOS and Windows.
Availability: Free from www.genetics.ucla.edu/software/mendel
Contact: klange@ucla.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
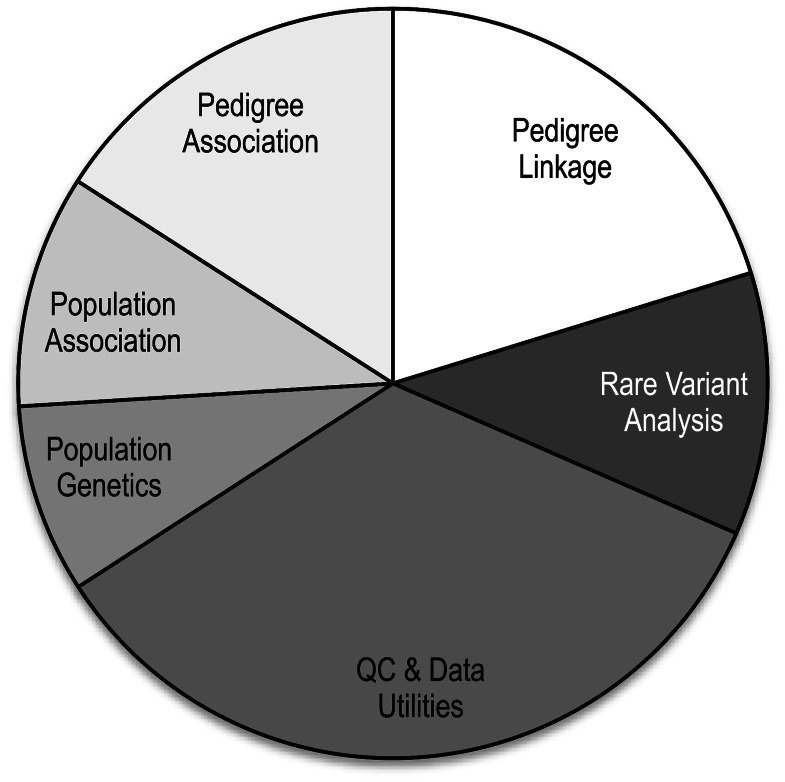
The statistical genetics package Mendel (Lange et al., 2001) has transitioned from its early emphasis on parametric linkage analysis (Lange et al., 1988) to become one of the most comprehensive packages available for the statistical analysis of genetic traits. Given the rapidly changing nature of genetic data analysis, from pedigree linkage to association in unrelated individuals (Risch and Merikangas, 1996) and back toward pedigrees (Ott et al., 2011), it seems timely to alert the genetics research community to the new features of Mendel. Table 1 lists its 29 current options in order of their introduction. These options cover most of the critical tasks of genetic epidemiology and population genetics including gene mapping by association and linkage (Fig. 1). Mendel can handle data from ascertained and randomly selected pedigrees, case-control studies, random samples and experimental crosses. Many of the latest options address genome-wide association study (GWAS) data and rare variant sequence data (Zhou et al., 2010, 2011). Finally, a number of Mendel options have features that deal with the chores of data manipulation and quality control (for example, Ayers and Lange, 2008; Lange and Sinsheimer, 2004; Sobel et al., 2002).
Table 1.
Mendel analysis options
| Option number | Analysis name | Option number | Analysis name |
|---|---|---|---|
| 1 | Mapping markers | 16 | Combining alleles |
| 2 | Linkage analysis | 17 | Gene dropping |
| 3 | Pedigree haplotyping | 18 | Combining loci |
| 4 | Non-parametric linkage (NPL) | 19 | Variance components |
| 5 | Mistyping | 20 | QTL association |
| 6 | Allele frequencies | 21 | Trim pedigrees |
| 7 | Risk calculation | 22 | Association given linkage |
| 8 | Gamete competition | 23 | SNP imputation (haplotyping) |
| 9 | Pedigree selection | 24 | GWAS |
| 10 | Kinship | 25 | File conversion |
| 11 | Genetic equilibrium (HWE/LE) | 26 | MFG incompatibility |
| 12 | Association by permutation | 27 | Inbred strains |
| 13 | TDT | 28 | Simulate traits |
| 14 | Penetrance estimation | 29 | Pedigree GWAS |
| 15 | Ethnic admixture |
Fig. 1.
Proportion of Mendel options devoted to various tasks
In designing Mendel, we pay particular attention to three issues: algorithm speed, statistical rigor and a clear user interface. For example, in computing pedigree likelihoods in linkage mapping, Mendel chooses pedigree-by-pedigree either the Elston–Stewart (Elston and Stewart, 1971) or the Lander–Green–Kruglyak algorithm (Kruglyak and Lander, 1998; Lander and Green, 1987), whichever is fastest. For some options, if there are multiple processor cores available, Mendel will parallelize the analysis to decrease the computation time. Mendel includes unique analysis options such as the gamete competition and maternal–fetal genotype incompatibility (MFG) tests. Even in common options, Mendel has many unique features. For example, Mendel will report outlier individuals and pedigrees, provide standard errors on most estimates and perform exact and permutation-based statistical tests. In some options, Mendel extends what is commonly available. For example, in GWAS, Mendel allows model selection via penalized regression with individual predictor and group penalties (Wu et al., 2009; Wu and Lange, 2008; Zhou et al., 2010, 2011). Also in GWAS, Mendel allows environmental predictors as well as single-nucleotide polymorphisms (SNPs), interaction analyses and pedigree-based association tests.
Mendel incorporates powerful optimization tools for maximum likelihood (ML) and penalized estimation (Lange, 2004, 2010). The ML engine accommodates parameter upper and lower bounds and linear constraints. The log-likelihood and parameter values output at each iteration are helpful in computing likelihood ratio and score statistics and diagnosing convergence failures
Although most analysis names in Table 1 are self-explanatory, a few require some elaboration. As one of the oldest, the Allele Frequencies option has evolved over time. It continues to estimate allele frequencies for specified groups and conducts tests of homogeneity across groups using likelihoods ratios. It can exploit pedigree data in estimating allele frequencies even when founders’ genotypes are unknown. If the designated groups are cases and controls, then the homogeneity test is a test of association between disease status and marker alleles. In the absence of group information, the Allele Frequencies option conducts a parsimonious test of Hardy–Weinberg Equilibrium (Zhou et al., 2009).
The Risk Calculation option uses phenotypes and genotypes of family members to calculate the genetic risk of disease for specified individuals in the pedigree. The Gamete Competition option is a parametric version of the transmission disequilibrium test (TDT) that accepts arbitrary pedigrees, quantitative traits and multiple linked markers (Sinsheimer et al., 2000, 2001). The Kinship option calculates the degree of relationship between two relatives, using pedigree information and, if desired, genotypes. This option can quickly estimate both global and local kinship coefficients from dense genome-wide SNP data (Day-Williams et al., 2011). The Ethnic Admixture option (Sinsheimer et al., 2008) estimates the ancestry fractions for each individual. This option has been recently updated to identify the most informative markers for ethnic discrimination and calculate principal components based on these markers.
The Combining Alleles option is helpful in conducting asymptotic tests of linkage or association when low-frequency alleles lead to biased inference. The Gene Dropping option simulates genotypes based on a given genetic map and either ethnic specific allele frequencies or fixed founder genotypes. The resulting data files with user-specified missing data patterns can be entered in subsequent analyses. The Combining Loci option constructs superloci from multiple unordered SNPs. The alleles at these superloci correspond to the possible haplotypes across the SNPs. Because phase is unknown, the superloci are no longer codominant systems. Many of Mendel’s options can leverage superloci and thus gain information over single SNP analyses.
We continue to add new options and refine existing ones. The 2012 version of Mendel added three options. The first of these, MFG Incompatibility, models interactions between maternal and child genotypes (Childs et al., 2010). The classic example is Rh maternal–fetal incompatibility. The second option, Inbred Strains, represents a new approach to quantitative trait locus (QTL) mapping with inbred strains (Zhou et al., 2012). This option implements a mixed effects model that correctly captures polygenic background, handles multivariate traits and copes with pedigrees of arbitrary complexity. The QTL effect is modeled at the mean level as a vector of regression coefficients on the strain origin pair (maternal–paternal) imputed for each individual at the current QTL location along the genome. Finally, the third new option, Simulate Traits, simulates either univariate traits determined by generalized linear models or multivariate Gaussian traits determined by variance components. Mean effects arising from genotypes at a major locus can be included. This new option works in tandem with our Gene Dropping option. Efficient simulation of phenotype and genotype data is key to the development and evaluation of new statistical genetic tests (Ritchie and Bush, 2010).
Two of the new additions to the 2013 version of Mendel provide fast association testing in pedigrees and improved genotype imputation through matrix completion. The association tests in the new Pedigree GWAS option are score tests rather than likelihood ratio tests, which avoids parameter estimation for each SNP in a GWAS. Multivariate traits with missing data can be analyzed. Pedigree structure may be explicitly provided, or the required kinship information can be estimated from the data. The Supplementary File describes the steps a user takes to perform this analysis. It took Mendel <90 s on a standard laptop computer to read, quality check and analyze a set of 46 pedigrees with 807 individuals and 547 458 SNPs. Our new genotype imputation method, implemented in the existing SNP Imputation option, uses matrix completion (Candès and Recht, 2009; Chi et al., 2013), which is a data mining technique that yields good imputation results with lower demands on memory and numerical processing. For both of these new additions to Mendel, we often achieve greater than 100-fold speedups over competing programs.
All of Mendel’s options are documented in a readable, comprehensive manual (www.genetics.ucla.edu/software/Mendel_doc.pdf). Each type of analysis is described in its own chapter and is best understood by consulting the sample input and output files accompanying that chapter. These examples illustrate virtually all facets of Mendel. The sample files also help in mastering the simple formats of the input data files and the keyword strategy of the control file. Putting the commands for a Mendel run in a control file rather than on the command line, eases editing and retention of program directives. Rather than creating or editing the control file directly, users can access Mendel through our Windows or Web frontends, which list each command, many with drop-down menus. The Windows frontend, Gregor, forms part of the Mendel package. The Web frontend, described at www.genetics.ucla.edu/software/MW, offers a rich environment combining data management and statistical analysis.
In practice, the sample input files posted with the documentation serve as templates for users’ input files. Data compression has proved critical in handling large SNP genotype files. Thus, the user can elect to input data via either text files or compressed files in PLINK format (Purcell et al., 2007). The Supplementary File to this Note describes the steps and files of a typical Mendel analysis. Each run of Mendel creates a summary and a detailed output file. The former displays the bottom-line results that most users will want to see first. The latter provides the details of ML estimation, and diagnostics such as summary statistics and outlier warnings.
Mendel is specifically designed to easily interact with other packages, including those for the visualization of analysis results. For example, Mendel is the computational engine within Mendel Enterprise, an enterprise-level clinical database and web application for genetic studies (www.genetics.ucla.edu/software/ME). With tools for collecting, cleaning, storing, mining, displaying and sharing phenotypic and genetic data, Mendel Enterprise is used in large collaborative studies across institutions. The server-based Mendel Enterprise uses the Software-as-a-Service construct and requires a contract.
We distribute the standalone Mendel statistical genetics software free of charge. Mendel, its documentation, installers, frontends and 75 sample analyses can be downloaded from our Web site. Supported platforms include, Linux, MacOS and Windows. During the 2012 calendar year, there were over 1300 unique downloads. We welcome users’ comments and suggestions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participants from our week-long Mendel courses and other users for their suggestions for improvements to the software.
Funding: NIH grants MH059490, GM053275, GM103774 and HG006139.
Conflict of interest: Although the statistical genetics package Mendel is freely available for non-commercial use, the clinical database Mendel Enterprise is provided via service contracts.
References
- Ayers KL, Lange K. Penalized estimation of haplotype frequencies. Bioinformatics. 2008;24:1596–1602. doi: 10.1093/bioinformatics/btn236. [DOI] [PubMed] [Google Scholar]
- Candès EJ, Recht B. Exact matrix completion via convex optimization. Found. Comput. Math. 2009;9:717–772. [Google Scholar]
- Chi EC, et al. Genotype imputation via matrix completion. Genome Res. 2013;23:509–518. doi: 10.1101/gr.145821.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs EJ, et al. Modeling maternal-offspring gene-gene interactions: the Extended-MFG Test. Genet. Epidemiol. 2010;34:512–521. doi: 10.1002/gepi.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day-Williams AG, et al. Linkage analysis without defined pedigrees. Genet. Epidemiol. 2011;35:360–370. doi: 10.1002/gepi.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston RC, Stewart J. A general model for the genetic analysis of pedigree data. Hum. Hered. 1971;21:523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES. Faster multipoint linkage analysis using Fourier transforms. J. Comput. Biol. 1998;5:1–7. doi: 10.1089/cmb.1998.5.1. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc. Natl Acad. Sci. USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K. Optimization. NY, USA: Springer; 2004. [Google Scholar]
- Lange K. Numerical Analysis for Statisticians. 2nd edn. NY, USA: Springer; 2010. [Google Scholar]
- Lange K, et al. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet. Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- Lange K, et al. MENDEL version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Amer. J. Hum. Genet. 2001;69(Suppl.):504. [Google Scholar]
- Lange K, Sinsheimer JS. The pedigree trimming problem. Hum. Hered. 2004;58:108–111. doi: 10.1159/000083031. [DOI] [PubMed] [Google Scholar]
- Ott J, et al. Family-based designs for genome-wide association studies. Nat. Rev. Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Bush WS. Genome simulation: approaches for synthesizing in silico datasets for human genomics. Adv. Genet. 2010;72:1–24. doi: 10.1016/B978-0-12-380862-2.00001-1. [DOI] [PubMed] [Google Scholar]
- Sinsheimer JS, et al. Gamete competition models. Am. J. Hum. Genet. 2000;66:1168–1172. doi: 10.1086/302826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsheimer JS, et al. SNPs and snails and puppy dogs' tails: analysis of SNP haplotype data using the gamete competition model. Ann. Hum. Genet. 2001;65:483–490. doi: 10.1017/S0003480001008843. [DOI] [PubMed] [Google Scholar]
- Sinsheimer JS, et al. Estimating ethnic admixture from pedigree data. Am. J. Hum. Genet. 2008;82:748–755. doi: 10.1016/j.ajhg.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, et al. Detection and integration of genotyping errors in statistical genetics. Am. J. Hum. Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Lange K. Coordinate descent algorithms for lasso penalized regression. Ann. Appl. Stat. 2008;2:224–244. [Google Scholar]
- Wu TT, et al. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25:714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. Association screening of common and rare genetic variants by penalized regression. Bioinformatics. 2010;26:2375–2382. doi: 10.1093/bioinformatics/btq448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. Penalized regression for genome-wide association screening of sequence data. Pac. Symp. Biocomput. 2011;2011:106–117. doi: 10.1142/9789814335058_0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, et al. A heterozygote-homozygote test of Hardy-Weinberg equilibrium. Eur. J. Hum. Genet. 2009;17:1495–1500. doi: 10.1038/ejhg.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, et al. Quantitative trait loci association mapping by imputation of strain origins in multifounder crosses. Genetics. 2012;190:459–473. doi: 10.1534/genetics.111.135095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.