Abstract
Type 1 pili, produced by uropathogenic Escherichia coli (UPEC), are multisubunit fibers that play crucial roles in recognition of and adhesion to host tissues1. During pilus biogenesis, subunits are recruited to an outer membrane assembly platform, the FimD usher, which catalyzes their polymerization and mediates pilus secretion2. The recent crystal structure of an initiation complex provided insights into the initiation step of pilus biogenesis resulting in pore activation, but very little is known about the elongation steps that follow3. To address this question, we determined the structure of an elongation complex whereby FimD is traversed by the tip complex assembly composed of FimC:FimF:FimG:FimH. This structure reveals the conformational changes required to prevent backsliding of the nascent pilus through the FimD pore and also reveals unexpected properties of the usher pore. We show that the usher pore is energetically designed to facilitate substrate passage within the pore and that the circular binding interface between the pore lumen and the folded substrate plays a role in transport by defining a low energy pathway along which the nascent pilus polymer is guided during secretion.
All pilus subunits exhibit an incomplete immunoglobulin (Ig)-like fold lacking β-strand G, leaving a large hydrophobic groove across the subunit surface4-7. Folding of pilus subunits is catalyzed by a periplasmic chaperone (FimC for type 1 pili)8 (Supplementary Fig. 1A). In a mechanism termed “donor-strand complementation (DSC)”, FimC donates the missing β-strand in trans, thereby complementing and stabilizing the pilus subunit’s fold (Supplementary Fig. 1B)4,6. The resulting binary chaperone:subunit complexes are next recruited to an outer membrane assembly nanomachine, the usher (FimD for type 1 pili), for pilus assembly and secretion. Subunit polymerization occurs through a mechanism termed “donor strand exchange (DSE)”, whereby the N-terminal extension (Nte) of the subunit next in assembly replaces the chaperone strand in the groove of the previously assembled (or receiving) subunit (Supplementary Fig. 1B)5.
Ushers consist of five functional domains: an N-terminal domain (NTD), a pore domain which, in the resting state, is occluded by a plug domain, and two C-terminal domains (CTD1 and CTD2). Recently, the crystal structure of the pilus biogenesis initiation complex FimD:FimC:FimH (Supplementary Fig. 1C) inspired a model for subunit recruitment, polymerization, and secretion by the usher (Supplementary Fig. 1D)3. FimH consists of two domains: a pilin domain (FimHP) similar to all other pilus subunits and a receptor binding domain, subsequently referred to as the lectin domain FimHL6. In the FimD:FimC:FimH structure, FimHL is located inside the FimD pore. FimHP is still in DSC interaction with the chaperone FimC bound to CTD1 and CTD2. The usher NTD, stabilized by the plug domain, is free and serves as a recruitment site for the next chaperone:subunit complex in assembly (FimC:FimG (Supplementary Fig. 1D)). Binding to the NTD positions the FimG Nte peptide in an ideal position for DSE to occur3, leading to the dissociation of FimC bound to FimHP. As FimC has no binding affinity for the usher CTDs on its own, it dissociates from the complex. At this point, the entire nascent pilus translocates up inside the pore and the chaperone:subunit complex bound to the NTD transfers to the CTDs in a hand-over step (Supplementary Fig. 1D). The usher NTD is now free to re-enter another cycle of subunit incorporation by recruiting the next chaperone:subunit complex for assembly (FimC:FimF). This model for the subunit-incorporation cycle remains however to be structurally documented. Here we describe the structure of the FimD:FimC:FimF:FimG:FimH elongation complex, a post-initiation state during pilus biogenesis.
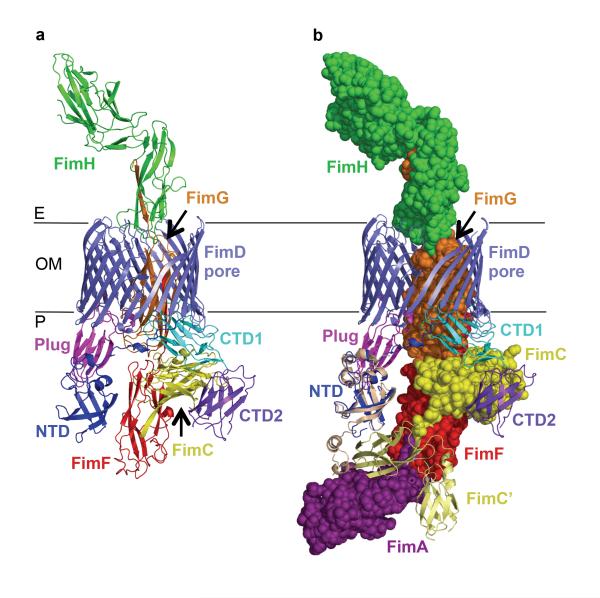
The FimD:FimC:FimF:FimG:FimH complex was purified to homogeneity, crystallized, and its structure determined to a resolution of 3.8 Å (Supplementary Tables 1 and 2 and Supplementary Figs. 2, 3 and 4). The crystal structure of FimD:FimC:FimF:FimG:FimH captures the secretion of the type 1 pilus tip consisting of pilus subunits FimF, FimG and FimH through the usher pore across the outer bacterial membrane (Fig. 1A). The adhesin FimH is incorporated at the distal end of the pilus tip and found on the extracellular side of the transporter. FimG occupies the lumen of the usher β-barrel and contacts the usher CTD1, plug and β-barrel domains. As the last assembled pilus subunit, FimF is found still in DSC interaction with its chaperone FimC, which is bound by the usher C-terminal domains CTD1 and CTD2 in the periplasm. Thus, the structure provides the remarkable view of a transporter entirely traversed by its protein polymer substrate, with one subunit having entirely emerged from the transporter and another still remaining on the cellular side of the transporter. The type 1 pilus tip subunits FimH, FimG and FimF are linked non-covalently by DSE, where the N-terminal extension of FimF is inserted into FimG, and that of FimG into FimHP. The usher NTD shares only an interface with the plug domain and is otherwise exposed. Modeling shows that indeed the NTD is available to bind the next incoming chaperone:subunit complex, FimC:FimA, bringing it into close proximity to FimF and in the appropriate orientation to undergo DSE (Fig. 1B). This is consistent with the DSE model for pilus subunit incorporation introduced by Phan et al.3
Figure 1. Structure of the FimD:FimC:FimF:FimG:FimH complex.
A) Crystal structure FimD:FimC:FimF:FimG:FimH. All proteins are in ribbon representation; FimD NTD, β-barrel, plug domain, CTD1 and CTD2 in blue, slate, magenta, cyan and purple, respectively; FimC, FimF, FimG and FimH in yellow, red, orange, and green, respectively. B) Recruitment of the next chaperone:subunit FimC:FimA complex in assembly by the FimD:FimC:FimF:FimG:FimH complex. FimD is in cartoon representation, coloured as in A. Chaperone FimC (yellow), pilus subunits FimF (red), FimG (orange) and adhesin FimH (green) are in sphere representation. Recruited chaperone:subunit complex FimC’:FimA (PDB ID 4DWH; pale yellow cartoon and purple spheres, respectively) is modelled at the NTD based on the crystal structure of the isolated FimD NTD domain bound to FimC:FimF (PDB ID 3BWU; see Supplementary Methods for details).
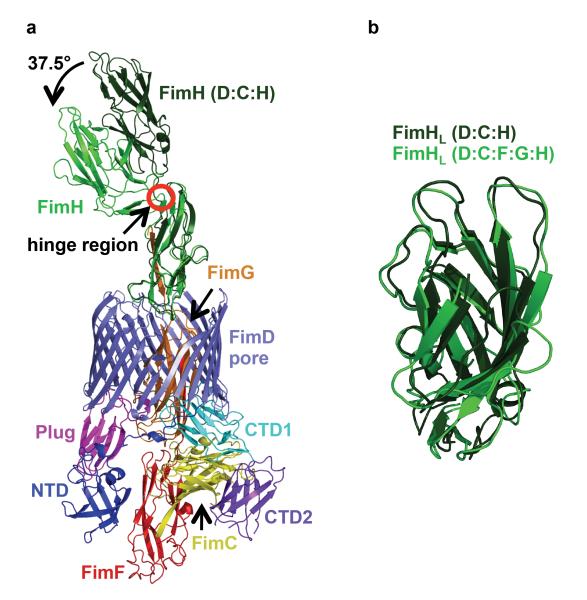
When the FimH proteins in the initiation FimD:FimC:FimH and elongation FimD:FimC:FimF:FimG:FimH complexes are superimposed, it becomes apparent that FimH undergoes a large conformational change when exiting the usher pore with the angle between the two domains closing in (Fig. 2A and Supplementary Fig. 5)9. We speculate that such a conformational change may provide the necessary energy to favor translocation of FimH outside the pore and possibly prevent inwards motion back to the periplasm. Another remarkable structural rearrangement affecting FimH during transport is the compression/decompression of the FimHL domain structure pre/post transport, respectively (Fig 2B).
Figure 2. Comparison of the structure of FimH before and after translocation.
A) Superposition of the structure of FimH from the FimD:FimC:FimH initiation complex (in dark green) with that of FimH from the elongation FimD:FimC:FimF:FimG:FimH complex (same color coding as in Figure 1, panel A). FimHp was used for the superposition. B) Superposition of the structure of FimHL from the initiation complex (dark green) with that of the same domain from the elongation complex (light green).
Using the physically realistic Rosetta energy function10,11, we next investigated the interactions between the usher pore and its inserted substrate, FimHL in the initiation FimD:FimC:FimH complex, and FimG, in the elongation FimD:FimC:FimF:FimG:FimH complex. In these calculations the protein assemblies were represented as essentially rigid components. Subunits were moved along a central pore axis (Supplementary Fig. 6A-C) with only minor deviations that can be sampled by rigid-body minimization, a reasonable assumption due to the close steric fit of subunits within the pore that would otherwise clash.
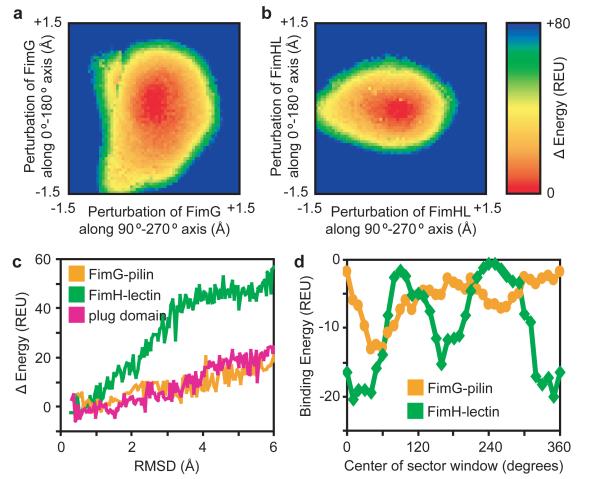
Translating FimG and FimHL subunits within the pore laterally along a fine grid and plotting the calculated energies reveals a steep energy well, with forces on all sides returning the perturbed subunits to their central ground state positions. (Methods; Fig. 3, A and B; Supplementary Fig. 6D). However, when translated parallel to the pore axis to mimic subunit entry or exit, the calculated energies with FimG in the barrel increase slowly (even for 6 Å root mean squared deviation (rmsd) perturbations), whereas with FimHL inside the pore there was a steep energy increase (Fig. 3C and Supplementary Fig. 6E). Thus, the subunit or domain inside the pore is more tightly held in the initiation complex than it is in the elongation complex.
Figure 3. Steep energy funnels and opposing binding surfaces position the translocating substrate at the centre of the pore.
A) The heat map is equivalent to looking down the pore axis, with the energy increase plotted as FimG is laterally translated (i.e. perpendicular to the pore axis) along a finely-spaced grid within the usher lumen. FimG located at any position on the grid will experience a force dependent on the slope of the potential energy well, returning FimG to its central ground state. REU, Rosetta Energy Units. B) As in (A), with FimHL now inside the usher lumen. C) Subunits/domains occupying the pore were randomly rotated within ± 8° about their geometric centres and translated up to 6 Å parallel to the pore axis (for the plug domain, displacement was only in the direction of the periplasm to mimic plug extrusion during activation). Calculated energies are plotted against the rmsd from the respective minimized crystal structures. 8000 perturbations were made for each subunit, with the outline of the lowest energy conformations shown. FimG, orange; FimHL, green; plug, magenta. D) Plot of binding energy as a 60°-sector window emanating from the pore axis is rotated around the respective structures. FimG, orange. FimHL, green.
In apo FimD, the plug domain is found inside the pore12,13 and must be expelled and replaced by FimHL in an activation process that might be triggered by engagement of FimH with FimD. When the plug domain (beginning with the structure of apo-FimD (PDB ID 3OHN)) was translated towards the periplasm to mimic its extrusion from the pore, the energy increased slowly similar to FimG (Fig. 3C). Hence while the plug domain and FimG may readily exit the usher pore along the pore axis, FimHL forms tight interactions. We hypothesise that this may be essential for activation, in which FimHL competes off the plug domain from inside the pore.
By sweeping a 60°-sector window emanating from the pore axis around the usher barrel and computing the binding energy within this window (Fig. 3D; Supplementary Figs. 7 and 9), we observe two binding energy troughs and two peaks that are roughly 180° apart for both FimHL inside the FimD pore of the FimD:FimC:FimH structure and FimG inside the FimD pore of the FimD:FimC:FimF:FimG:FimH structure. Regions of strong binding energy correlate with more intimate atomic contacts as measured by solvent accessibility (Supplementary Fig. 7B), and regions that tightly bind FimG are distinct from those that tightly bind FimHL, indicating specialized binding sites. Usher pores appear to have evolved binding sites that face opposite each other, resulting in the placement of the translocating substrate at the very center of the pore. Since pilin subunits share the same Ig-like fold and have high sequence similarity (compared to FimG, the pilin domains of FimH, FimF, and FimA share 19-25% sequence identity and 62-67% similarity), it is likely that all pilin subunits occupy the same central position in the usher pore, and our results for FimG are generalizable amongst these proteins.
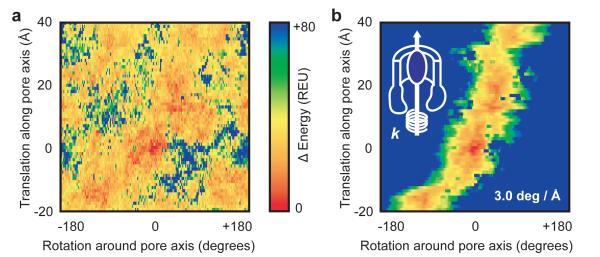
It has been previously proposed that incoming chaperone:pilin subunits are recruited to the usher NTD and then transfer post-DSE to the usher CTDs during assembly (see introduction above). This requires a rotation of the NTD-bound chaperone:subunit complex of about 100-120° after DSE has occurred (Supplementary Fig. 8A). The torque produced from the transfer of the incoming subunit from the NTD to the CTDs might be accommodated through rotations about linker residues between pilus building blocks. Alternatively, rigid body rotation of the entire tip assembly might necessitate a rotation of the translocating subunit within the pore. To investigate such a possibility, we calculated an energy landscape as FimG pilin is rotated by 2° steps and translated by 1 Å steps up/down the usher pore. The calculated landscape reveals a continuous low energy path for FimG as it enters and exits the FimD pore (visualized by following the low energy hot colours in Fig. 4A and Supplementary Fig. 8) that requires a counterclockwise twist. To predict a possible trajectory through the energy landscape, we imposed a torsional spring term to limit the extent to which FimG can rotate at each translation step (this ensures connectivity in the low energy path) and the entry-exit trajectory for FimG pilin is then derived from the lowest energy states (Fig. 4B). By repeating these calculations using input structures minimized with and without constraints from the crystallographic electron density, different spring constants, and structures refined to either 3.8 Å or 4.1 Å resolutions, a twist of 1.6-3.0 degrees per angstrom translation is observed in the trajectories (Supplementary Fig. 8). The presence of a low energy exit path requiring pilin rotation was further validated with an alternatively defined pore axis based on the membrane-like packing of FimD within the crystal lattice (Supplementary Fig. 9). Visual inspection of the FimD:FimC:FimF:FimG:FimH crystal structure reveals that the three assembled pilin subunits representing entering (FimF), inside (FimG) and exiting (FimHP) the usher pore are each separated by 53 Å translations and 110°/120° counterclockwise rotations (Supplementary Fig. 8A). Hence the twist of FimG pilin as it enters-exits the pore captured in the computed energy landscape matches the magnitude and direction of the rotations observed and anticipated from static crystallographic evidence. These results are likely generalizable to other pilin subunits due to high sequence and structural similarity.
Figure 4. A low energy pathway through the pore lumen facilitates translocation of subunits and their transfer from NTD to CTDs.
A) Starting from the native crystal structure with FimG and its complementing strand from FimF inside the FimD pore (subunits FimH, FimF and FimC were not considered), FimG was translated by 1 Å steps up to 40 Å out of the pore, and up to 20 Å back towards the periplasm, along the pore axis. At each translational step, FimG is rotated around the pore axis by 2° increments. Each sampled FimG rotation-translation conformation was minimized after rotamer repacking to resolve small clashes, and the calculated potential energy is plotted against the initial perturbation. Lower, favourable energies are hotter colours. B) The energy landscape calculated in panel A is modified by addition of a torsional spring potential (E = ½.k.Δθ2; represented by the coil with spring constant k twisted around the pore axis in the overlaid schematic at top-left) to derive a connected trajectory, with Δθ the angle from the lowest energy FimG conformation prior to each 1 Å step, and starting with FimG positioned at 0 Å, 0°.
Similar analysis was applied to FimHL within the FimD lumen from its respective crystal structure (FimD:FimC:FimH), but no single, clear low energy entry-exit path was observed (Supplementary Fig. 10). This is not surprising. Indeed, FimHL differs markedly from all other pilus subunits or domains: while FimHP, FimG, FimF or FimA have a classical pilin fold that consists of six β-strands complemented by the donor strand of the next subunit in assembly, FimHL has a β-barrel jelly roll fold. Furthermore, as shown here, the FimD pore forms tighter interactions with FimHL than with a representative pilin subunit. Thus, while all other pilus subunits and domains are easily extracted from and guided through the pore, unbinding and translocation of FimHL will require more energy, likely via conformational changes yet to be characterized.
The crystal structure presented here provides unprecedented structural evidence supporting the model first proposed by Phan et al.3 for the usher-mediated subunit-incorporation cycle during pilus biogenesis. It also demonstrates that usher pores form finely-tuned circular protein-protein interfaces specifically geared to i-actively maximize transport by forming diametrically-opposed binding sites that position substrates at the very center of the pore and ii- facilitate specific steps during transport by imposing rotational and translational constraints through defined energy paths. We anticipate that all transporters will demonstrate similar levels of sophistication by having evolved function-specific features facilitating defined steps during substrate translocation.
Methods
Expression and purification of the FimD:FimC:FimF:FimG:FimH complex
E. coli Tuner (Novagen) was transformed with plasmids pNH237 encoding fimCHis:F:G:H under arabinose control and pAN2 encoding fimDStrep under IPTG control3,13. Bacteria were grown in TB media containing 35mg.ml−1 kanamycin and 25mg.ml−1 chloramphenicol at 37°C. Protein overexpression was induced by addition of 100 μM IPTG and 0.1% (w/v) L-arabinose supplemented with 0.1% (v/v) glycerol at OD600 = 1.0 for 48 hours at 16°C. By not providing the FimA subunit to the assembly system, pilus production stalls after incorporation of the last tip subunit FimF, resulting in production of homogeneous FimD:tip complexes.
Protein purification of the FimD:FimC:FimF:FimG:FimH complex
The FimD:FimC:FimF:FimG:FimH complex was purified as described in Phan et al3 for the FimD:FimC:FimH complex.
Crystallization of the FimD:FimC:FimF:FimG:FimH complex
Plate-like crystals of FimD:FimC:FimF:FimG:FimH were grown in hanging drops at 20°C using the vapour diffusion method, where 1 μl of the complex (8mg.ml−1) was mixed with an equal volume of 1.2-2.0 M sodium formate and equilibrated against 600 μl reservoir solution. Crystals were transferred into mother liquor with 25% (v/v) glycerol as cryoprotectant and then flash frozen in liquid nitrogen.
Structure determination of the FimD:FimC:FimF:FimG:FimH complex
Data were collected at Diamond (beamline IO2) at 100 °K and were indexed, integrated and scaled to 3.8 Å resolution using the XDS software package14. High resolution cut off of the data was chosen according to Karplus and Diederichs15 (Supplementary Table 1). The space group was determined by POINTLESS16. Space group, cell dimensions, and data collection statistics are reported in Supplementary Table 1.
The phase problem was solved by molecular replacement (MR) using PHASER and the structures of FimD (PDB ID 3RFZ), FimC:FimF (PDB ID 3BWU) and FimG:FimH (PDB ID 3JWN) as search models17. The MR model for FimD:FimC:FimF:FimG:FimH obtained by PHASER was refined by alternating rounds of model building with COOT18 using B factor sharpened maps generated with PHENIX19 and refinement cycles with CCP4 REFMAC20, CNS21 and PHENIX19. Initial rigid body refinement (rigid bodies were assigned for subunits FimC, FimF, FimG and FimH and for the β-barrel, plug, N-terminal and the two C-terminal domains of FimD) was carried out by REFMAC and resulted in a Rfree of 45%. For the next round of refinement we followed the CNS protocol for low resolution refinement which includes torsion angle annealing, grouped B factor and deformable elastic network (DEN) refinement21,22. Two B factors were assigned to each amino acid, one to the main chain atoms and one to the side chain atoms. FimC (PDB ID 3BWU), FimD (PDB ID 3RFZ), FimF (PDB ID 3BWU), FimG (PDB ID 3JWN) and FimH (PDB 3JWN) were used as DEN reference files. After a two-dimensional grid search for the DEN parameters using the SBGrid Science Portal webservice, DEN parameters γ and ω were set 0.2 and 10023. DEN parameter γ balances the influences of the diffraction data and the reference model, whereas ω is the weighting factor for the DEN potentials. The starting annealing temperature was also optimized employing the SBGrid Science Portal DEN web service and set to 1500 K. In the final rounds of DEN refinement, the starting and reference model were the same. DEN refinement changed the positioning of the N- and C-terminal domains which appear to be quite flexible, as well as the shape of the usher β-barrel, and therefore improved Rfree considerably. DEN refinement converged at Rfree of 31%. The final refinement rounds were carried out with PHENIX. The refinement protocol included 3 rounds of cartesian and individual B factor refinement. The atomic displacement parameters and stereochemistry weights were optimized during the refinement and resulted in tight restraints. Finally, Rfree converged at 29.8%. The same Rfree flag was maintained for cross validation throughout the refinement process. Note that high-resolution data to a resolution of 3.8 Å were included because the resulting final model had a better Rfree value, improved stereochemistry and improved electron density (Supplementary Tables 1 and 2, Supplementary Figs. 3 and 4).
Computational Methods
For analysis of static structures, the refined crystal structures were relaxed in the Rosetta force field using 2 rounds of FastRelax and Cartesian space minimization (Frank DiMaio, manuscript in preparation) with electron density constraints24. B-factor sharpened 2mFo-DFc density maps were generated in PHENIX19 using deposited structure factors for the FimD:FimC:FimH complex or structure factors to 4.1 Å resolution for the FimD:FimC:FimF:FimG:FimH complex. Rosetta command line and score term weights are detailed in Supplementary Fig. 11. The electron density-constraining score term accounts for ~40% and ~55% of the total score for FimD:FimC:FimF:FimG:FimH and FimD:FimC:FimH, respectively. Relaxed decoys from 50 relaxation runs were well converged both in total score (scores ranged from -5964 to -5911 for FimD:FimC:FimF:FimG:FimH, and -6376 to -6326 for FimD:FimC:FimH) and structure (rmsd to an arbitrary reference decoy was ≤ 0.22 Å for FimD:FimC:FimF:FimG:FimH and ≤ 0.14 Å for FimD:FimC:FimH). The lowest scoring decoys (Supplementary. Fig. 12 shows fit of relaxed structures to electron density) were chosen for final analysis in Fig. 3, and Supplementary Figs. 7, 8G-H and 9E-H. The Rosetta program’s correlation coefficient for measuring agreement between coordinates and a density map24 decreased by 3% (FimD:FimC:FimF:FimG:FimH) and 2% (FimD:FimC:FimH) following relaxation, indicating a small trade-off between minimizing structural features within Rosetta’s force field and maintaining a close fit to the experimental electron density map.
To generate FimD sectors, residues were included if the Cα atom’s projection on a plane perpendicular to the pore axis fell within the sector’s desired angle range, with the Cα projection of FimD-Asp208 set to 0°. This creates a PDB file containing FimG-pilin (aa 12-144)/FimF-Nte (aa 1-12) or FimH-lectin (aa 1-157) bound to only a subset of FimD residues. Binding energy and buried solvent accessible surface area were calculated using Rosettascripts (Supplementary Fig. 13) without rotamer repacking in either the bound or unbound states (rotamer repacking at the edges of a FimD sector would be meaningless)25. As a ‘sanity check’, the combined binding energy from any set of sectors adding up to a full 360° sweep consistently approximates the binding energy calculated for the complete FimD protein bound to its transported substrate.
Translational and rotational perturbations were made using the convpdb.pl application in the MMTSB suite26. Perturbed structures were minimized in Rosetta using full atom score12 with rounds of rotamer repacking, and side chain and backbone gradient minimization (Supplementary Fig. 14-16)11,25. In these cases where large dynamic subunit motions were explored (Figs. 3C and 4, and Supplementary Figs. 6, 8B-F, 9I and 10), starting structures were also first pre-minimized in full atom score12, ensuring that any rmsd measurements to the initial ground state accurately reflected the applied perturbations, not simply differences from minimizing in different score functions.
For determining potential FimG trajectories through the FimD pore, noise in the energy landscape was dampened by averaging the energy at each sampled FimG rotation-translation with the energies calculated for FimG rotations within 4°. Starting with FimG at 0°-0 Å, a torsional spring potential (E = ½.k.Δθ2) was added to Rosetta’s calculated potential energy. Δθ is measured to the lowest energy conformation at the previous translational step.
Structure figures were generated with PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
Supplementary Material
Acknowledgments
This work was funded by Medical Research Council grant 85602 to GW. DB and EP are supported by grant P41 GM103533 from the National Institute of General Medical Studies at the US National Institutes of Health. We thank the staff of beamline ID23-1 at ESRF and IO2 at Diamond and Dr. Ambrose Cole for technical assistance during data collection.
Footnotes
Declaration of any competing interests. The authors have no competing interests.
Accession number. The atomic coordinates and structure factors of the FimD:FimC:FimF:FimG:FimH complex have been deposited in the PDB (PDB ID 4J3O).
References
- 1.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science. 2008;320:376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- 3.Phan G, et al. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer FG, et al. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 5.Sauer FG, Pinkner JS, Waksman G, Hultgren SJ. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell. 2002;111:543–551. doi: 10.1016/s0092-8674(02)01050-4. [DOI] [PubMed] [Google Scholar]
- 6.Choudhury D, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 7.Zavialov AV, et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113:587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- 8.Vetsch M, et al. Pilus chaperones represent a new type of protein-folding catalyst. Nature. 2004;431:329–333. doi: 10.1038/nature02891. [DOI] [PubMed] [Google Scholar]
- 9.Le Trong I, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β-sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das R, Baker D. Macromolecular modeling with Rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 11.Leaver-Fay A, et al. Rosetta3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Smith BS, Chen LX, Baxter RH, Deisenhofer J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc Natl Acad Sci U S A. 2009;106:7403–7407. doi: 10.1073/pnas.0902789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remaut H, et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 17.McCoy AJ, et al. PHASER crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of COOT. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 23.O’Donovan DJ, et al. A grid-enabled web service for low-resolution crystal structure refinement. Acta Crystallogr D Biol Crystallogr. 2012;68:261–267. doi: 10.1107/S0907444912001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using Rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleishman SJ, et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feig M, Karanicolas J, Brooks CL., 3rd MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.