Abstract
Large scale production of hepatocytes from a variety of genetic backgrounds would be beneficial for drug screening and to provide a source of cells to be used as a substitute for liver transplantation. However, fully functional primary hepatocytes remain difficult to expand in vitro and circumventing this problem by using an alternative source of cells is desirable. Here, we describe a 25 day protocol to direct the differentiation of human pluripotent stem cells into a near homogenous population of hepatocyte-like cells. As cells progress through this protocol they express genes in a chronological manner similar to that described during in-vivo hepatic development. The protocol relies on culture systems devoid of serum, feeders or complex extra-cellular matrices enabling molecular analyses without interference from unknown factors. This approach works efficiently with human embryonic stem cells and human induced pluripotent stem cells and was recently used to model liver diseases in vitro.
INTRODUCTION
The liver has a very broad spectrum of functions during adult life. It processes consumed substances, sustains reserves of iron, vitamins and minerals, stores glycogen and produces bile for digestion of lipids, albumin (Alb) and blood clotting factors. Finally, the liver detoxifies alcohol, drugs and other chemicals as well as removing inhaled poisons such as exhaust or smoke, all of which accumulate in the bloodstream. Most of these activities are managed by one cell type, the hepatocyte, which constitutes the main cellular unit of the liver.
End stage liver disease that targets one or several of these critical liver functions must be dealt with by orthoptic liver transplantation (OLT) since only a healthy donor liver can restore the missing metabolic function. However, this ultimate solution does not represent a cure since it has a high risk of surgical complications, indefinite immunosuppression associated with severe side effects, and eventually leads to organ rejection. Furthermore, whilst the number of organ donors has remained constant over the past 10 years the demand for liver transplantation has more than doubled. This situation is likely to continue to worsen in the foreseeable future due to a Hepatitis C pandemic and increases in cirrhosis associated with obesity. Thus the development of alternative therapies to OLT has become a major objective in the field of regenerative medicine.
A cell based therapeutic approach involving transplantation of healthy hepatocytes into the livers of affected patients may facilitate a complete correction of all aspects of the clinical syndrome associated with liver failure. Such an approach already carries precedence within clinical practice since nearly 100 patients suffering Inherited Metabolic Diseases (IMDs) have to date received donor hepatocytes to treat a variety of inherited liver diseases including A1ATD, Glycogen storage disorders (GSD), OCTD, Crigler Najjar, and factor VII deficiency 1. Similarly, the development of bio-artificial liver (BAL) fuelled with primary hepatocytes could represent an advantageous alternative for bridge therapy in patients suffering of acute liver failure. Primary hepatocytes are also needed for diverse in vitro applications including drug development and toxicology screening by the pharmaceutical industry.
Unfortunately, all these applications are limited or impaired by the scarce number and quality of available donor cells. Indeed, primary hepatocytes are fully differentiated cells that are impossible to expand in vitro. As a consequence, hepatocytes can only be obtained from organ donation and often only livers of poor quality are available for cell purification. For all these reasons, an alternative source of cells to generate fully functional hepatocytes is urgently needed.
Human pluripotent stem cells (hPSCs) derived from embryos at the blastocyst stage (human Embryonic Stem Cells or hESCs)2 or from reprogrammed fibroblasts by overexpression of pluripotency factors (human Induced Pluripotent Stem Cells or hIPSCs)3 could represent an advantageous system to produce hepatocytes like cells from a wide diversity of genetic backgrounds. Indeed, these pluripotent cells share the unique characteristic to self renew in-vitro while maintaining the capacity to differentiate into a broad number of cell types. By combining these unique properties, hESCs / hIPSCs could enable the generation of a large quantity of cells for clinical applications, including hepatocytes4.
Experimental Design
Here, we describe the protocol we used to derive the cells described in references 5 & 6. This protocol directs differentiation of hPSCs into a near homogenous population of foetal like hepatocytes which can undergo functional maturation following prolonged culture (Figure 1). This protocol has been validated with 5 hESCs (H9, Val9, FES22/29, and hSF-6) and 20 hIPSCs to model IMDs including Alpha-1-Antytripsin Deficiency, Glycogen Storage Disorder 1 alpha, and hypercholesterolemia 5,6 (See Table 1). The procedure uses chemically defined media (CDM) devoid of serum and complex extra-cellular matrices such as Matrigel. Thus, this culture system could easily be transferred to GMP conditions and avoids the presence of unknown factors which could interfere with molecular analyses.
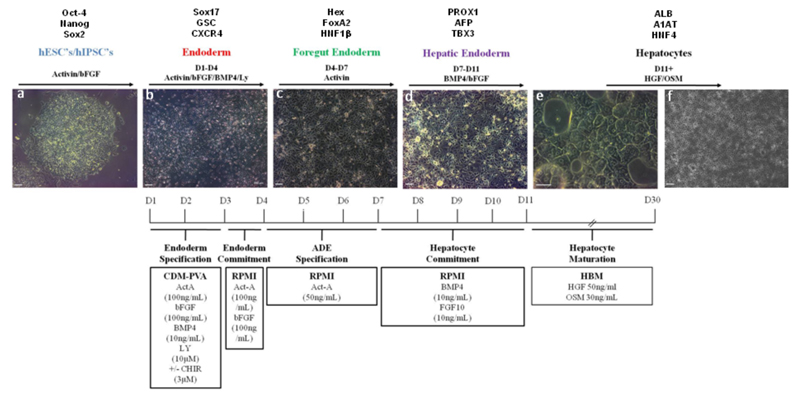
Figure 1. Protocol to differentiate hPSCs into hepatocytes.
Bright field pictures showing differentiation of hPSCs (H9, WiCell) into hepatocytes following a natural path of development. Panel a: Pluripotent cells are organised in tightly packed colonies with well-defined borders and display a high nuclear to cytoplasmic ratio. Panel b: Definitive endoderm specification starts with the migration of cells away from the original colony and with an increase in cell size. Panel c: Foregut endoderm cells have a polygonal / cobblestone shape while being organised into an epithelium. Panel d: Hepatic endoderm display canaliculi-like structures with a dark cytoplasm. As cells differentiate further, lipid vesicles can be observed and multi-nucleated cells appear. Panel e and f: hepatocyte-like cells generated after 25 days of differentiation . Differentiation time line detailing developmental stages with expected gene expression, basal medium and growth factor combinations and concentrations are provided (Text above and below panels). Scale bar: 200 μm.
Table 1.
hESC and hIPSC lines differentiated into artificial hepatocytes.
| Cell Line | Origin | Ref. |
|---|---|---|
| H9 | hESCs (WiCell, US) | 2 |
| hSF6 | hESCs (UCSF, US) | 13 |
| Val9 | hESCs (CIPF, Spain) | 14 |
| FES22 | hESCs (University of Helsinki, Finland) | 15 |
| FES29 | hESCs (University of Helsinki, Finland) | 15 |
| BBHX6 BBHX7 BBHX8 |
hIPSC lines from dermal fibroblast / Patient Caucasian female 28 yo (Cambridge University, UK) |
5 |
| IPS40 IPS48 IPSC54 |
hIPSC from foreskin fibroblast (Cambridge University, UK) | 5 |
| MRC5.1 | hIPSC from embryonic lung fibroblast (Cambridge University, UK) |
16 |
| Bi.1 Bi.2 Bi.3 |
hIPSC from embryonic dermal fibroblast / Patient Caucasian 70 yo (Cambridge University, UK) |
16 |
| A1AT-1.1 A1AT-1.2 A1AT-1.3 |
hIPSC line from dermal fibroblast /patient Caucasian male 65 yo with A1AT deficiency (Cambridge University, UK) |
5 |
| A1AT-2.1 | hIPSC line from dermal fibroblast /Caucasian male 2 mo with A1AT deficiency (Cambridge University, UK) |
5 |
| GSD 1.1 GSD 1.2 GSD 1.3 |
hIPSC line from dermal fibroblast /patient Caucasian male 25 yo with glycogen storage disorder. (Cambridge University, UK) |
5 |
| LDL1 | hIPSC line from dermal fibroblast /patient Caucasian with familial hypercholesteremia. (Kremlin Bicetre, France) |
5 |
| CN1 | hIPSC line from dermal fibroblast /patient Caucasian 2 mo with Crigler Najjar syndrome. (Cambridge University, UK) |
5 |
| HT1 | hIPSC line from dermal fibroblast /patient Caucasian 2 mo with hereditary tyrosinaemia. (Cambridge University, UK) |
5 |
The protocol comprises four stages which mimic the embryonic development of the liver and enable the production of differentiated cells following a natural path of development. The procedure starts at the beginning of the differentiation protocol, with splitting and experimental plate set-up. This day is designated Day 0 (D0). In the first stage, hPSCs are differentiated into definitive endoderm (DE) cells which represent the earliest precursors of all endodermal organs (Liver, Pancreas, Lung, Gut, Thyroid). The second stage differentiates these DE cells into anterior definite endoderm (ADE) cells, which represent a common progenitor between Liver, Pancreas, Lung, and Thyroid. During the third stage ADE cells differentiate into hepatic progenitors. During the fourth and final stage of the protocol the resulting hepatocytes like cells undergo functional maturation. Each stage is marked by the expression of specific genes while the hepatocytes like cells obtained after 25 days of differentiation display functional characteristics shared by both foetal and adult hepatocytes including Albumin and A1AT secretion, Glycogen storage, Cholesterol up take, and inducible detoxification activity of cytochrome P450 enzymes including CYP3A7, CYP1A1. As maturation is continued foetal characteristics begin to diminish while functions associated with adult functionality increase to reach a maximum level at Day 35.
The distinct gene expression profile exhibited at each stage of the protocol can be used to determine the developmental time point of hepatic cells generated in vitro. For example, Alpha-Fetoprotein (AFP)7 and CYP3A7 represent respectively the major serum protein and cytochrome enzyme expressed by the liver during foetal life8 while high levels of albumin and CYP3A4 activity are found in hepatocytes from the adult liver8. The cells resulting from the protocol described below still express significant levels of AFP and CYP3A7 while also expressing markers of adult hepatocytes such as alpha-1-antitrypsin, apolipoprotein-F, CYP1A1, CYP3A4, CYP1A2 and CYP1B1. Furthermore, the cells generated by this protocol display functional activity including uptake indocyanine green and cholesterol, inducible cytochrome P450 activity and secretion of Albumin and Alpha-1-Antitrypsin5,6. Therefore, hepatic cells produced using the protocol described here could be intermediate between adult and foetal hepatocytes. However, only a systematic comparison between primary and foetal hepatocytes obtained at different stages of development will enable definition of the precise level of maturity of hepatocytes generated in-vitro. For these reasons, these cells are best described as an hepatocyte like cell model. Despite the uncertainty surrounding their precise developmental identity, the hepatocytes like cells derived using this protocol can be useful to model in vitro a broad number of inherited metabolic disorders5 and are able to colonise the liver of animal model for 8 weeks while maintaining the expression of Albumin and ATT6. Identification of the appropriate maturation period should be determined using primary hepatocytes as a positive control for the desired hepatic functionality and output, similar to the controls detailed below. With regard to genetic modification, genes can be easily overexpressed or knocked down in hepatocyte like cells (approximately Day20-Day35) using gene therapy approach. As an example, we have shown that a lentivirus vector containing an APOF promoter driving the expression of the GFP reporter gene can be useful to purify artificial hepatocytes. Flow cytometry analyses showed that 85% of cells expressed GFP 3 days after transduction9.
Whilst this method has worked with the majority of hESCs and hIPSCs grown in our laboratory around 20% of hIPSC lines appear to be resistant to endoderm differentiation and thus we have not been able to generate hepatic cells from these lines. The variability between lines remains difficult to explain and can be partially corrected by optimising the concentration of each growth factor especially during the first stage of the protocol. Indeed, DE differentiation appears to be the most critical part of our approach and thus, we recommend this stage is optimised to ensure successful differentiation toward the hepatic pathway. Furthermore, day 25 of this protocol represents the earliest time point at which these cells can be used to model basic hepatic metabolic function. However, cells differentiated at day 35 systematically display highest level of Albumin secretion and CyP activity and thus represent the optimal stage for functions analyses such as toxicology screen. Finally, we recommend to use primary hepatocytes (available from Lonza, Biopredict and Invitrogen) as positive control for the basic characterisations performed on Hepatocyte like cells especially Q-PCR analyses, albumin secretion and CYP3A4 activity (representative analyses and expected outcomes are described in detail later.)
MATERIALS
Critical
- All the tissue culture reagents listed are stored, reconstituted and used as specified in the product sheet supplied by the manufacturer unless specifically stated.
- Recombinant proteins and Chemical inhibitors/activators should be stored at −20°C after reconstitution.
- Kits used for functional validations should be used as specified by the manufacturer on or after D25 of differentiation.
REAGENTS
hPSC,(either hESCs or hIPSCs)
Ca2+ and Mg2+ - free Dulbecco’s PBS (PBS−/−) (Gibco 14190)
RPMI 1640 + GLUTAMAX (Gibco 61870)
IMDM (Gibco 21980)
F12 + GlutaMAX −1 (Gibco 31765)
B27 Supplement containing insulin (Gibco 17504-044)
Hepatocyte Basal Medium (Lonza CC-3199)
MEM Non-Essential Amino Acids (MEM-NEAA) (Gibco 1140)
Bovine Serum Albumin (BSA) fraction V (Europa Bioproducts, SIGMA or PAA)
Gelatine – Porcine (Sigma G1890)
β-Mercaptoethanol (Sigma M6250)
Foetal Bovine Serum (Biosera S1818)
Glutamine (Invitrogen 25030024)
Thioglycerole >=97% (Sigma M6145)
Dulbecco’s Modified Eagle’s Medium (DMEM)
DMSO (Sigma D2650).
Poly Vinyl Alcohol (Sigma P8136)
Activin-A (R&D System 338-AC)
Basic Fibroblast Growth Factor (bFGF) (R&D Systems 233-FB)
Bone Morphogenetic Factor 4 (BMP-4) (R&D Systems 314-BP)
Oncostatin M (R&D Systems 295-OM)
Hepatocyte Growth Factor (Peprotech 100-39)
LY294002 (Promega V1201)
CHIR99021 (StemGent 04-0004)
Fibroblast Growth Factor 10 (FGF10) (Autogen Bioclear ABC144)
Transferrin (Roche 10652202001)
Insulin (Roche 11376497001)
Concentrated Lipids – Chemically Defined (Gibco 11905)
Penicillin/Streptomycin (Pen/Strep) (Gibco 15140122)
Collagenase Type IV (Invitrogen 17104-019)
Dispase (Gibco 17105-041)
4% Paraformeldeyhide (PFA) (Alfa Aesar 16% w/v aq. Sol. Methanol free 43368)
Donkey Serum (Serotec CO6SB)
Reagents required for characterisation of cells
CRITICAL protocols for immunohistochemistry and Q-PCR are not given here; see refs 5, 6, 9 &16
 Alpha-Feto-Protein Primary Antibody (DAKO A0008)
Alpha-Feto-Protein Primary Antibody (DAKO A0008) Albumin Primary Antibody (R&D 188835)
Albumin Primary Antibody (R&D 188835) Alpha-1-Antitrypsin Primary Antibody (DAKO A0012)
Alpha-1-Antitrypsin Primary Antibody (DAKO A0012) CXCR4 (R&D MAB173)
CXCR4 (R&D MAB173) SOX17 (R&D AF1924)
SOX17 (R&D AF1924) HNF4a (C-19) (SantaCruz sc6556)
HNF4a (C-19) (SantaCruz sc6556) CK18 (ab82254)
CK18 (ab82254) Alexa Flour 488 – secondary Antibody
Alexa Flour 488 – secondary Antibody Alexa Flour 647 – secondary Antibody
Alexa Flour 647 – secondary Antibody Cell Dissociation Buffer – Enzyme Free Hanks-based (13150-016)
Cell Dissociation Buffer – Enzyme Free Hanks-based (13150-016) Periodic Acid-Schiff (PAS) Kit (Sigma 395B-1Kit)
Periodic Acid-Schiff (PAS) Kit (Sigma 395B-1Kit) Indocyanine Green (Sigma 21980)
Indocyanine Green (Sigma 21980) LDL-Kit (Cayman Chemicals 10011125)
LDL-Kit (Cayman Chemicals 10011125) Albumin Blue (Active Motif 15002)
Albumin Blue (Active Motif 15002) A1AT ELISA Kit (AbCam AB108799)
A1AT ELISA Kit (AbCam AB108799) P450 Glo Assay (Promega V9920, V9770, V9790)
P450 Glo Assay (Promega V9920, V9770, V9790) Primers Sequence for Q-PCR are given in Supplementary Table 1
Primers Sequence for Q-PCR are given in Supplementary Table 1
EQUIPMENT
Inverted Microscope – required for characterisation of cells
Horizontal Laminar Flow Hood or biosafety cabinet suitable for tissue culture
CO2 incubator controlling temperature (37°C), humidity, and CO2 (5%)
Cell Culture Centrifuge
10cm Tissue Culture Plates
12-well Tissue Culture Plates
6-well Tissue Culture Plates
Gelatine/MEF coated tissue culture plates
15mL Tissue Culture Tubes
50mL Tissue Culture Tubes
REAGENT SET UP
Serum containing medium for coating tissue culture plates (500mL)
Combine 450mL DMEM/F12, 50mL FBS, 5mL Glutamine, 5mL Pen/Strep, 3.5μL β-mercaptoethanol. Media can be stored at 4°C for up to 1 month after opening.
CRITICAL STEP
- Sterilize Serum-containing media using an 0.22μm filtration device.
- Serum coating can be replaced by human fibronectin 15g/ml (Millipore, cat 341635). However, this alternative is less cost effective and might be less efficient with specific hIPSC lines. Thus, we recommend serum coating for routine use and for media optimisation with new hPSC lines.
CDM-BSA medium for maintenance of human embryonic stem cells (500mL)
BSA 2.5 grams, DMEM-F12 250mL, IMDM 250mL, Concentrated lipids 5mL, Thioglycerole 20μL, Insulin 350μL Transferrin 250μL, Pen/Strep 5mL. Media can be stored at 4°C for up to 1 month after opening.
CRITICAL STEP
- Sterilize CDM-BSA media using an 0.22μm filtration device.
- Medium should be warmed to 37°C prior to use.
- BSA batches must be screened for ability to support the undifferentiated growth on hESCs. Not all batches of BSA fraction V are suitable and must therefore be pre-screened. We found suitable batches from SIGMA, PAA and Europa-bioproducts. Quality of the resulting CDM-BSA should be compared against standard feeder cultures with particular emphasis on maintenance of colony morphology and undifferentiated growth characteristics as well as continued expression of POU5f1, NANOG and SOX2 with early differentiation markers such as CDX2, T, MIXL1, SOX17 at levels comparable to the feeder cultures.
CDM-PVA medium for maintenance of human induced pluripotent stem cells and differentiation to endoderm (500mL)
PVA 0.5 grams, DMEM-F12 250mL, IMDM 250mL, Concentrated lipids 5mL, Thioglycerole 20μL, Insulin 350μL, Transferrin 250μL, Pen/Strep 5mL. Media can be stored at 4°C for up to 1 month after opening.
CRITICAL STEP
- PVA needs to be dissolved in 50mL of IMDM by placing 0.5g PVA in a 50ml tube with IMDM and placed on a tube roller overnight at 4C.
- Sterilize CDM-PVA media using an 0.22μm filtration device.
- Medium should to be warmed to 37°C prior to use.
RPMI-B27 differentiation medium for differentiation of ADE and hepatoblast specification (500mL)
RPMI 1640 490mL, B27 10mL, NEAA 5mL, Pen/Strep 5mL. Medium should to be warmed to 37°C prior to use. Media can be stored at 4°C ; use within 3 weeks.
Gelatine Solution for coating tissue culture plates (500mL)
Gelatine 0.5g, Embryo transfer water 500mL, heat to 56°C until gelatine dissolves, cool to room temperature (18-25°C).
CRITICAL STEP
- Sterilize Gelatine solution using an 0.22μm filtration device
- Gelatine may be stored for 4-8 weeks
Collagenase for dissociation of hESCs and hIPSCs colonies (500mL use within 4 weeks)
DMEM-F12 400mL, Knockout Serum Replacer Medium 100mL, Glutamine 5mL, β-mercaptoethanol 3.5μL, collagenase Type IV 500mg, mix by inversion.
CRITICAL STEP
- Sterilize Collagenase using an 0.22μm filtration device and then aliquot into 50mL tubes.
- Collagenase should to be warmed to 37°C prior to use.
Hepatocyte Basal Medium (500mL)
Prepared as per the manufacturers specifications including bullet kit. Medium should to be warmed to 37°C prior to use. Media can be stored at 4°C for up to 1 month after opening.
Dispase for dissociation of hIPSCs (500mL)
Mix and filter 500mg dispase with 500mL DMEM. Media can be stored at 4°C for up to 1 month after opening.
CRITICAL STEP
- Sterilize Dispase using an 0.22μm filtration device and then aliquot into 50mL tubes.
- Dispase should to be warmed to 37°C prior to use.
Dispase:Collagense (1:1) for passaging hIPSCs or hESCs (Use within 2 weeks)
Mix dispase and collagenase at a ratio of 1:1 to obtain the required volume for passaging cells. Dispase:Collagenase (1:1) should to be warmed to 37°C prior to use.
Small molecules
Dissolve LY294002 and CHIR99021 in DMSO at a stock concentration of 10 mM.
CRITICAL STEP
- Check that small molecules are well dissolved and then store at −20C.
Maintenance of hPSCs
Maintain hPSCs in CDM containing Activin-A (10ng/mL) and bFGF (12ng/mL) as described in references 5, 6 & 9 in a humidified tissue culture incubator at 37°C, 5% CO2 and atmospheric O2 10. Medium should be replaced daily and all growth factors added directly to the medium prior to use.
CRITICAL STEP
- Differentiation efficacy can be increased by splitting hPSCs grown on feeders or in CDM directly to culture conditions inductive for mesoderm differentiation described below. (CDM-PVA + Activin + FGF2 + BMP4 + LY + CHIR). See Figure 1 for differentiation schematic.
- FACS analyses can be performed to confirm that 80-90% of the cells express the pluripotency markers Oct-4 and Tra-1-60.
- Growth factors cannot be kept at 4C for longer than 1 week and basal medium cannot be kept more than 4 weeks at 4°C.
- All growth factors should be added directly to fresh medium each day – making bulk batches of medium containing growth factors is not advisable unless all medium will be used within 2-3 days.
EQUIPMENT SETUP
Gelatine/ serum coated tissue culture plates
Coat desired numbers of plates for maintenance and differentiation using gelatine solution at room temperature (approximately 18-25°C) for 30 minutes. Aspirate gelatine and replace with serum containing medium, place at 37°C for at least 24hrs.
CRITICAL STEP
Serum coated plates must be prepared at least 24 hours prior to use and can be stored at 37C for 7-14 days.
PROCEEDURE
Passaging hPSCs for differentiation
1) Day 0: Take hPSCs that are 80% confluent, aspirate medium and wash with 1 volume of Ca2+/Mg2+-free PBS (PBS−/−). Minimal volumes will depend on the plate used, however, we recommend 0.5mL per well of a 12-well plate, 1mL per well of a 6 well plate and 5mL per 10cm dish.
2) Aspirate the PBS and replace with a minimal volume of collagenase:dispase solution. Incubate at 37°C until 90% of colonies entirely lift from the bottom of the plate (>30mins). Typically most colonies will have detached from the bottom of the plate after 60 minutes.
3) Transfer cells to a 15mL tube. Wash colonies by centrifuging at 180g for 2 minutes, removing supernatant, and resuspending in CDM-PVA without cytokines. Repeat this wash but resuspend the pellet in CDM-PVA containing Activin and bFGF. The volume of CDM containing Activin and bFGF should be appropriate to facilitate the appropriate splitting ratio (see critical step).
CRITICAL STEP
hPSCs should be split so that after 6-8 days of culture cells will be 80% confluent and ready for passaging again. This is usually obtained if cells are split at a ratio of between 1:6-10. For example a 1x 10cm tissue culture plate could be split into 6-10x 10cm tissue culture plates, 6-10x 6-well plates or 6-10x 12-well plates as needed. However this ratio will vary depending on the hESC or hIPSC line and must be determined by the user. For the cells we have used, a typical cell plating density is 200,000 cells per 10cm plate.
4) Resuspend the colonies using a 10ml serological pipette to facilitate breakup of large colonies into smaller pieces. For 12-well plates take 1mL of cell suspension and add to 17 ml of CDM-PVA containing Activin-A and bFGF and then distribute 1.5 ml of this cell suspension to each well of the 12-well plate.
5) Incubate cells under normal conditions overnight.
6) Day 1: Replace medium with fresh CDM-PVA with Activin and bFGF and incubate cells a further day.
?TROUBLESHOOTING
Endoderm differentiation of hPSCs
7) Day 2: Induce differentiation by replacing medium with freshly prepared CDM- PVA containing Activin-A (100ng/mL), bFGF (100ng/mL), BMP-4 (10ng/mL), LY294002 (10μM), CHIR99021 (3μM) and incubate for 24 hours only. Typical morphology of cells undergoing differentiation can be seen in Figure 2 panel a.
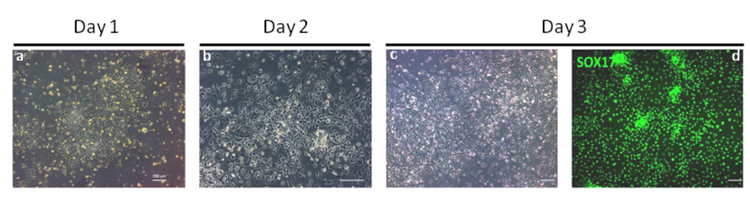
Figure 2. Microphotographs showing endoderm differentiation of hPSCs.
This stage is the most critical of the differentiation protocol as the production of homogenous population of DE cells is necessary for the efficient production of artificial hepatocytes. At Day 1 hESCs begin to undergo EMT marked by migration of cells away from the pluripotent colonies and start to express the primitive streak markers T, MIXL1 and EOMES17. By Day 2 significant migration of cells has occurred and the original colony is no longer visible. The expression of T, and MIXL1 is downregulated while the expression of SOX17 and CXCR4 is induced17. Panel a and b are bright field images of cell morphology on days 1 and 2 respectively. Panel c and d: By Day 3, endoderm cells homogenously express SOX17 (Panel c, bright field image; Panel d, Antibody: R&D AF1924) as shown by immunostaining4. Under optimal conditions >90% of cells are CXCR4+/SOX17+ by FACS analyses17. Scale bar: 200 μm.
CRITICAL STEP
- CHIR99021 is included in the first day of differentiation only. It should not be included in any subsequent days of differentiation at any stage of the protocol.
?TROUBLESHOOTING
8) Day 3: Replace medium with freshly prepared CDM-PVA with Activin-A (100ng/mL), bFGF (100ng/mL), BMP-4 (10ng/mL), LY294002 (10μM). This change in media should induce endoderm specification over the next 24 hours. Incubate cells for 24 hours only. Typical morphology of cells undergoing differentiation can be seen in Figure 2 panel b.
CRITICAL STEP
- CHIR99021 should not be included on this or any subsequent day’s medium changes.
9) Day 4: Replace medium with freshly prepared RPMI-B27 medium containing Activin-A (100ng/mL), bFGF (100ng/mL). This change in media should lead to endoderm commitment over the next 24 hours. Incubate cells for 24 hours Typical morphology of cells undergoing differentiation can be seen in Figure 2 panel c.
CRITICAL STEP
- FACS analyses can be performed at the end of the 24h incubation (on Day 5) to confirm that 60-90% of the cells express the endoderm marker CXCR4 and/or SOX17.
- Q-PCR may also be performed to demonstrate SOX17, CXCR4 expression and absence of mesoderm, MIXL1, T gene expression as well as extraembryonic, CDX2, and neurectoderm, PAX6.
Differentiation to Anterior Definitive Endoderm (ADE)
10) Days 5 - 7: Aspirate medium and replace with RPMI-B27 differentiation medium with Activin-A (50 ng/ml). Incubate cells for 3 days, replacing media every 24hrs (three times). ADE is specified over a three day period with medium changes every 24 hours until the third day (Day 8) when Hepatic differentiation may be induced as described in Step 7. Typical morphology of cells undergoing differentiation can be seen in Figure 2 panel b.
CRITICAL STEP
- Anteriorising the endoderm takes three days and is counted from D5, including D5, D6 and all of D7. On D8 cells are then specified to the hepatic lineage as described in Step 11.
Specification of Hepatoblast
11) Days 8 - 12: Aspirate medium from cells and replace with RPMI-B27 medium containing BMP-4 (20ng/mL) and FGF10 (10ng/mL). Incubate cells for 3 days, replacing media every 24hrs (three times). Then incubate for a further 24h. Typical morphology of cells undergoing differentiation can be seen in Figure 3 panels a-c.
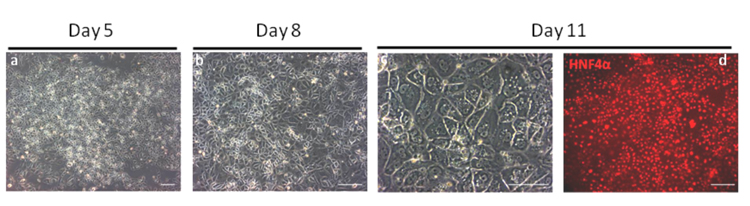
Figure 3. Microphotographs showing Hepatic Specification.
By day 5 of the protocol, differentiated cells starts mesenchymal to epithelium transition and express anterior foregut marker such as Sox17, FOXA2 and HHEX. At day 8, cells form a monolayer and express FOXA1, FOXA2 and HHEX. Panel a and b are bright field images of cell morphology on days 5 and 8 respectively. Panel c and d: At Day 11, hepatic cells display a specific appearance including a dark cytoplasm (Panel c, bright field image), light nucleus, and canaliculated borders. These cells express homogenously hepatic markers such as HNF4a (Panel d, Antibody SantaCruz sc6556) as shown by immunsotaining4. Under optimal conditions >90% of cells are HNF4+ by FACS analyses17. Scale bar: 200 μm.
CRITICAL STEP
- Hepatic specification takes four days and is counted from the beginning of D8, including D8, D9 D10 and all of D11. The maturation phase starts on D12 as described in Step 12.
- Successful Hepatic Specification can be monitored by analysing the the expression of HNF4, PROX1, HHEX, AFP and TBX3 by Q-PCR. HNF4 expression should be expressed in 90% of the cells and day 10 of the differentiation.
? TROUBLESHOOTING
Maturation of hepatoblast into artificial hepatocytes
12) Day 12 onward: Replace medium with an appropriate volume of hepatocyte basal medium supplemented with OSM (30ng/mL) and HGF (50ng/mL). Incubate cells, replacing media every 2 days. Typical morphology of Hepatocytes like cells can be seen in Figure 4 panel a.
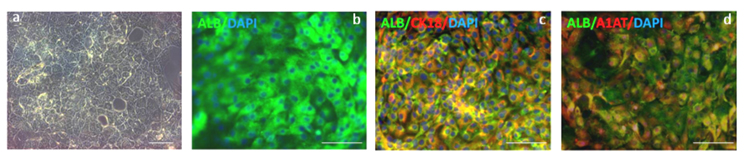
Figure 4. Hepatocytes like cells generated after 20 days of differentiation.
Cells expressing markers specific to hepatocytes such as albumin and α-1-antitrypsin are obtained after 20 days of differentiation. Panel a: The resulting cells display a distinct morphology common to primary human hepatocytes including multinuclei, distinct canaliculated border, and polygonal appearance. Panel b-d: Immunostaining analyses showing that under optimal conditions >85% ALB+/A1AT+ cells can be obtained (Panel b: immunostaining for Albumin (green; antibody R&D 188835); Panel c, co-immunostaining for Alb (green; antibody R&D 188835) and CK18 (red; antibody ab82254) Panel d, co-immunostaining for Alb (green; antibody R&D 188835) and A1AT(red; antibody DAKO A0012))4. Panels b,c and d are all also counterstained with DAPI (blue) to show nuclei. Scale bar: 200 μm.
CRITICAL STEP
- Medium should be changed every second day during the maturation phase of the differentiation protocol. Morphological characteristics of hepatocytes such as polygonal shape, multi-nucleated cells, canaliculi-like structures appear at approximately at Day 15 of the protocol along with gene expression of immature foetal hepatocytes such as AFP and CYP3A7. Albumin, APOF and α-1-antitrypsin can be detected in the medium as early as Day 15 and inducible cytochrome P450 activity can be observed by Day 20. Protein secretion, cytochrome activity, LDL uptake, and glycogen storage are reliably detected and assayed between Day 20-Day 30. Cell survival beyond this time can vary depending on the cell type and cell line, therefore we recommend screening several maturation media. Finally, FACS analyses can be performed to confirm that at least 80% of the cells co-express AFP, ALB and A1AT after 25 days of differentiation. Longer maturation will result in a decrease in the number of AFP expressing cells and an in crease in ALB/AAT expression.
- Cell proliferation during maturation is very limited and thus, splitting hepatocytes like cells is not possible.
? TROUBLESHOOTING
TIMING
Day 0 (steps 1-5)
Approximately 2 hour will be required to passage and plate cells for maintenance and experimental purposes.
Day 1 (steps 6)
Only a medium change is required and should take approximately 10 - 20 minutes to complete.
Day 2 (step 7)
The first day of differentiation will require approximately 30 minutes to prepare the medium, aspirate previous medium and replace with fresh medium.
Day 3 (step 8)
Approximately 20-30 minutes to prepare medium with cytokines, aspirate previous days medium and add to tissue culture plates.
Day 4 (step 9)
Approximately 10 minutes to add cytokines to medium, aspirate old medium and add fresh medium.
Day 5 - 7 (step 10)
Approximately 10 minutes to add cytokines to medium, aspirate old medium and add fresh medium.
Day 8 - 11 (step 11)
Approximately 10 minutes to add cytokines to medium, aspirate old medium and add fresh medium.
Day 12 onwards (step 12)
Approximately 10 minutes every second day of the protocol will be required in order to add fresh cytokines to medium, aspirate old medium and add fresh medium.
TROUBLESHOOTING
For Troubleshooting guidance see Table 2.
Table 2.
TROUBLESHOOTING
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| 4 and 5 | Poor Attachment of hPSCs to tissue culture plates |
Variability between lines | Add Rho kinase inhibitor Y- 27632 in the medium during passaging and for the next 24 hours after passaging. |
| 6 | Decrease of pluripotency following passaging |
Break-up of hPSC colonies is not optimal |
If clumps are too big (>3000 cells), the middle of the colony differentiates disturbing the downstream endoderm specification. This can be easily avoided by vigorous pipeting of the clumps during passaging. If clumps are too small (<200 cells), hPSC will differentiate becoming unsuitable for downstream liver differentiation. This can be avoided by decreasing enzymatic treatment and by gently dissociating clumps during passaging. |
| 7, 8 and 9 | Suboptimal differentiation | Variability between lines | DE differentiation is the most important part of the protocol. Indeed, artificial hepatocytes are systematically obtained as long this step of the protocol works efficiently. Thus optimisation of the first part of the protocol is essential.. For that, we recommend to define the most efficient concentration of FGF and CHIR in step 1. For that, cells should be differentiated in increasing/decreasing dose of CHIR and / or bFGF and then DE differentiation can be monitored using QPCR/Immuno and flow cytometry analyses. At least 60% of the cells should express those markers to ensure an efficient hepatic differentiation |
| 11 | Suboptimal differentiation | BMP 4 effect | BMP4 might have a negative effect on hepatoblast specification in some hPSC lines. Thus, step 7 can be replaced by extending step 6 for 3 additional days (i.e. 3 additional days in RPMIB27 supplemented with Activin). Importantly, increasing or decreasing dose of BMP4 has no effect on hepatoblast specification in vitro. |
| 12 | Suboptimal maturation | Variability between lines | Hepatoblast derived hPSCs respond differentially to a variety of basal cell culture media and thus optimisation might be necessary for specific cell lines. Hepatozyme can be used instead of HBM. |
Table 2 / Troubleshooting Table.
ANTICIPATED RESULTS
Following the protocol above, approximately 15 million immature hepatocytes can be generated from 600 000 hPSCs plated on a 10cm plate. Typical results that are obtained from following this protocol can be seen in Figures 2,3,4 and Supplementary Figure 1 and Supplementary Figure 2. Importantly, cell death can be observed during Step 7-8 and we recommend decreasing the dose of LY294002 if this issue becomes too important. Q-PCR analysis should demonstrate that the cells generated at day 27 of this protocol express ALB, A1AT, APOF, TAT, TDO2, TTR and CYP3A7 (Supplementary Figure 1 and see previous report4); Expression of HNF4, A1AT and ALB should be seen by immunostaining (Figure 3 and 4) and FACS4. ELISAs can be used to demonstrate secretion of Albumin and A1AT (Supplementary Figure 1). The cells should show inducible detoxifying function associated with cytochrome P450 activity (Supplementary Figure 1C), store glycogen4 as shown by PAS staining, up take Low Density Lipoprotein (LDL)4 and indocyanin Green (Supplementary Figure 1D). The methods we used to characterise hPSCs derived hepatocytes (FACS / ICC / Q-PCR / CYP3A4 / PAS staining / LDL up take / Albumin and ATT ELISA protocols) have been described elsewhere 4,11,5,10,12,9. Importantly, at Day 25, we expect that 80% of the cells generated with this protocol express key markers such as ALB and A1AT4. These cells also continue to express foetal hepatocytes markers such as AFP and CYP3A7 while the expression of adult markers such as CyP3A4 remains limited (Supplementary Figure 1). However, functional characteristics of hepatocyte like cells (Alb/A1AT secretion, CYP450 activity) continue to increase after Day 25 and reach their maximal level between Day 31-Day 35 (Supplementary Figure 1 ). Furthermore, the expression of foetal hepatocytes markers such as AFP and Cyp3A7 decreases progressively between Day 25 and Day 35 (Figure 5A). After Day 35, hepatocytes like cells progressively dye while their performance start to decrease. We recommend using primary hepatocytes as positive control for characterising hepatocyte like cells (Supplementary Figure 1).
Supplementary Material
Supplementary Table 1: Primer sequences for Q-PCR analyses of gene expressed during differentiation of artificial hepatocytes.
Supplementary Figure 1: Characterisation of hESCs derived Hepatocytes like cells and Comparison with Primary Hepatocytes. (A) Expression of hepatic markers during differentiation of hESCs. Q-PCR analyses showing the progressive decrease in AFP expression as cells start to mature and at the same time increase expression of the hepatic makers (Albumin and A1AT) during maturation. Primary Hepatocytes (Biopredicts) were used as positive control. Data are presented as the average of 3 biological replicates and error bars indicate standard deviation. (B) ELISA analyses showing increase in Albumin and A1AT secretion during maturation of hepatocytes like cells. Primary Hepatocytes (Biopredicts) were used as positive control. Data are presented as the average of 3 biological replicates and error bars indicate standard deviation. Medium only was used as negative control (C) hPSCs derived hepatocytes cells also display detoxifying activity associated with cytochrome P450 family members such as CyP3A4. This activity can be further induced by chemical such as dexamethasone. (D) Cells also display the ability to take up and metabolise indocyanine green as described by Yamada et el 200218 . Scale bar: 200 μm
Supplementary Figure 2: Comparison of hPSC derived hepatocytes to human primary fetal hepatocytes. Foetal hepatocytes differentiated from hPSC’s share an expression profile very similar to in-vivo derived primary human foetal hepatocytes. AFP, ALB, APOF and CYP3A4 show very similar expression levels when compared between the two cell types, while A1AT does appear to be generally higher along with several other mature metabolic genes such as such as tyrosine amino-transferase (TAT), tryptophan 2,3-dioxygenase (TDO2) and transthyretin (TTR).
Acknowledgement
This work was funded by MRC Senior non-clinical Fellowship (T.T. and L.V.), the Cambridge Hospitals National Institute for Health Research Biomedical Research Center (L.V.), the EU grant LivES and the Evelyn Trust (N.H.), and the Children’s Liver Disease Foundation (CPS).
Footnotes
Author Contributions statement:
NRFH: Protocol concept, design, interpretation, validation and optimisation
CPS: Protocol Optimisation & Validation
TT: Protocol optimisation and validation
LV: Protocol concept, design, interpretation, validation and optimisation
The authors declare that they have no competing financial interests
REFERENCES
- 1.Fisher RA, Strom SC. Human hepatocyte transplantation: worldwide results. Transplantation. 2006;82:441–449. doi: 10.1097/01.tp.0000231689.44266.ac. [DOI] [PubMed] [Google Scholar]
- 2.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Rashid ST, Vallier L. Induced pluripotent stem cells--alchemist’s tale or clinical reality? Expert Rev Mol Med. 2010;12:25. doi: 10.1017/S1462399410001596. [DOI] [PubMed] [Google Scholar]
- 5.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusa K, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bader D, et al. Alpha-fetoprotein in the early neonatal period--a large study and review of the literature. Clin Chim Acta. 2004;349:15–23. doi: 10.1016/j.cccn.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Stevens JC, et al. Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther. 2003;307:573–582. doi: 10.1124/jpet.103.054841. [DOI] [PubMed] [Google Scholar]
- 9.Touboul T, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 10.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, et al. The E47 transcription factor negatively regulates CD5 expression during thymocyte development. Proc Natl Acad Sci U S A. 2004;101:3898–3902. doi: 10.1073/pnas.0308764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez RT, et al. Manipulation of OCT4 levels in human embryonic stem cells results in induction of differential cell types. Exp Biol Med (Maywood) 2007;232:1368–1380. doi: 10.3181/0703-RM-63. [DOI] [PubMed] [Google Scholar]
- 14.Aguilar-Gallardo C, et al. Derivation, characterization, differentiation, and registration of seven human embryonic stem cell lines (VAL-3, -4, -5, -6M, -7, -8, and -9) on human feeder. In Vitro Cell Dev Biol Anim. 2010;46:317–326. doi: 10.1007/s11626-010-9285-3. [DOI] [PubMed] [Google Scholar]
- 15.Mikkola M, et al. Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Dev Biol. 2006;6:40. doi: 10.1186/1471-213X-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallier L, et al. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells. 2009;27:2655–2666. doi: 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- 17.Teo AK, et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada T, et al. In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells. 2002;20(2):146–54. doi: 10.1634/stemcells.20-2-146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Primer sequences for Q-PCR analyses of gene expressed during differentiation of artificial hepatocytes.
Supplementary Figure 1: Characterisation of hESCs derived Hepatocytes like cells and Comparison with Primary Hepatocytes. (A) Expression of hepatic markers during differentiation of hESCs. Q-PCR analyses showing the progressive decrease in AFP expression as cells start to mature and at the same time increase expression of the hepatic makers (Albumin and A1AT) during maturation. Primary Hepatocytes (Biopredicts) were used as positive control. Data are presented as the average of 3 biological replicates and error bars indicate standard deviation. (B) ELISA analyses showing increase in Albumin and A1AT secretion during maturation of hepatocytes like cells. Primary Hepatocytes (Biopredicts) were used as positive control. Data are presented as the average of 3 biological replicates and error bars indicate standard deviation. Medium only was used as negative control (C) hPSCs derived hepatocytes cells also display detoxifying activity associated with cytochrome P450 family members such as CyP3A4. This activity can be further induced by chemical such as dexamethasone. (D) Cells also display the ability to take up and metabolise indocyanine green as described by Yamada et el 200218 . Scale bar: 200 μm
Supplementary Figure 2: Comparison of hPSC derived hepatocytes to human primary fetal hepatocytes. Foetal hepatocytes differentiated from hPSC’s share an expression profile very similar to in-vivo derived primary human foetal hepatocytes. AFP, ALB, APOF and CYP3A4 show very similar expression levels when compared between the two cell types, while A1AT does appear to be generally higher along with several other mature metabolic genes such as such as tyrosine amino-transferase (TAT), tryptophan 2,3-dioxygenase (TDO2) and transthyretin (TTR).