Abstract
Ci-MRF is the sole myogenic regulatory factor (MRF) of the ascidian Ciona intestinalis, an invertebrate chordate. In order to investigate its properties we developed a simple in vivo assay based on misexpressing Ci-MRF in the notochord of Ciona embryos. We used this assay to examine the roles of three structural motifs that are conserved among MRFs: an alanine-threonine (Ala-Thr) dipeptide of the basic domain that is known in vertebrates as the myogenic code, a cysteine/histidine-rich (C/H) domain found just N-terminal to the basic domain, and a carboxy-terminal amphipathic α-helix referred to as Helix III. We show that the Ala-Thr dipeptide is necessary for normal Ci-MRF function, and that while eliminating the C/H domain or Helix III individually has no demonstrable effect on Ci-MRF, simultaneous loss of both motifs significantly reduces its activity. Our studies also indicate that direct interaction between CiMRF and an essential E-box of Ciona Troponin I is required for the expression of this muscle-specific gene and that multiple classes of MRF-regulated genes exist in Ciona. These findings are consistent with substantial conservation of MRF-directed myogenesis in chordates and demonstrate for the first time that the Ala/Thr dipeptide of the basic domain of an invertebrate MRF behaves as a myogenic code.
Keywords: chordate, Ciona, evolution, MRF, muscle, myogenic code
Introduction
Myogenic regulatory factors (MRFs) are basic helix-loop-helix (b-hlh) transcription factors that play important roles in metazoan muscle development (reviewed by Baylies and Michelson, 2001; Pownall et al., 2002; Buckingham etal ., 2003; Tajbakhsh, 2005; Berkes and Tapscott, 2005; Tapscott, 2005). Vertebrates have four MRFs with distinct but overlapping functions that are essential for myogenesis and that are distinguished from other b-hlh transcription factors by their ability to induce muscle in non-muscle cell types (Weintraub et al., 1989; Rudnicki et al., 1992; Venuti et al., 1995; Kassar-Duchossoy et al. 2004; Tapscott, 2005; Bryson-Richardson and Currie, 2008). Invertebrates also possess MRFs that induce myogenesis when expressed in non-muscle cells (Venuti et al., 1991; Krause et al., 1992; Meedel et al., 2007), but differ from vertebrates in usually having only a single MRF. Additionally, in some invertebrates such as Drosophila and C. elegans myogenesis occurs in the absence of MRF activity (Chen et al., 1994; Balagopalan et al., 2001), whereas in others such as Ciona intestinalis it does not (Meedel et al., 2002; 2007). These differences are consistent with the possibility that the roles of MRFs in myogenesis have changed significantly during evolution (but see Olson and Klein, 1998; Fukushige et al., 2006).
Detailed studies of MRF structure/function relationships and gene regulatory mechanisms in vertebrates have focused on three conserved structural motifs: an alanine-threonine (Ala-Thr) dipeptide of the basic region often referred to as the myogenic code, a cysteine/histidine (C/H) rich domain just N-terminal to the b-hlh domain, and an amphipathic α-helix near the carboxyl terminus known as Helix III. All three motifs play important roles in regulating muscle-specific gene activity in vertebrates. Individually, however, their importance varies depending on the target gene thus indicating that vertebrate MRFs regulate different muscle genes by distinct mechanisms (Brennan et al., 1991; Davis and Weintraub 1992; Schwarz et al., 1992; Rawls et al., 1995; Gerber et al., 1997; Kablar et al., 1997; Wang and Jaenisch, 1997; Bergstrom and Tapscott, 2001; de la Serna et al., 2001; Myer et al., 2001; Berkes et al., 2004; Cao et al., 2006 Heidt et al., 2007). Consistent with this idea, vertebrate MRFs have been shown to bind to consensus E-box motifs of some genes and non-consensus E-boxes of others (Blackwell and Weintraub, 1990; Heidt et al., 2007), to bind non-E-box motifs (Shklover et al., 2007), and to interact directly or even indirectly with different muscle genes via associations with a variety of transcription factors (Molkentin et al., 1995; Groisman et al., 1996; Berkes et al., 2004; Ohkawa et al., 2006; Albini and Puri, 2010; Liu et al., 2010; Delgado-Olguín et al., 2011).
Structure/function studies with vertebrates have provided important insights into the mechanisms by which muscle gene activity is regulated by MRFs, but invertebrate systems such as Ciona intestinalis have much to offer as well. Ciona has only a single MRF gene and smaller families of many MRF target genes (Meedel et al., 1997; Dehal et al., 2002; Chiba et al., 2003). Thus, MRF-regulated myogenesis in Ciona offers the advantage of simplicity when compared to vertebrates whose multiple MRFs regulate both common and distinct sets of genes, and that in some cases have different roles at common gene targets (Rawls et al., 1995; Kablar et al., 1997; Wang and Jaenisch, 1997; Bergstrom and Tapscott, 2001; Myer et al., 2001; Cao et al., 2006). However, like vertebrates, Ciona is a chordate that requires MRF activity for muscle development so its analysis is likely to provide insights relevant to understanding the properties of vertebrate MRFs. Ciona also has a number of other features that make it ideally suited for studying developmental gene regulatory mechanisms. These include ease of obtaining large numbers of gametes, simple methods of fertilization and embryo culture, rapid and synchronous development that can be studied at single cell resolution, and the availability of efficient gene introduction techniques (Corbo et al., 2001; Kumano and Nishida, 2007). We took advantage of these attributes to carry out detailed studies of Ci-MRF and here report the first functional analysis of the C/H, Helix III, and Ala-Thr motifs of an invertebrate MRF. As in vertebrates, all three motifs were found to be necessary for normal Ci-MRF activity. Also similar to vertebrates, our studies identified multiple classes of MRF-regulated genes in Ciona and provided evidence for a direct interaction between CiMRF and an essential E-box of a muscle-specific gene. These findings extend our understanding of the properties of conserved MRF motifs and establish Ciona as a useful experimental system for further exploring MRF regulatory mechanisms.
Materials and Methods
Plasmid Construction
A vector containing approximately 3.3kb of the cis-regulatory region and the 5′ untranslated region of the Ciona intestinalisBrachyury gene (Ci-Bra) was constructed to drive Ci-MRF expression in the notochord. ~3kb of this sequence was obtained as an XhoI/PciI fragment from the plasmid T3.5m5GFP (gift of R. Zeller); we obtained the remaining ~0.3kb cis-regulatory region and 5′ untranslated region by PCR of T3.5m5GFP using the primers 5′TTTTGACATGTCAATCAAAATCGG3′ and 5′CGACTGCAGTATAGGTTTGTAACTCGCACT3′. This smaller fragment was digested with PciI and PstI and cloned into XhoI/PstI digested pSP72 (Promega) along with the larger 3kb XhoI/PciI fragment to create pTReg, which in addition to containing Ci-Bra cis-regulatory sequences contained several restriction sites of pSP72 that were suitable for cloning. We chose this large fragment of Ci-Bra for our studies because it has been shown to give robust and faithful expression of reporter genes in the notochord, with only occasional misexpression in the mesenchyme lineage (Corbo et al., 1997).
Our studies required the use of plasmids that express full-length Ci-MRF transcripts and since no cDNAs encoding the 5′ termini of these mRNAs were available we used PCR to prepare a 0.35kb fragment from genomic DNA that encoded the 5′ untranslated region and N-terminal coding sequences common to both Ci-MRF mRNAs. The primers used for PCR were 5′CGATCTGCAGAAATCCAGCCGGTAGTTTGAC3′ and 5′CAACCAGACGCCATATTACTGAGC3′ and the resulting product was digested with PstI and SacI and cloned into pBluescript II KS (+) to create pCiMRF5′. A plasmid encoding full-length CiMRFa, designated pTCiMRFa, was constructed by excising the insert of pCiMRF5′ with PstI and SacI and cloning it into PstI/BamHI digested pTReg together with a 1.5kb SacI/BamHI fragment from plasmid pMD6.3 that contained the remainder of CiMRFa (Meedel et al., 1997). pTCiMRFb, encoding full-length CiMRFb, was constructed in a similar manner by ligating together PstI/SacI digested pCiMRF5′, PstI/SalI digested pTReg and a 2.3kb SacI/SalI fragment from plasmid pc9m3.5 that contained the remainder of the CiMRFb coding sequence (Meedel et al., 1997).
We used the QuickChange Site-directed Mutagenesis Kit (Stratagene) to introduce mutations in pMD6.3 and pc9m3.5 that replaced the sequence encoding the alanine398-threonine399 dipeptide encoded by Ci-MRF with a sequence coding for an asparagine dipeptide. Primers were designed using Stratagene’s web-based PrimerDesign software for mutagenesis and were: 5′ACACGACCGGCGGAGGGCAAACAATCTACGAGAGAGACGACGCC3′ and 5′GGCGTCGTCTCTCTCGTAGATTGTTTGCCCTCCGCCGGTCGTGT3′. The resulting cDNA clones were sequenced to verify that only the desired changes were incorporated, and the strategy described above to construct pTCiMRFa and pTCiMRFb was used to prepare misexpression plasmids pTCiMRFaNN and pTCiMRFbNN that encoded the alanine398-threonine399 to asparagine-asparagine mutation.
pTCiMRFaΔCH and pTCiMRFbΔCH are plasmids from which Ci-MRF sequences encoding amino acids 365–385 (HYHH·····CKAC) are deleted (this deletion corresponds to bases 1093–1155 beginning with the ATG start codon of CiMRFb; Genbank accession number U80080). Epoch Life Sciences supplied a plasmid construct (pCiMRFΔCH) with a 927 base pair insert spanning bases 277–1256 of CiMRFb but with bases 1093–1155 deleted and containing an EcoRI site followed by four random bases at the 3′ end to facilitate cloning. To create pTCiMRFaΔCH this 927 base pair insert was digested with SacI and AflII and inserted into SacI/AflII digested pMd6.3. The resulting plasmid (pMd6.3ΔCH) was digested with SacI and SalI and together with the PstI/SacI insert of pCiMRF5′ was cloned into PstI/SalI digested pTReg to create pTCiMRFaΔCH. pTCiMRFbΔCH was constructed in a similar manner, except that the insert from pBSKCiMRFΔCH was excised with SacI and BsmBI and inserted into SacI/BsmBI digested pBSCiMRFb (created by ligating together the PstI/SacI insert of pCiMRF5′, the 2.3 kb SacI/SalI insert of pc9m3.5, and PstI/SalI digested pBluescript KS II+) to create pBSCiMRFbΔCH. The insert of pBSCiMRFbΔCH was excised with PstI and SalI and cloned into PstI/SalI digested pTReg to create pTCiMRFbΔCH.
We constructed a negative control plasmid, pTLacZ, by subcloning a 3.6kb BamHI/BglII fragment from pSP72.127βgal (gift of R. Zeller; Corbo et al., 1997) into BamHI/BglII digested pTReg. pSP72.127βgal was derived by Corbo et al. (1997) from pPD1.27 (Fire et al., 1990) and in addition to the LacZ coding region contains an SV40 nuclear localization signal and an SV40 polyadenylation sequence that were also incorporated into pTLacZ.
For coelectroporation experiments two plasmids containing regulatory sequences of the Ciona intestinalisTnI gene driving LacZ expression were constructed. Ci500nZ contained wild-type TnI regulatory sequences necessary and sufficient to drive robust expression of LacZ in the muscle lineage (Khare et al., 2011); details of its construction have been presented previously (Khare et al., 2011), where it was referred to as CiTnI(-836/-335)nZ. A second plasmid, Ci500EboxSDMnZ, containing a mutated E-box sequence (CAGCTG → acGCgt) was constructed from Ci500nZ by overlap extension PCR as described by Horton (1997) using 5′ATTGGTACCGTAGGTGCTTGTGAC3′ (P1) and 5′GATAAacGCgtCAGTATGACGTCAC3′ (P2) as primers to make the “left” half of the construct, and 5′ATACTGacGCgtTTATCGCCTGAGCA3′ (P3) and 5′AATAGGCCTCCCTTCAGAAATCTAA3′ (P4) to make the “right” half of the construct (the lower case bases in P2 and P3 indicate the E box mutation that we introduced). All constructs used in this study were confirmed by sequencing.
Animals and Electroporation
Adult Ciona intestinalis were collected from the Sandwich Marina in Sandwich, MA and Point Judith Marina in Snug Harbor, RI. Eggs were obtained by dissection of the oviduct, and fertilized in vitro with sperm of several individuals. Embryos were dechorionated immediately after fertilization using the methods described by Mita-Miyazawa et al. (1985). After electroporation, embryos were raised on Petri dishes coated with 1% agarose in 0.2 μm filtered seawater at 18°C.
Misexpression plasmids were electroporated into Ciona embryos as described in Corbo et al. (1997). Embryos were collected in 200μL seawater and added to 600μL 0.77 M mannitol. Approximately 25μg of misexpression plasmid was electroporated into embryos ~25–35 minutes after fertilization. Beginning at the 8-cell stage normally cleaving embryos were isolated and then treated with cytochalasin B at a final concentration of 1μg/mL to arrest cleavage at the 64-cell stage (~4.25 hours post-fertilization). Embryos were fixed for in situ hybridization at 11–12 hours post fertilization, when normally developing embryos reached the early tail-formation stage. Typically, a single experiment with a given plasmid yielded 50–200 cleavage-arrested embryos that were suitable for in situ hybridization.
In Situ Hybridization and Enzyme Histochemistry
Embryos electroporated with a given plasmid were divided into groups containing a minimum of 8 embryos (typically groups consisted of 15 or more embryos; see Tables S1, S2, S3) and subjected to in situ hybridization using digoxigenin-labeled antisense RNA probes essentially as described by Wada et al. (1995). Incubation times for color development ranged from 3 to 48 h depending on the probe. Embryos for acetylcholinesterase histochemistry were fixed for 30–40 minutes on ice in seawater containing 4% paraformaldehyde. Acetycholinesterase activity was localized using the method of Karnovsky and Roots (1964). Incubation times for color development were 2–4 hours at room temperature. In some experiments, electroporation efficiency was evaluated by measuring β-galactosidase activity in pTLacZ electroporated embryos. Embryos for β-galactosidase histochemistry were fixed for 30 minutes on ice in seawater containing 1.5% paraformaldehyde, 0.1% Tween 80; they were then washed in phosphate buffered saline containing 0.1% Tween 80 and incubated in staining solution (0.04% XGal, 2mM MgCl2, 0.06 M Na2HPO4, 0.04 M NaH2PO4, 4mM potassium ferrocyanide, 4mM potassium ferricyanide, 0.1% Tween 80) at room temperature for 1–4 hours.
Analysis, Statistics, and Photography
An embryo was considered to be expressing a muscle gene in the notochord if cells reacting with a given probe were observed at two opposite poles corresponding to the primary muscle and notochord lineages. Experiments to evaluate the effects of misexpressing Ci-MRF always included pTLacZ electroporated embryos as a negative control, and experiments to evaluate mutated forms of Ci-MRF included both pTLacZ and a positive control (pTCiMRFa or pTCiMRFb). Chi-square analysis was used to evaluate whether differences in the misexpression of muscle markers observed in control and experimental embryos were significant; a p-value of less than 0.05 was considered significant. A Mantel-Haenszel test for repeated tests of independence was applied to each chi-square analysis to ensure that the data from individual experiments were sufficiently homogenous to be combined (Sheskin 1997; Ott 1993).
Images in Figures 1A and B and 5 A and B were obtained using a Leica MZ16 dissecting microscope with a Leica DFC 290HD camera and were digitized with LAS V3.5 software. All other photographs were taken using an Olympus System BHS model microscope with a Pixelink 6.6 megapixel camera and were digitized with Pixelink Image Capture software. All image cropping and annotation was done using ImageJ software.
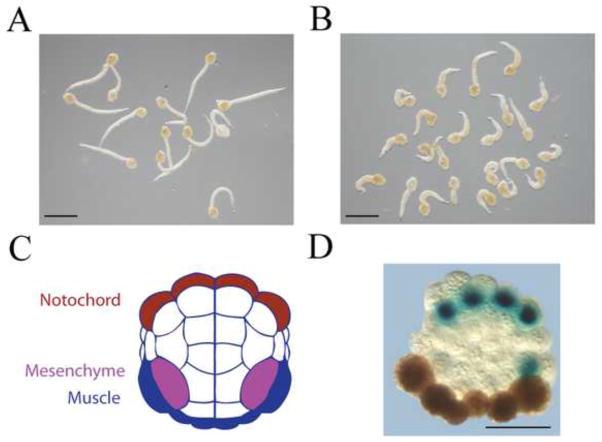
Figure 1. Expression of Ci-MRF in the notochord disrupts tail development.
A. Embryos at 14 hours post fertilization electroporated as zygotes with pTLacZ. Scale bar is 500μm. B. Embryos at 14 hours post fertilization electroporated as zygotes with pTCiMRFb. Scale bar is 500μm. C. Diagram of Ciona intestinalis embryo at the 64-cell stage highlighting the primary notochord and muscle lineages, and the mesenchyme lineage. D. Cleavage-arrested 64-cell embryo at 14 hours post fertilization electroporated with pTLacZ and assayed by histochemical methods for acetylcholinesterase (brown-stained cells), a highly specific marker of muscle differentiation in Ciona intestinalis (Meedel and Whittaker, 1979) and β-galactosidase (blue-stained cells). Adobe Photoshop was used to enhance image background. Scale bar is 50μm.
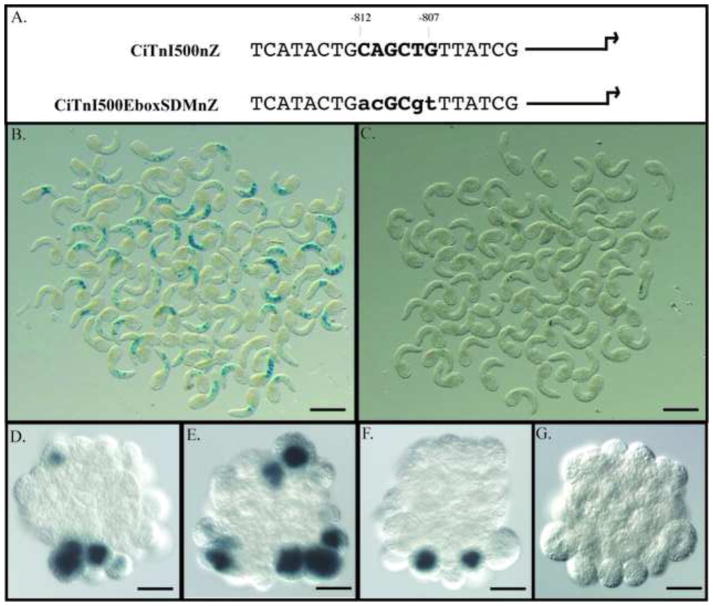
Figure 5. TnI Reporter activity requires an intact E-box.
A. Diagram illustrating the location of an essential E-box and its mutated counterpart (both shown in bold) in the two TnI reporter plasmids. Arrow indicates the position of the normal translation start site. B. C. Expression of β-galactosidase in early tail formation stage embryos electroporated with CiTnI500nZ and CiTnI500EboxSDMnZ respectively. Scale bars are 500μm. D. E. Activity of β-galactosidase in cleavage-arrested embryos co-electroporated with CiTnI500nZ and pTCiMRFb. F. Activity of β-galactosidase in a cleavage-arrested embryo co-electroporated with CiTnI500nZ and pSP72. G. Example of an embryo co-electroporated with CiTnI500EboxSDMnZ and pTCiMRFb showing no β-galactosidase activity. Scale bars in D–G are 50μm.
Results
Assaying the Myogenicity of Ci-MRF
In order to examine the properties of Ci-MRF, we developed an assay that uses cis-regulatory sequences of the Brachyury gene to drive expression of plasmids encoding synthetic and naturally occurring variants of Ci-MRF proteins in the notochord of developing embryos. An important feature of this assay is that electroporated embryos are obtained in sufficient numbers to allow us to identify by statistical analysis Ci-MRF variants that have relatively subtle differences in myogenic activity.
The first two plasmids tested, pTCiMRFa and pTCiMRFb, encode the small and large transcript of Ci-MRF, respectively (Meedel et al., 1997; Figure S1). Initial studies with either plasmid resulted in embryos with severely abnormal tails (Figure 1A, B). This effect was similar to, though more pronounced than what was observed when Macho-1 was expressed in the notochord (Kugler et al., 2010) and indicates that muscle gene regulatory factors may interfere with notochord gene expression, or that activating muscle-specific genes may interfere in some way with notochord development. Because this effect limited our ability to distinguish between the muscle and notochord lineages, we blocked cell division at the 64-cell stage with cytochalasin B, and assayed myogenesis in the resulting embryos several hours later by in situ hybridization. Such an approach is possible because ascidian embryos undergo tissue-specific differentiation when cleavage-arrested (Whittaker, 1973; Crowther and Whittaker, 1986), and their highly stereotypical and invariant pattern of cell division makes it possible to distinguish the muscle and notochord lineages in cleaving embryos (Nishida, 1987; Figure 1C). Figure 1D illustrates the utility of this method. The notochord and muscle lineages are easily distinguished in this embryo, and muscle differentiation clearly took place under these conditions. We note, however, that the use of cleavage arrest in this assay allowed us to detect muscle gene expression only in the A-line notochord lineage, which gives rise to 32 of the 40 notochord cells, but not in the B-line notochord cells because their proximity to the muscle lineage prevented their unambiguous identification after cleavage arrest (Figure 1C).
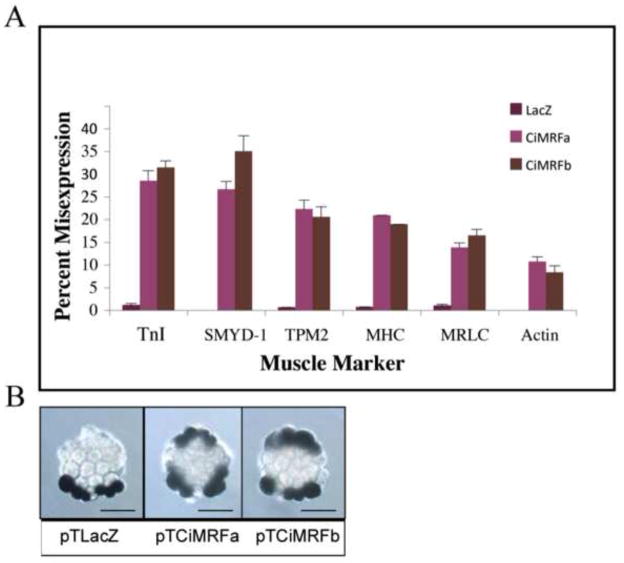
The ability of Ci-MRF to elicit myogenesis in the notochord of electroporated embryos was assessed by in situ hybridization using nine different muscle-specific probes and by using a histochemical assay to detect the activity of acetylcholinesterase. Transcripts encoding actin, TnI, TPM2, MHC, MRLC, and SMYD-1 were detected in the notochord lineage of embryos electroporated with either pTCiMRFa or pTCiMRFb; no transcripts encoding CKM, MLC, or TnT were detected in the notochord in these experiments nor was there any evidence of acetylcholinesterase activity in the notochord (Table 1; Table S1; Figure 2). Although pTCiMRFa and pTCiMRFb encode proteins that differ in a motif of known functional importance in vertebrate MRFs (i.e. Helix III), no significant differences were noted in the ability of the two plasmids to elicit the expression of these muscle markers.
Table 1.
Muscle Genes Assayed
| Gene/Gene Family | Abbreviation | Feature | Clone ID* | Accession # |
|---|---|---|---|---|
| Actin | Actin | Thin Filament | citb095F03 | XM_002126220 |
| Myosin Heavy Chain | MHC | Thick Filament | cilv003k12 | AK115565.1 |
| Myosin Regulatory Light Chain | MRLC | Thick Filament | citb104p01 | AK116716.1 |
| SET-MYND Domain | SMYD-1 | Gene Regulatory† | citb009d08 | AK112854.1 |
| Tropomyosin 2 | TPM2 | Thin Filament | cilv034e06 | AK174927.1 |
| Troponin I | TnI | Thin Filament | pcTp2 | U55261 |
| Acetylcholinesterase | Ache | Cholinergic | NC | NM_001128877 |
| Creatine Kinase | CKM | Metabolic | citb072f10 | NW_001955200 |
| Myosin (alkali) Light Chain | MLC | Thick Filament | cilv022011 | AK174821.1 |
| Troponin T | TnT | Thin Filament | citb012e12 | NW_001955435 |
Clone ID refers to the cDNA clone that was used for preparing probes for in situ hybridization. Transcripts of the six genes in bold type were detected in the notochord when Ci-MRF was expressed in that lineage; transcripts of the four genes in regular type were not detected in the notochord under those conditions.
SMYD-1 also appears to play a role in sarcomere assembly (Li et al., 2011). No clone (NC) was used for Ache, which was assayed using a histochemical method. Except for pcTp2 (MacLean et al., 1997) all clones were obtained from the National Institute of Genetics of Japan.
Figure 2. Ci-MRF activity in the notochord elicits expression of markers of terminally differentiated muscle.
A. Expression of TnI, MHC, SMYD-1, TPM2, MRLC, and actin in the notochord of embryos electroporated with pTCiMRFa and pTCiMRFb. Results are indicated as percentages of the total number of embryos in all experiments that misexpress a given muscle marker. All muscle markers listed above showed a significant level of expression in the notochord when compared to control (pTLacZ) embryos (p <0.01). Error bars represent standard deviation. Neither pTCiMRFa nor pTCiMRFb elicited the expression of Ache, CKM, MLC, or TnT in this assay. B. The expression of TnI in 64-cell cleavage arrested embryos electroporated with pTLacZ, pTCiMRFa and pTCiMRFb. pTLacZ electroporated embryos express TnI only in the primary muscle lineage. pTCiMRFa and pTCiMRFb electroporated embryos express TnI in the primary muscle and primary notochord lineages. Scale bar is 50μm.
Four of the markers we studied (actin, MHC, MLC, and MRLC) are members of highly conserved gene families in Ciona (Chiba et al., 2003), which raises the possibility that our probes recognized transcripts produced by multiple members of each of these gene families. Indeed, we expect that this is the case because the probes we used contained a substantial portion of the coding region of MHC, and essentially the entire coding regions of actin, MLC and MRLC. For convenience we will use the terms actin, MHC, MLC, and MRLC when referring to these four multi-gene families; later we will discuss how their inclusion in this study affects our interpretation of the mechanisms by which Ci-MRF regulates muscle gene expression. Multiple tropomyosin genes also exist in Ciona but they do not show the same high degree of conservation as the actin, MHC, MLC, and MRLC gene families (Chiba et al., 2003). In addition, genomic blots under reduced stringency conditions using probes complementary to the entire coding sequence of TPM1 (which is the tropomyosin whose sequence is the most similar to TPM2) failed to detect the existence of any other tropomyosin genes (Meedel and Hastings, 1993). Thus, we are confident that the TPM2 probe we used does not recognize transcripts of any other tropomyosin gene. The other five genes used as markers of muscle development (Ache, CKM, SMYD-1, TnI and TnT) are not members of multigene families.
CiMRFa and CiMRFb have Different Requirements for the C/H Domain
The experiments described above indicate that Helix III is not required for the functional activity of Ci-MRF in our assay, at least for regulating the activity of the genes we examined. This result was not unexpected considering the studies of Berkes et al. (2004) who showed that the expression of many MyoD-regulated genes is controlled independently of both Helix III and the C/H domain. In order to examine the role of the C/H domain in Ci-MRF, we created mutant plasmids in which its entire coding sequence was deleted. This resulted in a plasmid lacking the sequences encoding the C/H domain and Helix III (pTCiMRFaΔCH), and in a plasmid that was missing only the sequence encoding the C/H domain (pTCiMRFbΔCH). It should be noted that pTCiMRFaΔCH encodes a protein that lacks not only the C/H domain and Helix III but also a 54 amino acid region that begins 75 amino acids downstream of the b-hlh domain and extends to Helix III. While we cannot exclude the possibility that this region is critical for Ci-MRF function, the observation that its absence from pTCiMRFa did not impair the myogenicity of this plasmid relative to pTCiMRFb argues against this possibility.
pTCiMRFbΔCH elicited ectopic expression of all six of the muscle markers that we tested at levels similar to pTCiMRFb; conversely, the ability of pTCiMRFaΔCH to elicit ectopic muscle gene expression was significantly lower than pTCiMRFa for all genes tested (Figure 3; Table S2). Interestingly, we identified no gene in this study whose expression was directed by Ci-MRF for which the C/H domain and Helix III were required independently. The presence of the C/H domain in the absence of Helix III elicited target gene expression; similarly, the presence of Helix III in the absence of the C/H domain drove target gene expression. Only when both domains were absent did we see an effect on the ability of Ci-MRF protein to direct muscle gene expression, and in all cases this ability was reduced as compared to wild-type Ci-MRF, but not eliminated.
Figure 3. Effect of ΔC/H mutation on myogenicity of Ci-MRF.
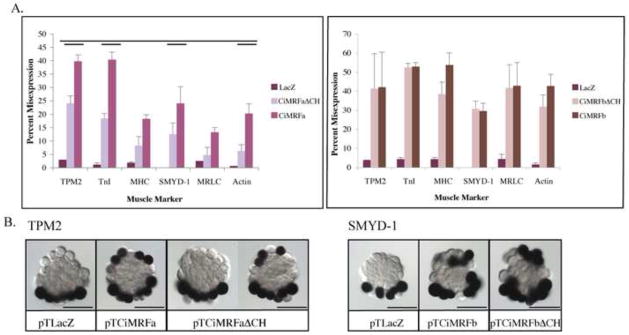
Panel A (left): ΔC/H mutant CiMRFa drove the expression of all six muscle markers in the notochord at levels significantly lower than wild type CiMRFa (single bar denotes p<0.05, double bar denotes p<0.01). Error bars represent standard deviation. Panel A (right): ΔC/H mutant transcripts of CiMRFb drove the expression of all six muscle markers in the notochord in embryos at levels that were not significantly different from embryos electroporated with wild type CiMRFb. Error bars represent standard deviation. Panel B (left): In situ hybridization showing TPM2 expression in embryos electroporated with the designated plasmid. Panel B (right): In situ hybridization showing SMYD-1 expression in embryos electroporated with the designated plasmid. Scale bars are 50μm.
The Ala-Thr Dipeptide of the Basic Region is Essential for Normal CiMRF Activity
All proteins encoded by MRF genes have an alanine and a threonine at positions 13 and 14 of their basic domains respectively; although many b-hlh proteins contain an alanine at position 13, only MRFs have alanine and threonine residues at these positions (Olson and Klein, 1994; Müller et al., 2003). Moreover, the Ala-Thr dipeptide was shown to be critical for the myogenic activity of vertebrate MRFs, hence it has been referred to as the “myogenic code” (Brennan et al., 1991; Davis and Weintraub, 1992; Heidt et al., 2007). In order to test the role of this dipeptide in Ciona myogenesis, we mutated the sequence encoding it to a sequence encoding an asparagine dipeptide in pTCiMRFa and pTCiMRFb to create pTCiMRFaNN and pTCiMRFbNN, respectively. This particular mutation was chosen because an asparagine dipeptide occurs at the corresponding position of the basic domain of related, but non-myogenic b-hlh proteins such as E12 and E47 (Murre et al., 1989). The ability of mutated and un-mutated plasmids to direct myogenesis was then compared as described above, using the six muscle markers that were expressed in the notochord lineage of embryos electroporated with pTCiMRFa or pTCiMRFb.
Mutating the Ala-Thr dipeptide eliminated the ability of both pTCiMRFa and pTCiMRFb to elicit the expression of actin,SMYD-1, and TPM2 in the notochord lineage. Both mutant plasmids directed expression of MHC, MRLC, and TnI in the notochord, but in significantly fewer embryos than the un-mutated plasmids (Figure 4; Table S3). These findings support the idea that the Ala-Thr dipeptide of the basic domain is a crucial feature of Ci-MRF, thereby providing the first experimental evidence that the myogenic code is a valid concept that extends at least to an invertebrate chordate. Our findings also indicate that the importance of the Ala-Thr dipeptide differs among the genes regulated by Ci-MRF.
Figure 4. Effect of mutating the myogenic code on Ci-MRF activity.
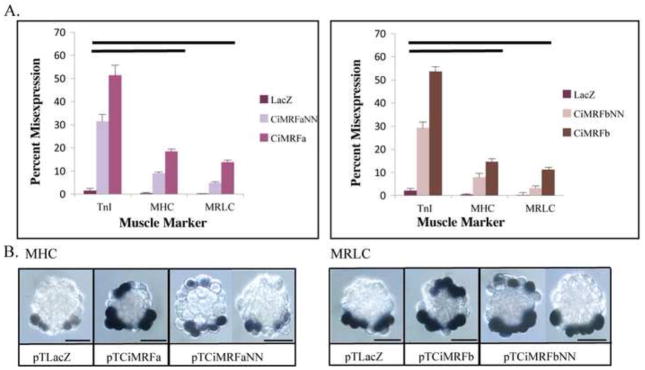
Panel A: Myogenic code mutants drove MHC, MRLC and TnI expression in the notochord at levels significantly lower than wild type Ci-MRF (single bar denotes p<0.05, double bar denotes p<0.01). Neither myogenic code mutant elicited the expression of TPM2, SMYD-1 or actin in the notochord (data not shown). Error bars represent standard deviation. Panel B (left): In situ hybridization showing MHC expression in embryos electroporated with the designated plasmid. Panel B (right): In situ hybridization showing MRLC expression in embryos electroporated with the designated plasmid. For both pTCiMRFaNN and pTCiMRFbNN examples of embryos that misexpressed the given marker and did not misexpress the given marker are shown. Scale bars are 50μm.
An upstream E-box is Essential for CiMRF-directed Expression of Ci-TnI
Previous studies demonstrating the importance of E-boxes for expression of muscle genes in Ciona (Johnson et al., 2004; Brown et al., 2007) and our demonstration that Ci-MRF plays a crucial role this process (Meedel et al., 2002; 2007; this communication) are consistent with the possibility that CiMRF functions, at least in part, to directly activate muscle target genes. We investigated this possibility by doing a series of co-electroporation experiments in which either pTCiMRFb or its parental plasmid, pSP72, was electroporated into embryos together with one of two Ci-TnI LacZ reporter plasmids. Both reporters contained Ci-TnI sequence extending from 335 to 836 base pairs upstream of the translation start site (i.e. -836 to -335) and were identical except that CiTnI500nZ had a wild-type E-box at nucleotides -812 to -807, and CiTnI500EboxSDMnZ had a mutated E-box at this site (Figure 5A).
CiTnI500nZ-electroporated embryos exhibited robust reporter gene activity in embryonic muscle (Khare et al., 2011; Figure 5B), whereas CiTnI500EboxSDMnZ-electroporated embryos showed significantly lower levels of reporter activity (Figure 5C). These results demonstrate that the E-box at -812/-807 is critical for the expression of Ci-TnI. We then compared the ability of pTCiMRFb and pSP72 to drive expression of CiTnI500nZ, and found that only pTCiMRFb was able to elicit CiTnI500nZ expression in the notochord (Figure 5D–F; Table 2). Consistent with this finding, the number of β-galactosidase positive cells was also higher in pTCiMRFb electroporated embryos than in those electroporated with pSP72 (Table 2). In addition, the percentage of embryos expressing LacZ was significantly higher in embryos electroporated with pTCiMRFb than in those electroporated with pSP72. A likely explanation of this result is that pTCiMRFb directed the expression of LacZ in the notochord of embryos that did not express this gene in muscle. These results show that CiMRF was necessary for the expression of the TnI reporter. Finally, we examined the effect of pTCiMRFb on the activity of CiTnI500EboxSDMnZ and found no embryos expressing LacZ in the notochord (Figure 5G; Table 2). Collectively, these findings demonstrate that an intact E-box is required for CiMRF-directed expression of the TnI reporter and they support our claim that CiMRF acts directly on the E-box at -812/-807 rather than functioning indirectly by, for example, stimulating the expression of another transcription factor that then targets Ci-TnI.
Table 2.
LacZ expression in co-electroporated embryos
| CiTnI500nZ | CiTnI500nZ | CiTnI500EBMnZ | CiTnI500EBMnZ | |
|---|---|---|---|---|
| pTCiMRFb | pSP72 | pTCiMRFb | pSP72 | |
| Exp. 1 | ||||
| # E | 64 | 119 | X | 91 |
| # NoE | 5 (8%) | 22 (18%) | X | 78 (86%) |
| # MisEN | 23 (36%) | 0 (0)% | X | 0 (0%) |
| # EC/E | 4.40±2.42 | 1.75±1.28 | X | 0.14±0.35 |
| Exp. 2 | ||||
| # E | 54 | 92 | 71 | X |
| # NoE | 6 (11%) | 58 (63%) | 70 (99%) | X |
| # MisEN | 20 (37%) | 0 (0%) | 0 (0%) | X |
| # EC/E | 3.50±2.45 | 0.50±0.75 | 0.01±0.12 | X |
| Exp. 3 | ||||
| # E | 69 | X | 118 | X |
| # NoE | 4 (6%) | X | 91 (77%) | X |
| # MisEN | 38 (55%) | X | 0 (0%) | X |
| # EC/E | 6.75±3.86 | X | 0.29±0.60 | X |
Plasmid combinations that were co-electroporated into embryos are shown at the top. Numbers in parenthesis indicate the percentage of embryos in a given category relative to the number of embryos examined. Abbreviations used are: # E (number of embryos examined); # NoE (number of embryos not expressing LacZ); # MisEN (number of embryos expressing LacZ in the notochord); #EC/E (mean number of cells expressing LacZ per embryo), and “X” (not done).
Discussion
Muscle Gene Activity in Response to Ci-MRF Expression in the Notochord
Six of the ten muscle markers we tested were expressed in the notochord when Ci-MRF was active in that tissue. The genes that were misexpressed represented a spectrum of features associated with the terminal muscle phenotype including a transcription factor involved in gene regulation and thin and thick filament proteins of the contractile apparatus. The ability of Ci-MRF to positively regulate myofibrillar protein expression is consistent with the absence of contractile structures in the muscle cells of Ci-MRF knockdown embryos (Meedel et al., 2007).
Some, if not all, of the muscle-specific genes that were expressed in the notochord assay are probably directly regulated by Ci-MRF. The best evidence of this was provided by co-electroporation experiments showing that Ci-MRF directed TnI expression depends on a GC-core E-box upstream of the TnI translation start site. SMYD-1 may also be a direct target of Ci-MRF since together with TnI it was routinely the most highly misexpressed gene examined in this study and it was associated with CiMRF in CHIP assays (as was TnI; Kubo et al., 2010). Although the presence of functional E-boxes in its upstream regulatory region has not been investigated, SMYD-1 does possess a GC-core E-box at base pairs -838/-833 (unpublished observation). No essential E-box was found in TPM2 (Brown et al., 2007), but TPM2 was associated with CiMRF in CHIP assays (Kubo et al., 2010), consistent with it being a direct Ci-MRF target. We speculate that TPM-2 may be regulated by Ci-MRF through interaction with an undiscovered E-box, or through binding to guanine-rich tetraplex structures (Etzioni et al., 2005; Shklover et al., 2007). Conversely CiMRF may not directly bind with TPM-2 DNA, but may elicit its expression more indirectly through interacting with other chromatin-associated factors. Interpreting the mechanisms by which Ci-MRF regulates the activity of actin, MHC, and MRLC is complicated because our probes undoubtedly recognize multiple members of each of these families, which may be regulated by different mechanisms (e.g. Kusakabe et al., 1995; 2004 and Brown et al., 2007). However, some members of the MRLC and actin gene families are known to contain GC-core E-boxes important for their expression (Johnson et al., 2004; Brown et al., 2007), and many were associated with CiMRF in CHIP assays (Kubo et al., 2010) as were several members of the MHC family, which have not been tested for the presence of functional E-boxes. Therefore, it is likely that at least some members of these gene families are direct targets of Ci-MRF.
Four of the markers we examined, Ache, CKM, MLC, and TnT were not expressed in the notochord, although this does not mean that none of them is a target of Ci-MRF. For example, at least two MLC genes were associated with CiMRF in CHIP assays (Kubo et al., 2010) and some family members were shown to have E-boxes that confer a low level of activity on the genes (Brown et al., 2007). Ache was not examined by Brown et al. (2007) nor was it associated with CiMRF in CHIP assays (Kubo et al., 2010), but in a previous study both Ache activity and MLC family transcripts occurred ectopically in embryos injected with Ci-MRF mRNA indicating that they are positively regulated by Ci-MRF (Meedel etal ., 2007). We suspect that the results obtained with Ache and MLC in the present study were due to our inability to assess myogenesis in the B-line notochord, which seems to be more readily transformed to muscle than A-line notochord (Meedel et al., 2007). Notably, of the four genes/gene families that were ectopically expressed in embryos reared from eggs injected with Ci-MRF mRNA two were expressed in the current study in the A-line notochord (actin and TnI) and two were not (Ache and MLC) demonstrating that there are complex and variable requirements for the expression of these markers that are met for the former pair but not the latter pair when Ci-MRF is expressed in the A-line notochord.
CKM is probably not regulated by Ci-MRF as no functioning E-boxes were found in its promoter, which did contain Tbx6 binding motifs necessary for activity (Brown et al., 2007), and it was not associated with CiMRF in CHIP assays (Kubo etal ., 2010). Functioning E-boxes were not found in TnT either (Brown et al., 2007) but it was associated with CiMRF in CHIP assays, indicating that if it is regulated by Ci-MRF the conditions necessary for its expression in the A-line notochord are not met by expressing Ci-MRF in those cells. Table 3 summarizes the results of the Brown et al. (2007) and Kubo et al. (2010) studies.
Table 3.
Characteristics of Muscle Genes Assayed
| Gene/Gene Family | CHIP Assay | Functional E-Box Detected |
|---|---|---|
| Actin | + | + |
| MHC | + | X |
| MRLC | + | + |
| SMYD1 | + | X |
| TPM2 | + | − |
| TnI | + | + |
| Ache | − | X |
| CKM | − | − |
| MLC | + | + |
| TnT | + | − |
Transcripts of the six genes/gene families in bold type were detected in the notochord when Ci-MRF was expressed in that lineage; transcripts of the four genes/gene families in regular type were not detected in the notochord under those conditions. CHIP assay data are from Kubo et al., 2010; “+” signifies an association of CiMRF with chromatin of the indicated gene and “−” signifies no association of CiMRF with chromatin of the indicated gene. E-box data are from Brown et al., 2007; “+” signifies the presence of at least one E-box that is important for expression, “−” signifies that no E-box important for expression was found and “X” signifies that no member of in the indicated gene/gene family was examined.
A Requirement for the C/H Domain or Helix III
The C/H domain and Helix III of vertebrate MyoD have been implicated in initiating muscle gene expression through their ability to remodel the chromatin of target genes (Gerber et al., 1997; Bergstrom and Tapscott, 2001). Berkes et al. (2004) found that mutating either element of MyoD individually affected a group of genes that was similar to the genes affected by mutating both simultaneously, indicating that the majority of the genes that rely on these elements of MyoD for their expression require both independently. In contrast, our analysis of Ci-MRF indicated that muscle gene expression was not typically affected when the C/H domain or Helix III were individually deleted, but was always affected when both were deleted concurrently. Because expression of the majority of the Ci-MRF-regulated genes that we studied was satisfied by the presence of either the C/H domain or Helix III we suggest that these two elements are likely to have roles in ascidian myogenesis that overlap to some degree. Such redundancy was not noted in MyoD indicating that the C/H domain and Helix III have evolved distinct functions in this vertebrate MRF (Berkes et al., 2004). Precedents exist for evolutionary changes in these motifs. For example, hlh-1 the MRF of C. elegans does not encode a C/H domain motif (Krause et al., 1990); in addition, replacing Helix III of MyoD with Helix III of myogenin disrupts the function of the resulting protein demonstrating that this motif has distinct roles in these two MRFs (Bergstrom and Tapscott, 2001). Evolutionary conservation has also been documented in Helix III since substituting this motif in MyoD with Helix III from the MRFs of C. elegans, Drosophila, or S. purpuratus (Figure S2) does not impair the ability of the resulting proteins to initiate gene expression (Bergstrom and Tapscott, 2001). Similar motif swapping experiments between vertebrate MRFs and CiMRF could be done to assess potential evolutionary changes in the roles of the C/H domain and Helix III in the chordates.
Our study also reveals a much more significant role for the C/H domain and Helix III in CiMRF than did the study of Berkes et al. (2004) for mouse MyoD. All six markers that were positively regulated by Ci-MRF were affected by mutating these two elements, whereas only 16 of 109 genes that were regulated by MyoD were affected by such mutations. It is unlikely that studying more genes would alter this trend; instead it seems that functional changes have occurred in these elements. Of the two motifs the sequence of Helix III is much more similar in CiMRF and MyoD than is the C/H domain (Figure S2) indicating that its function may be less diverged, a possibility that is supported by the Helix III swapping studies of Bergstrom and Tapscott (2001).
The Myogenic Code is Critical for Ci-MRF Activity
Mutation of the myogenic code of vertebrate MyoD results in decreased binding to DNA due to a combination of reduced ability to dimerize, reduced affinity for the E-boxes of target genes, and increased off rate from DNA (Heidt et al., 2007). These authors also concluded that the myogenic code of MyoD is necessary for efficient binding to canonical E-boxes (i.e. CANNTG) and that it is essential for binding to non-canonical E-boxes (e.g. CAACAGCTT) of genes such as myogenin whose myogenic code has also been shown to be important for its activity (Brennan et al., 1991). Despite its conservation in MRFs from worms to vertebrates and its importance as a determinant of myogenic specificity in vertebrate MRFs, a role for the myogenic code in muscle development has previously not been demonstrated in any invertebrate.
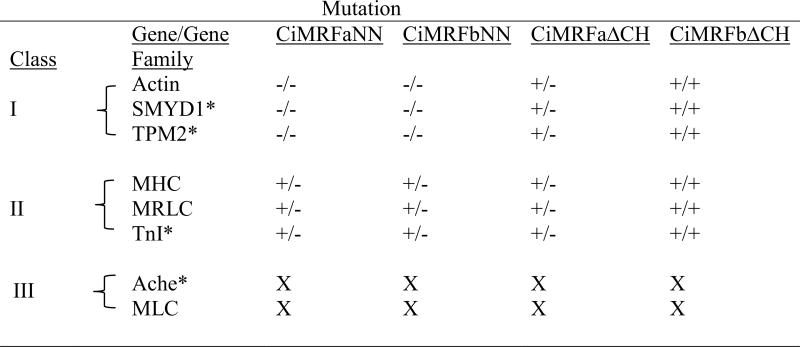
Our studies reveal that the myogenic code is necessary for Ci-MRF to function normally, although its role differs among the markers examined (Table 4). This difference is most easily interpreted when comparing the response of single copy genes such as TnI and SMYD-1 to mutating the myogenic code, which decreased the activity of the former gene and eliminated the activity of the latter. The presence of an essential canonical E-box in TnI (Brown et al., 2007; this study) is consistent with the interpretation that in CiMRF, as in MyoD, the myogenic code is required for efficient binding to canonical E-boxes, so that its mutation would be expected to reduce transcriptional output as we observed. The role of E-boxes in SMYD-1 expression has not been examined, but as noted earlier, a GC-core E-box does exist in this gene at approximately the same position as the E-box that is essential for TnI expression. Possible explanations for the different responses of TnI and SMYD-1 to mutating the myogenic code include: (1) the GC-core E-box identified in SMYD-1 may not be important for its expression, but a noncanonical E-box may be; (2) sequences near E-boxes may differentially modify their response to CiMRF (Yutzey and Konieczny, 1992; Fisher and Goding, 1992); (3) the types of trans-regulatory factors associated with the two genes may modify their responses to CiMRF in different ways (Molkentin et al., 1995; Groisman et al., 1996; Berkes et al., 2004; Albini and Puri, 2010; Liu et al., 2010; Delgado-Olguín et al., 2011); (4) or some combination of the latter two explanations.
Table 4.
Summary of Responses to Ci-MRF mutations
|
Data show the effects of each Ci-MRF mutation versus the corresponding un-mutated version of Ci-MRF; for example the column headed CiMRFaNN compares the response of the indicated gene or gene family in embryos electroporated with pTCiMRFaNN to its response in embryos electroporated with pTCiMRFa; in the case of actin, mutating the myogenic code eliminated its expression. Classes correspond to the three groups of Ci-MRF-regulated genes whose properties are further described in the text. Single copy genes are denoted with an *. Note that the genes in Class III did not respond to Ci-MRF in this assay, so the effects of these mutations were not determined as indicated by “X”. Other symbols: −/−, expression eliminated; +/− statistically significant reduction of expression; +/+, no statistically significant effect on expression.
TPM2 expression was eliminated when we mutated the CiMRF myogenic code. This result was somewhat surprising because Brown et al. (2007) did not find any E-boxes necessary for TPM-2 activity but they did identify other transcription factor binding sites that conferred significant activity to the gene. Several possible explanations exist for this result, three of which we mentioned above when discussing how Ci-MRF may regulate TPM-2 expression (see first section of the Discussion). Here we offer the additional possibility that the myogenic code may also function to confer an appropriate conformation on MRFs that is necessary for their interaction with other regulators of muscle gene transcription as suggested elsewhere (Heidt et al., 2007).
Expression of the MRLC marker was reduced when the myogenic code was mutated. At least some members of the MRLC family possess functionally important GC-core E-boxes (Brown et al., 2007), which is consistent with the possibility that the myogenic code of Ci-MRF is necessary for efficient binding to canonical E-boxes of MRLC genes. However, we cannot rule out other possibilities such as that mutating the myogenic code of CiMRF eliminated the expression of some MRLC genes, while having little or no effect on other genes of this family. At least some members of the actin gene family also contain functional GC-core E-boxes (Brown et al., 2007), but in this case mutating the myogenic code eliminated the expression of this marker. We suggest that this result may indicate a relatively weak interaction of CiMRF with E-boxes of actin family members that it regulates and that this interaction is particularly sensitive to mutating the myogenic code. The cis-regulatory regions of MHC genes have not been evaluated, so speculating on roles that the myogenic code may play in their expression is premature. In summary, while our results do not address the precise mechanism by which the myogenic code of Ci-MRF functions, they do provide clear evidence for its critical importance during Ciona myogenesis, the first time this has been demonstrated in any animal other than a vertebrate.
Multiple Classes of Ci-MRF Regulated Genes in Ciona
Our studies reveal the existence of three classes of Ci-MRF-regulated genes in Ciona. Genes in Classes I and II are distinguished by the degree of their response to mutation of the myogenic code and genes in Class III by their lack of expression in the notochord assay (Table 4). Notably, each of the classes of genes that we identified contains at least one single copy gene (Table 4). Thus, inclusion of multi-gene families in our study does not alter the conclusion that three distinct classes of MRF-regulated genes exist in Ciona. Indeed, because individual members of multi-gene families in ascidians, including actin and MRLC, are known to be regulated by distinct mechanisms (e.g. Kusakabe et al., 1995, 2004; Brown et al., 2007), it is likely that the number of MRF-regulated classes in Ciona will exceed the three we have identified, and that modifications will be necessary to the classification system shown in Table 4. Nevertheless, our results indicate a degree of muscle gene regulatory pathway complexity in ascidians that is reminiscent of that seen in vertebrates.
Evolution of MRF Regulated Myogenesis
A variety of different approaches including studies using transfected mammalian cells (e.g. Yutzey et al., 1990; Krause et al., 1992; Venuti et al., 1991) and in vivo rescue of null mutations (Zhang et al., 1999) are consistent with idea that MRF function is evolutionarily conserved. The present study adds another dimension to those analyses that reinforce this idea by establishing that the myogenic code dipeptide, the C/H domain, and Helix III are crucial for myogenic activity of Ci-MRF, as they are for vertebrate MRFs. Two additional observations support the idea that chordate MRFs are functionally conserved: first, multiple MRF-regulated pathways exist in both the vertebrates and in Ciona, and second, as in the vertebrates where a direct interaction between MRFs and E-boxes occurs to drive target gene expression, a direct interaction between CiMRF and an essential E-box appears to be required for the expression of the muscle-specific gene TnI. That the three motifs of Ci-MRF we examined did not always function in precisely the same manner as do their vertebrate counterparts was not surprising considering the variety of assays used in the different studies, the well documented functional divergence of different vertebrate MRFs (Rawls et al., 1995; Kablar et al., 1997; Wang and Jaenisch, 1997; Bergstrom and Tapscott, 2001; Myer et al., 2001; Cao et al., 2006; Hinits et al., 2009), and the Darwinian notion of “descent with modification”. The notochord assay described here will allow us to further compare these three motifs of the different vertebrate MRFs with their counterparts in Ciona using a uniform set of conditions that should mitigate these issues. The assay will also be useful for examining questions about evolutionary relationships between chordate MRFs and the MRFs of more distantly related metazoans.
Supplementary Material
Highlights.
A simple, functional assay for studying CiMRF-the Ciona myogenic regulatory factor.
The C/H domain and the C-terminal Helix III of CiMRF have partly redundant roles.
As in vertebrate MRFs the myogenic code dipeptide is crucial for CiMRF activity.
As in vertebrates multiple classes of MRF-regulated genes exist in Ciona.
MRF-directed myogenesis is complex and highly conserved in the chordates.
Acknowledgments
We thank Jane Loescher, and Jamina Oomen-Hajagos for their editorial suggestions and Ken Hastings (P.K.1s MSc research supervisor) for his critical comments and many suggestions for improving the paper. This work was supported by awards from the National Institutes of Health (2R15HD047357-02; 3R15HD047357-02S1; 3R15HD47357-02S2; 3R15HD047357-02S3) to THM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albini S, Puri PL. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it’s time to exchange! Exp Cell Res. 2010;316:3073–3080. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L, Keller CA, Abmayr SM. Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability. Developmental biology. 2001;231:374–382. doi: 10.1006/dbio.2001.0162. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Michelson AM. Invertebrate myogenesis: looking back to the future of muscle development. Current opinion in genetics & development. 2001;11:431–439. doi: 10.1016/s0959-437x(00)00214-8. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Molecular and cellular biology. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Molecular cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brennan TJ, Chakraborty T, Olsen EN. Mutagenesis of the myogenic basic region identifies an ancient protein motif critical for activation of myogenesis. Proceedings of the National Academy of Science, USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nature reviews Genetics. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. The EMBO journal. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- Chiba S, Awazu S, Itoh M, Chin-Bow ST, Satoh N, Satou Y, Hastings KE. A genomewide survey of developmentally relevant genes in Ciona intestinalis. IX. Genes for muscle structural proteins. Development genes and evolution. 2003;213:291–302. doi: 10.1007/s00427-003-0324-x. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Corbo JC, DiGregorio A, Levine M. The ascidian as a model organism in developmental and evolutionary biology. Cell. 2001;106:535–538. doi: 10.1016/s0092-8674(01)00481-0. [DOI] [PubMed] [Google Scholar]
- Crowther RJ, Whittaker JR. Differentiation without cleavage: multiple cytospecific ultrastructural expressions in individual one-celled ascidian embryos. Developmental biology. 1986;117:114–126. doi: 10.1016/0012-1606(86)90354-4. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992;256:1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nature genetics. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delgado-Olguín P, Brand-Arzamendi K, Scott IC, Jungblut B, Stainier DY, Bruneau BG, Recillas-Targa F. CTCF promotes muscle differentiation by modulating the activity of myogenic regulatory factors. J Biol Chem. 2011;286:12483–12494. doi: 10.1074/jbc.M110.164574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J Biol Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fisher F, Goding CR. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. The EMBO journal. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Groisman R, Masutani H, Leibovitch M-P, Robin P, Soudant I, Trouche D, Hrel-Bellan A. Physical interactin between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J Biol Chem. 1996;271:5258–5264. doi: 10.1074/jbc.271.9.5258. [DOI] [PubMed] [Google Scholar]
- Heidt AB, Rojas A, Harris IS, Black BL. Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Molecular and cellular biology. 2007;27:5910–5920. doi: 10.1128/MCB.01700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somatic, cranial, and fin muscle fibre populations. Development. 2009;136:403–414. doi: 10.1242/dev.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM. In: PCR Cloning Protocols: From Molecular Cloning to Genetic Engineering. White BA, editor. Vol. 67. Humana Press; Totowa, New Jersey: 1997. pp. 141–150. [Google Scholar]
- Johnson DB, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Karnovsky MJ, Roots L. A “Direct-Coloring” Thiocholine Method for Cholinesterases. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society. 1964;12:219–221. doi: 10.1177/12.3.219. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Khare P, Mortimer SI, Cleto CL, Okamura K, Suzuki Y, Kusakabe T, Nakai K, Meedel TH, Hastings KEM. Cross-validated methods for promoter/transcription site mapping in SL trans-spliced genes, established using the Ciona intestinalis troponin I gene. Nucleic Acids Res. 2011;39:2638–2648. doi: 10.1093/nar/gkq1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Fire A, Harrison SW, Priess J, Weintraub H. CeMyoD accumulation defines the body wall muscle cell fate during C. elegans embryogenesis. Cell. 1990;63:907–919. doi: 10.1016/0092-8674(90)90494-y. [DOI] [PubMed] [Google Scholar]
- Krause M, Fire A, White-Harrison S, Weintraub H, Tapscott S. Functional conservation of nematode and vertebrate myogenic regulatory factors. Journal of Cell Science Supplement. 1992;16:111–115. doi: 10.1242/jcs.1992.supplement_16.13. [DOI] [PubMed] [Google Scholar]
- Kubo A, Suzuki N, Yuan X, Nakai K, Satoh N, Imai KS, Satou Y. Genomic cis-regulatory networks in the early Ciona intestinalis embryo. Development. 2010;137:1613–1623. doi: 10.1242/dev.046789. [DOI] [PubMed] [Google Scholar]
- Kugler JE, Gazdoiu S, Oda-Ishii I, Passamaneck YJ, Erives AJ, Di Gregorio A. Temporal regulation of the muscle gene cascade by Macho1 and Tbx6 transcription factors in Ciona intestinalis. J Cell Sci. 2010;123:2453–2463. doi: 10.1242/jcs.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano G, Nishida H. Ascidian embryonic development: an emerging model system for the study of cell fate specification in chordates. Dev Dynamic. 2007;236:1732–1747. doi: 10.1002/dvdy.21108. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Hikosaka A, Satoh N. Coexpression and promoter function in two muscle actin gene complexes of different structural organization in the ascidian Halocynthia roretzi. Developmental Biology. 1995;169:461–472. doi: 10.1006/dbio.1995.1161. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Yoshida R, Ikeda Y, Tsuda M. Computational discovery of DNA motifs associated with cell type-specific gene expression in Ciona. Developmental Biology. 2004;276:563–580. doi: 10.1016/j.ydbio.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Li H, Xu J, Bian YH, Rotllant P, Shen T, Chu W, Zhang J, Schneider M, Du SJ. Smyd1b_tv1, a key regulator of sarcomere assembly, is localized on the M-line of skeletal muscle fibers. PloS one. 2011;6:e28524. doi: 10.1371/journal.pone.0028524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 2010;38:6857–6871. doi: 10.1093/nar/gkq585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean D, Meedel TH, Hastings KEM. Tissue-specific alternative splicing of ascidian troponin I isoforms: redesign of a protein isoform-generating mechanism during chordate evolution. J Biol Chem. 1997;272:32115–32120. doi: 10.1074/jbc.272.51.32115. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Whittaker JR. Development of acetylcholinesterase during embryogenesis of the ascidian Ciona intestinalis. The Journal of Experimental Zoology. 1979;210:1–10. doi: 10.1002/jez.1402100102. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Hastings KEM. Striated muscle-type tropomyosin in a chordate smooth muscle, ascidian body-wall muscle. J Biol Chem. 1993;268:6755–6764. [PubMed] [Google Scholar]
- Meedel TH, Farmer SC, Lee JJ. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: implications for the evolution of chordate muscle gene regulation. Development. 1997;124:1711–1721. doi: 10.1242/dev.124.9.1711. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Lee JJ, Whittaker JR. Muscle development and lineage-specific expression of CiMDF, the MyoD-family gene of Ciona intestinalis. Developmental biology. 2002;241:238–246. doi: 10.1006/dbio.2001.0511. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Chang P, Yasuo H. Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Developmental biology. 2007;302:333–344. doi: 10.1016/j.ydbio.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita-Miyazawa I, Ikegami S, Satoh N. Histospecific acetylcholinesterase development in the presumptive muscle cells isolated from 16-cell-stage ascidian embryos with respect to the number of DNA replications. J Embryol Exp Morphol. 1985;87:1–12. [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bhlh proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Müller P, Seipel K, Yanze N, Reber-Muller S, Streitwolf-Engel R, Stierwald M, Spring J, Schmid V. Evolutionary aspects of developmentally regulated helix-loop-helix transcription factors in striated muscle of jellyfish. Developmental biology. 2003;255:216–229. doi: 10.1016/s0012-1606(02)00091-x. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Myer A, Olson EN, Klein WH. MyoD cannot compensate for the absence of myogenin during skeletal muscle differentiation in murine embryonic stem cells. Developmental biology. 2001;229:340–350. doi: 10.1006/dbio.2000.9985. [DOI] [PubMed] [Google Scholar]
- Nishida H. Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. III. Up to the tissue restricted stage. Developmental biology. 1987;121:526–541. doi: 10.1016/0012-1606(87)90188-6. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. Embo J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Klein WH. bHLH factors in muscle development: deadlines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- Olson EN, Klein WH. Muscle minus myoD. Developmental biology. 1998;202:153–156. doi: 10.1006/dbio.1998.9020. [DOI] [PubMed] [Google Scholar]
- Ott RL. An introduction to statistical methods and data analysis. Duxbury Press; N. Scituate: 1993. [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annual review of cell and developmental biology. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, Olson EN. Myogenin’s functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Developmental biology. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Schwarz JJ, Chakraborty T, Martin J, Zhou JM, Olson EN. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Molecular and cellular biology. 1992;12:266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. CRC Press; Boca Raton, FL: 1997. [Google Scholar]
- Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic acids research. 2007;35:7087–7095. doi: 10.1093/nar/gkm746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. Skeletal muscle stem and progenitor cells: Reconciling genetics and lineage. Exp Cell Res. 2005;306:364–372. doi: 10.1016/j.yexcr.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Venuti JM, Goldberg L, Chakraborty T, Olson EN, Klein WH. A myogenic factor from sea urchin embryos capable of programming muscle differentiation in mammalian cells. Proceedings of the National Academy, USA. 1991;88:6219–6223. doi: 10.1073/pnas.88.14.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuti JM, Morris JH, Vivian JL, Olson EN, Klein WH. Myogenin is required for late but not early aspects of myogenesis during mouse development. The Journal of cell biology. 1995;128:563–576. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Katsuyama Y, Yasugi S, Saiga H. Spatially and temporally regulated expression of the LIM class homeobox gene Hrlim suggests multiple distinct functions in development of the ascidian, Halocynthia roretzi. Mechanisms of development. 1995;51:115–126. doi: 10.1016/0925-4773(95)00359-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jaenisch R. Myogenin can substitute for Myf5 in promoting myogenesis but less efficiently. Development. 1997;124:2507–2513. doi: 10.1242/dev.124.13.2507. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proceedings of the National Academy, USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker JR. Segregation during ascidian embryogenesis of egg cytoplasmic information for tissue-specific enzyme development. Proceedings of the National Academy of Sciences, USA. 1973;70:2096–2100. doi: 10.1073/pnas.70.7.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Rhodes SJ, Konieczny SF. Differential trans activation associated with the muscle regulatory factors MyoD1, myogenin, and MRF4. Molecular and cellular biology. 1990;10:3934–3944. doi: 10.1128/mcb.10.8.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Konieczny SF. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic acids research. 1992;20:5105–5113. doi: 10.1093/nar/20.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Chen L, Krause M, Fire A, Paterson BM. Evolutionary conservation of MyoD function and differential utilization of E proteins. Developmental biology. 1999;208:465–472. doi: 10.1006/dbio.1999.9218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.