Abstract
All salmonid species investigated to date have been characterized with a male heterogametic sex-determination system. However, as these species do not share any Y-chromosome conserved synteny, there remains a debate on whether they share a common master sex-determining gene. In this study, we investigated the extent of conservation and evolution of the rainbow trout (Oncorhynchus mykiss) master sex-determining gene, sdY (sexually dimorphic on the Y-chromosome), in 15 different species of salmonids. We found that the sdY sequence is highly conserved in all salmonids and that sdY is a male-specific Y-chromosome gene in the majority of these species. These findings demonstrate that most salmonids share a conserved sex-determining locus and also strongly suggest that sdY may be this conserved master sex-determining gene. However, in two whitefish species (subfamily Coregoninae), sdY was found both in males and females, suggesting that alternative sex-determination systems may have also evolved in this family. Based on the wide conservation of sdY as a male-specific Y-chromosome gene, efficient and easy molecular sexing techniques can now be developed that will be of great interest for studying these economically and environmentally important species.
Keywords: sdY, salmonids, sex-determining gene, sex-determination locus, molecular sexing
Introduction
Master sex-determining genes are the main genetic switches controlling the gonadal sex differentiation cascade leading to the development of ovaries or testes. In mammals, which harbor an XX/XY system, the Sry (sex-determining region of the Y chromosome) gene was identified in the early 1990s (Sinclair et al. 1990) as the first vertebrate master sex-determining gene. In chickens, which have a female heterogametic (ZZ/ZW) sex-determination system, the Z-linked Dmrt1 (doublesex and mab-3 related transcription factor 1) gene is a strong candidate for the chicken sex-determining gene (Smith et al. 2009), and this gene triggers gonad masculinization by a double dosage mechanism in males compared with females. Interestingly, dmrt1 has also been shown to be conserved on the Z chromosome in the majority of bird species (Stiglec et al. 2007) including in the ancient ratite emu species (Shetty et al. 2002), suggesting a conservation of this gene as the master sex-determining gene in all bird species. In amphibians, the only currently known sex-determining gene is the dm-W gene in the African clawed frog, Xenopus laevis (Yoshimoto et al. 2008; Yoshimoto and Ito 2011). As this gene is located on the W female heterochromosome and evolved through the duplication of a dmrt1 autosomal gene that lost its transactivation domain, this suggests that dm-W acts as a dominant negative regulator of dmrt1 (Yoshimoto and Ito 2011). This evolution was, however, relatively recent because dm-W is conserved only in some Xenopus species and was most likely acquired after the divergence of the genera Silurana and Xenopus (Bewick et al. 2011). In teleost fish, an extremely high diversity in sex-determination systems and sex chromosomes has been found (Mank and Avise 2009), even within the same genus, as in the medaka (Oryzias spp.) (Takehana et al. 2007). Different master sex-determining genes have been recently characterized in teleosts including the dmrt1 gene duplication, dmrt1Y in Oryzias latipes and O. curvinotus (Matsuda et al. 2002, 2007), the gsdfY (gonadal soma- derived growth factor on the Y chromosome) gene in O. luzonensis (Myosho et al. 2012), the amhy (anti-Müllerian hormone on the Y) gene in the Patagonian pejerrey, Odontesthes hatcheri (Hattori et al. 2012), the amhr2 in the Tiger Pufferfish, Takifugu rubrupes (Kamiya et al. 2012), and the sdY (sexually dimorphic on the Y-chromosome) gene in the rainbow trout (Yano et al. 2012). To date, the evolution of these teleost master sex-determining genes has only been characterized in medaka species, in which the switch from one sex determinant to another has been found to be an evolutionarily recent event (Matsuda et al. 2003; Volff et al. 2003).
Salmonids sex-determination has been a long-standing mystery and also an important applied challenge, as many of these species are important both for aquaculture, in which sex control would be often beneficial, and for environmental purposes because of their emblematic value. Their sex-determination system is often described as always being male heterogametic (XX/XY) [reviewed in (Davidson et al. 2009)]. However, this assumption has been based only on the knowledge gained from the examination of a small subset of species, all belonging to the Salmoninae subfamily, i.e., in the genera Oncorhynchus, Salmo, and Salvelinus [reviewed in (Davidson et al. 2009)], whereas very little is currently known about sex-determination in the Coregoninae and nothing is known about sex-determination in the Thymallinae. In the majority of these species, the sex-determining (SEX) locus has been identified on different chromosomes (Woram et al. 2003; Li et al. 2011), leaving an open question about the uniqueness of a master sex-determining gene in this family (Davidson et al. 2009).
The recent discovery of sdY as the master sex-determining gene in rainbow trout (Yano et al. 2012) now allows this issue to be revisited. This is especially relevant because the sdY gene is included in the Y-chromosome genomic sequence of the rainbow trout (OmyY1), which shares strong similarities with that of the Chinook salmon, Oncorhynchus tshawytscha (OtY3) (Brunelli et al. 2008), suggesting that sdY may be conserved as a Y-chromosome-specific sequence, at least within the Oncorhynchus genus. In this study, we thus investigated whether sdY is conserved as a male-specific Y-chromosome gene in other salmonids. Our results demonstrated that sdY is a male-specific conserved sequence found only in salmonid species and is tightly linked to the sex-determining (SEX) locus both in rainbow trout and brown trout, Salmo trutta, two salmoninae that are found in different genera. These results shed new light on salmonid sex-determination by demonstrating that the majority of salmonids do share a common sex-determination system (XX/XY) and a common SEX locus and that sdY may be this common master sex-determining gene.
Members of the salmonid family are present worldwide and many of them are species of major importance for aquaculture, wild stock fisheries, or recreational sport fisheries. Salmonids are also emblematic fish species of the Northern hemisphere and much concern has been raised about their preservation as many salmonid species are considered as threatened with extinction in many countries. This identification of a conserved master sex-determining gene in salmonids will then have important practical outcomes for molecular sexing. For instance, this will be of great interest for a better control of sex determination in aquaculture (and therefore improved artificial selection practices). Being able to unambiguously assess the sex of most salmonids (except coregonines) at any life stages will also contribute to facilitate and improve wild population surveys as well as stocking practices.
Materials and methods
Isolation of genomic DNA
Genomic DNA was extracted from fin clips of Coregonus lavaretus, Coregonus clupeaformis, Stenodus leucichthys, Thymallus thymallus, Oncorhynchus mykiss, Oncorhynchus masou, Oncorhynchus tshawytscha, Salvelinus malma malma, Salvelinus alpinus, Salvelinus fontinalis, Salvelinus namaycush, Salmo trutta, Salmo salar, Hucho hucho, Parahucho perryi, and Esox lucius preserved in 95% ethanol or dried on silica beads. All these animals were sexed based on macroscopic examination of the gonads or the emission of gametes during the active reproductive season. The number of samples per species, the locations of the collected samples, and the names of the collectors are listed in Table 1. Genomic DNA extractions were performed in 96-well plates using the Chelex® method as previously described (Gharbi et al. 2006). For each animal, a small portion of fin clip (2 mm × 2 mm) was placed in 100 μL of digestion solution, containing 5% of Chelex® 100 resin (Bio-Rad, Marnes-la-Coquette, France) and 0.5 μg/μL of proteinase K (Roche Diagnostics, Meylan, France) in distilled water. The samples were incubated at 55°C for 2 h, and then the proteinase K was inactivated at 95°C for 10 min. The plates were then briefly centrifuged to pellet the Chelex® resin, and the supernatant containing genomic DNA was stored at 4°C or −20°C.
Table 1.
Origins of the samples and presence/absence of the sdY genotype in several salmonids
| Subfamilies | Species | Common name | Location | Collector | sdY positive animals | Total | |
|---|---|---|---|---|---|---|---|
| Males | Females | ||||||
| Coregoninae | Coregonus lavaretus | European whitefish | Lake Leman, Rhone, France | Alexis Champigneulle | 8/8 | 9/9 | 17 |
| Coregonus clupeaformis | Lake whitefish | Lake Ontario, Ontario, Canada | Vance Trudeau | 14/14 | 14/14 | 28 | |
| Stenodus leucichthys | See fish | Innoko River, Alaska, United States | John Burr, Jeff Olsen, April Behr, Randy Brown | 41/41 | 0/13 | 54 | |
| Thymallinae | Thymallus thymallus | Grayling | Pisciculture Saumon du Rhin, Obenheim, France | Martin Gerber | 27/27 | 0/27 | 54 |
| Salmoninae | Oncorhynchus mykiss | Rainbow trout | PEIMA, Sizun, France | A.Y and collaborators | 218/218 | 0/207 | 425 |
| Oncorhynchus masou | Masu salmon | Field Science Center Oizumi Station, Yamanashi, Japan | Masaru Yagisawa, Goro Yoshizaki | 5/5 | 0/5 | 10 | |
| Oncorhynchus tshawytscha | Chinook Salmon | Big creek, Alaska, United States | Ora Schlei | 27/27 | 1/41 | 68 | |
| Salvelinus malma malma | Dolly Varden trout | Salmon River, Alaska, United States | Mark Lisac | 20/20 | 0/20 | 40 | |
| Salvelinus alpinus | Arctic char | Salmoniculture des Monts d'Arrée, Huelgoat, France | A.Y and collaborators | 9/9 | 0/12 | 21 | |
| Salvelinus fontinalis | Brook trout | Salmoniculture des Monts d'Arrée, Huelgoat, France | A.Y and collaborators | 13/13 | 0/15 | 28 | |
| Salvelinus namaycush | Lake char | Pisciculture Fédérale de Cauterets, Cauterets, France | Cauterets' Fish Hatchery Technicians | 19/20 | 0/19 | 39 | |
| Salmo trutta | Brown trout | PEIMA, Sizun, France | A.Y and collaborators | 73/73 | 0/76 | 149 | |
| Salmo salar | Atlantic salmon | River Loir, France | Guillaume Evanno | 20/20 | 0/20 | 40 | |
| Hucho hucho | Huchen | Polsh Anglers Association Hatchery, Lopuszna, Polland | Tomasz Mikolajczyk | 10/10 | 0/10 | 20 | |
| Parahucho perryi | Sakhalin taimen | Field Science Center Oizumi Station, Yamanashi, Japan | Masaru Yagisawa, Goro Yoshizaki | 7/7 | 0/3 | 10 | |
| Esocidae | Esox lucius | Northern pike | Domaine de Lindre, Moselle, rue principale - 57260 Lindre-Basse, France | Thibaut Glasser, Julien Periz | 0/10 | 0/10 | 20 |
Cloning of the sdY sequences in various salmonid species
RNA extraction and reverse transcription reactions were performed as previously described (Govoroun et al. 2001) using testis samples collected from O. masou, S. trutta, S. salar, P. perryi, T. thymallus, S. alpinus, S. fontinalis and C. lavaretus. PCR reactions were performed using 0.5 μm of each primer, 1 μL of cDNA diluted 1:30, 200 μm dNTP mixture, and 2 μL 10× PCR Buffer (Sigma Aldrich Chimie s.a.r.l., Lyon, France) with 0.5 units of JumpStart Taq DNA Polymerase (Sigma Aldrich) in a total volume of 20 μL. Thermal cycling consisted of denaturation for 60 s at 94°C followed by 30 amplification cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension of 3 min at 72°C. The primers used for RT-PCR are listed in Tables 2 and 3. These PCR products were directly used for sequencing. In H. hucho, the sdY gene was cloned and sequenced directly from genomic DNA using the Long PCR Product Sequencing (LoPPS) approach (Emonet et al. 2007). In S. leucichthys, the sdY gene was also PCR amplified from genomic DNA using several of the primer combinations that are listed in Tables 2 and 3. These PCR products were sequenced directly. Sequence alignments using partial sequences of sdY were processed using ClustalW. The sequence accession numbers for the proteins retained in the analyses are C. auratus Irf9, AFL69828; S. salar Irf9a, ACN11040; O. mykiss SdY, AB62689; T. thymallus SdY, JF826018; S. trutta SdY, JF826019; S. salar SdY, JF826020; S. fontinalis SdY, JF826021; S. alpinus SdY, JF826022; O. masou SdY, JF826023; P. perryi SdY, JF826024; H. hucho SdY, JF951962; C. lavaretus SdY, JF826025; and S. leucichthys SdY, JX196650. Rainbow trout Irf9a and b and Atlantic salmon Irf9b proteins were deduced from the following nucleotide sequences: CX258512, GE783444, EZ806353, CA346150, and CA375227.
Table 2.
Names and combinations of the primers
| Primers used to characterize the sdY sequence in several salmonid species | |||
| Species | Primer Pair | ||
| Oncorhynchus masou | sdY E1Sb | sdY E4AS1a | |
| Salmotrutta | sdY E1Sb | sdY E4AS1a | |
| Salmosalar | sdY E1Sb | sdY E4AS1a | |
| Parahuchoperryi | sdY E1Sb | sdY E4AS1a | |
| Salvelinus alpinus | sdY E1Sb | sdY E4AS1a | |
| Salvelinus fontinalis | sdY E1Sb | sdY E4AS1a | |
| Coregonus lavaretus | sdY E1Sb | sdY E4AS1a | |
| Huchohucho | sdY E1Sa | sdY E4AS1a | |
| Parahuchoperryi | sdY E1Sa | sdY E4AS1a | |
| Thymallusthymallus | sdY E1Sb | sdY E4AS1a | |
| Stenodus leucichthys (E1) | OmyY1Fw11022 | OmyY1Rv11199 | |
| Stenodus leucichthys (E2) | OmyY1Fw11178 | OmyY1Rv11918 | |
| Stenodus leucichthys (E3) | OmyY1Fw13771 | OmyY1Rv13914 | |
| Stenodus leucichthys (E4) | OmyY1Fw15916 | sdY E4AS1a | |
| Primers for genotyping sdY in several species | |||
| Species | Primer Pair | ||
| Coregonus lavaretus | sdY E1S1 | sdY E2AS4 | |
| Coregonus clupeaformis | sdY E1S1 | sdY E2AS4 | |
| Stenodus leucichthys | sdY E2S1 | sdY E2AS4 | |
| Thymallus thymallus | sdY E1S1 | sdY E2AS4 | |
| Oncorhynchus mykiss | sdY E2S1 | sdY E2AS2 | |
| Oncorhynchus masou | sdY E1S1 | sdY E2AS5 | |
| Oncorhynchus tshawytscha | sdY E2S1 | sdY E2AS4 | |
| Salvelinus malma malma | sdY E2S1 | sdY E2AS4 | |
| Salvelinusalpinus | sdY E2S1 | sdY E2AS4 | |
| Salvelinus fontinalis | sdY E2S1 | sdY E2AS2 | |
| Salvelinus namaycush | sdY E2S1 | sdY E2AS1 | |
| Salmo trutta | sdYE1S1 | sdY E2AS4 | |
| Salmo salar | SS sdY S | SS sdY AS | |
| Hucho hucho | HH sdY S | HH sdY AS | |
| Parahuchoperryi | sdY E1S1 | sdY E2AS4 | |
| all salmonid samples | 18S S | 18S AS | |
| Primers tested for amplification of sdY in Northern pike (Esox lucius) | |||
| Species | Primer Pair | ||
| Esox lucius | sdY E2S2 | sdY E2AS2 | |
| Esox lucius | sdY E2S1 | sdY E2AS1 | |
| Esox lucius | sdY E2S1 | sdY E2AS3 | |
Table 3.
Primers sequence
| Primers | Sequence |
|---|---|
| sdY E1S1 | ATGGCTGACAGAGAGGCCAGAATCCAA |
| sdY E1Sa | CTGCCCTTCAATGGCTGACAGAGAG |
| sdY E1Sb | TTCAATGGCTGACAGAGAGGCCAGA |
| sdY E2S1 | CCCAGCACTGTTTTCTTGTCTCA |
| sdY E2S2 | GTGGAGTACTGCGAAGAGGAGGT |
| sdY E2AS1 | TGCTCTCTGTTGAAGAGCATCAC |
| sdY E2AS2 | CTGTTGAAGAGCATCACAGGGTC |
| sdY E2AS3 | AGGAGACTGTGGCTTGGCTATG |
| sdY E2AS4 | CTTAAAACCACTCCACCCTCCAT |
| sdY E2AS5 | AGAGCATCACAGGGTCCACATCACG |
| sdY E4AS1a | GGGAGGACTCAAGCCAGATCCTGAA |
| HH sdY S | CCATGTCTGATCGTTTGAGGAAA |
| HH sdY AS | GCATAGATGCCTTCCTCCCTAGA |
| SS sdY S | GGCCTATGCATTTCTGATGTTGA |
| SS sdY AS | AGAGGATTGAACGGTCAGAGGAG |
| 18S S | GTYCGAAGACGATCAGATACCGT |
| 18S AS | CCGCATAACTAGTTAGCATGCCG |
| OmyY1Fw11022 | TAKTTGAGTCCATCTGCCCTTCA |
| OmyY1Rv11199 | TCAGACATGGAAATACCACAT |
| OmyY1Fw11178 | TGTGGTTATTTCCATGCTGAT |
| OmyY1Rv11918 | CACCRKTTTYTCAGGCATTAC |
| OmyY1Fw13771 | TATTACTGACTCTGTGTGTGTCC |
| OmyY1Rv13914 | TGAGTAAGAGAATCTGTACCG |
| OmyY1Fw15916 | TGGTGTTGATTATAATTARATG |
PCR analysis for population and mapping family genotyping using the sdY sequence
Generally, PCR was performed using 0.1 μm of each primer, 40 ng of DNA, 200 μm dNTP mixture, and 2 μL of 10× PCR Buffer (Sigma Aldrich) with 0.5 units of JumpStart Taq DNA Polymerase (Sigma Aldrich) in a total volume of 20 μL. Thermal cycling consisted of denaturation for 60 sec at 94°C followed by 40 amplification cycles for sdY and 30 cycles for 18S:94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension of 3 min at 72°C. PCR products were electrophoresed on a 2% agarose gel. In addition, we tested the multiplex PCR analysis developed for rainbow trout (Yano et al. 2012) in S. leucichthys, S. malma malma, and O. tshawytscha as an easier molecular sexing tool allowing the simultaneous detection of both the sdY and 18S genes. These multiplex PCR were performed using sdY primers, with 18S primers as a positive amplification control. Each reaction contained 2 μL of a 1:5 dilution of Chelex®-extracted genomic DNA, 0.4 μm of each sdY primer (sdY E2S1 and sdY E2AS4; see Table 3 for the primer sequences), 0.1 μm of each 18S primer (18S Fw and 18S Rv; see Table 3 for the primer sequences), 250 μm dNTP mixture, 1.5 mm MgCl2, 5× Green GoTaq Reaction Buffer (Promega France, Lyon, France), and 0.3 units of GoTaq polymerase (Promega), in a final volume of 12.5 μL. The following temperature cycling regime was applied: denaturation for 3 min at 95°C followed by 35 amplification cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension of 3 min at 72°C. The PCR products were electrophoresed on 2% agarose gels.
Characterization of sdY sex linkage groups in salmonids
In rainbow trout, two full-sib families of 46 individuals each were genotyped with sdY and markers previously assigned to RT01 (Guyomard et al. 2006). In brown trout, sdY mapping was performed on the same family panels as in (Gharbi et al. 2006). In these two species, centromeres were mapped using gynogenetic lines; DNA extraction, genotyping methods, marker accession numbers, map constructions, and graphic representation were all as previously described (Gharbi et al. 2006; Guyomard et al. 2006).
Results
sdY is a conserved sequence in salmonids
Using homology-based cloning with primers designed in the coding region of the rainbow trout sdY cDNA (Tables 2 and 3), we obtained partial sdY sequences in 10 additional salmonid species representative of the three salmonid subfamilies, i.e., Salmoninae (O. masou, S. salar, S. trutta, H. hucho, P. perryi, S. alpinus, and S. fontinalis), Thymallinae (T. thymallus), and Coregoninae (C. lavaretus and S. leucichthys). However, we were unable to obtain any sdY-related PCR products in the Northern pike (E. lucius), which belongs to the Esocidae family and is considered to be the closest salmonid sister group (Lopez et al. 2004). ClustalW alignment of the deduced protein sequences revealed that SdY proteins are highly conserved (more than 87.1% identity) among salmonids (Fig. 1). As SdY shares some homology with the C-terminal domain of interferon regulatory factor 9 (Irf9) proteins (Yano et al. 2012), we also compared these SdY sequences with several fish Irf9 sequences. This comparison confirmed the relatively low identity of SdY with the C-terminal sequences of Irf9 (34.7–47.2%) but highlighted the fact that certain stretches of highly conserved amino-acids of the SdY proteins are also conserved in the Irf9 proteins (Fig. 1).
Figure 1.
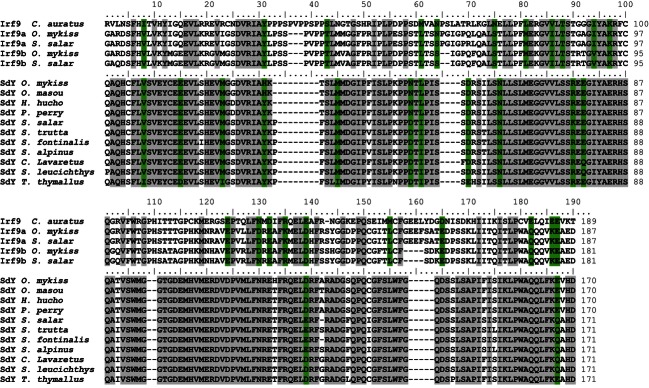
Alignment of Irf9 and SdY amino acid sequences in different salmonids. Using homology-based cloning, partial sequences of SdY were characterized in 11 representative species from the three salmonid subfamilies. ClustalW alignment of SdY deduced protein sequences reveals that SdY proteins are highly conserved in salmonids. Gray and green shading identify identical and similar amino acids, respectively. SdY sequences also show high homology with Irf9 sequences of different teleosts, and certain stretches of highly conserved amino-acids of the SdY proteins were also conserved in the Irf9 proteins. For SdY, identity and similarity are shown with respect to SdY sequences only and for Irf9 with respect to Sdy and Irf9 sequences. Sequence accession numbers for the proteins retained in the analyses are described in the Materials and methods section.
The sdY gene is conserved as a male-specific Y-chromosome sequence in many salmonids
sdY was strictly conserved as a male-specific sequence in nearly all Salmoninae members studied (O. mykiss, O masou, S. salar, S. trutta, H. hucho, P. perryi, S. alpinus, S. fontinalis, and S. malma malma) and in Thymallinae (T. thymallus); see Fig. 2. The only exceptions to this clear-cut distribution were one sdY-positive female out of 41 in O. tshawytscha (Fig. 3, Table) and one sdY-negative male out of 20 in S. namaycush (Table 1). However, in the Coregoninae subfamily, two different patterns emerged: in one species, S. leucichthys, sdY was clearly male specific; in two other species, C. lavaretus and C. clupeaformis, all males and all females were sdY positive (Fig. 2, Table 1). To facilitate the use of sdY as a potential molecular sexing tool, we tested a simple multiplex PCR assay that was initially developed for rainbow trout (Yano et al. 2012) to co-amplify sdY and a positive control, the 18S gene. This multiplex PCR approach was tested in three different salmonid species (S. leucichthys, S. malma malma, and O. tshawytscha) and was found to be efficient and reliable (Fig. 3).
Figure 2.
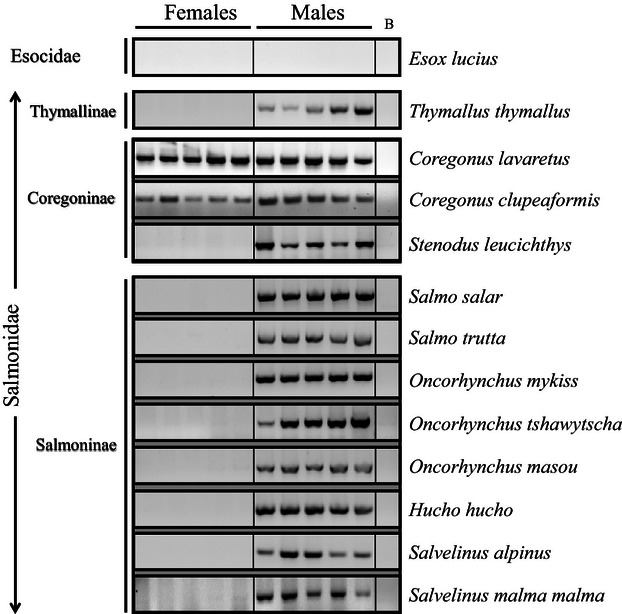
sdY sequence conservation in the male genomes of different salmonids.PCR amplification of sdY in male and female (n = 5) genomic DNA samples from different salmonid species showing a male-specific signal in Salmoninae and Thymallinae. In the Coregoninae species C. lavaretus and C. clupeaformis, sdY was detected both in male and female genomic DNA. No PCR products were obtained in Northern pike, Esox Lucius (Esocidae), which was chosen as an outgroup species closely related to salmonids.
Figure 3.
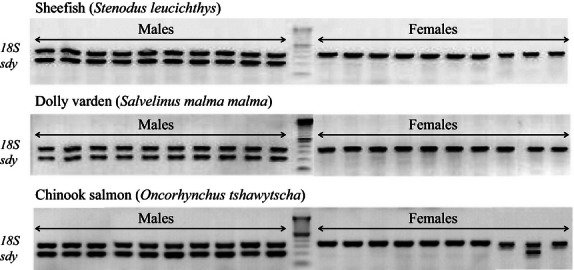
Multiplex PCR in Stenodus leucichthys, Salvelinus malma malma, and Oncorhynchus tshawytscha.PCR co-amplification of sdY and 18S (positive amplification control) in males and females genomic DNA samples (n = 10 animals/sex) from sheefish (S. leucichthys), dolly varden (S. malma malma) and Chinook salmon (O. tshawytscha). In the sheefish and dolly varden, sdY was strictly detected in males, whereas in Chinook salmon, sdY was detected in all males and also in a single female. The positive control 18S was detected in all PCR reactions.
sdY co-localizes with the phenotypic sex on the SEX linkage groups in both rainbow trout and brown trout
Genetic mapping of sdY along with several microsatellite SEX-linked markers in both rainbow trout (O. mykiss) and brown trout (S. trutta) demonstrates that sdY is tightly linked with the SEX locus on both rainbow trout linkage group 01 (RT01) and brown trout linkage group 18 (BT18); see Fig. 4. In brown trout, the 2.2 cM distance obtained between sdY and the phenotypic sex likely reflects a phenotyping error, most likely because the brown trout mapping families were less than 1-year old. This hypothesis was further confirmed when no recombination was found between sdY and the sexual phenotype in 150 individuals in a wild panmictic brown trout population. A comparison of the sex linkage groups BT18 in brown trout and RT01 in rainbow trout (Yano et al. 2012) demonstrated that RT01 and BT18 share no conserved markers, with the notable exception of sdY, and that the SEX locus surrounding markers from one species are all mapped on an autosomal group in the other species, BT28 with RT21 and RT01 with BT13 (Fig. 4).
Figure 4.
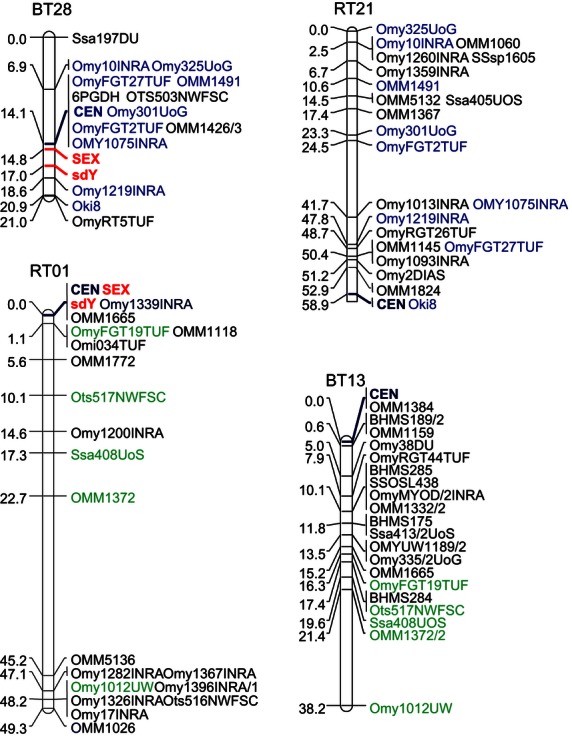
sdY and SEX-associated markers in rainbow trout (Oncorhynchus mykiss) and brown trout (Salmo trutta). Sex linkage groups (BT18, RT01) and their homologous counterparts resulting from the salmonid-specific whole-genome duplication (RT21 and BT13) are given in two Salmoninae species, i.e., brown trout (BT) and rainbow trout (RT). RT01 and BT18 share no conserved markers, with the notable exception of sdY, which is tightly linked with SEX in these two species. Common markers between BT28 and RT21 are shown in blue, and common markers between RT01 and BT13 are shown in green; CEN = centromere.
Discussion
It has been often suggested that genes involved in the sex differentiation pathway may have evolved independently as new master sex-determining genes in the course of vertebrate evolution (Schartl 2004; Marshall Graves and Peichel 2010). This idea has been mainly supported by the functional conservation of dmrt1 as a master sex determinant in diverse vertebrate lineages (Yoshimoto et al. 2008; Herpin and Schartl 2011; Mawaribuchi et al. 2012). However, the recent finding that the rainbow trout master sex-determining gene, sdY (sexually dimorphic on the Y-chromosome) resulted from the de novo evolution of an immune-related gene demonstrated that there is likely an unexpected evolutionary plasticity in the mechanisms underlying vertebrate sex-determination (Yano et al. 2012).
In this study, we investigated the extent of the conservation and evolution of sdY in the salmonid family. Based on a previously described similarity between two Salmoninae Y-chromosome-specific sequences, i.e., the rainbow trout OmyY1 genomic sequence containing the sdY gene and the OtY2 sequence found in Chinook salmon (Brunelli et al. 2008), we hypothesized that sdY may be conserved as a Y-chromosome gene, at least within the Oncorhynchus genus. Quite surprisingly, our results demonstrate that sdY is not only conserved as a male-specific Y-chromosome sequence in Oncorhynchus but also in many different salmonid species belonging to the Salmoninae, Thymallinae, and Coregoninae subfamilies (Fig. 5). We further confirmed this male-specific linkage in both rainbow trout and brown trout (Salmo trutta) by gene mapping experiments that demonstrated that sdY maps to the SEX linkage group in these two species. sdY is therefore the first and only sex marker known to be conserved among different salmonids. This conserved and tight male-specific linkage of sdY in salmonids contrasts with previous genetic mapping analyses showing that sex-linked markers in a given species always map to autosomal homologous linkage groups in other species (Woram et al. 2003; Li et al. 2011). This general lack of Y-chromosome synteny has often been used to suggest that salmonids most likely do not share the same master sex-determining gene (Davidson et al. 2009). Our results now provide clear evidence that the majority of salmonids do share the same master sex-determining locus and potentially also the same master sex-determining gene. These results also support one of the two hypotheses proposed by Woram and collaborators (Woram et al. 2003) suggesting that this gene has jumped into different ancestral autosomes during the evolution of salmonids, resulting into new Y sex chromosomes. Whether these chromosomal transitions were driven by transposition or translocation mechanisms is yet unknown, but the existence of several degenerate transposable elements found in the OmyY1 sequence (Brunelli et al. 2008) a few kb upstream of sdY strongly suggests that the sdY/SEX locus has been transposed by moveable elements.
Figure 5.
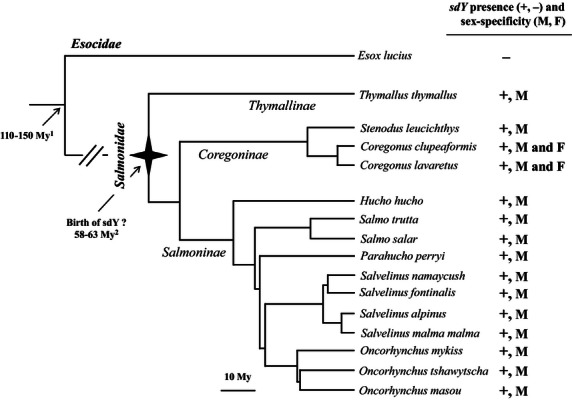
Phylogeny of salmonids and evolution of sdY. Phylogram of the species investigated in this study plotted with the presence (+)/absence (−) of sdY and its sex-specificity (M: male specific, M, F: present both in males and females. The salmonid phylogeny and the timing of divergence of species were based on Crête-lafrenière and collaborators (Crête-Lafreniere et al. 2012). Divergence time between Esocidae and Salmonidae was based on Near and collaborators (Near et al. 2012).
Our results provide an additional evidence that salmonid sex chromosomes are extremely ‘young’ (i.e., homomorphic) sex chromosomes in term of evolution and strongly support the jumping sex locus hypothesis proposed by Woram and collaborators (Woram et al. 2003). According to the ‘fountain of youth’ hypothesis (Perrin, 2009) young sex chromosomes would escape from degeneration by maintaining rare recombination events, that would be promoted by occasional spontaneous temperature-dependent sex inversions. The high-turnover model is an alternative hypothesis that postulates that the high turnover of master sex-determining genes in fish would enable the replacement of sex chromosomes before they begin to degenerate (Schartl 2004; Volff et al. 2007). In salmonids occasional spontaneous temperature-dependent sex inversion have been reported but most of them involved masculinization of genetic females (Craig et al. 1996; Azuma et al. 2004), thus not promoting sex inversion toward the female phenotype that has the highest rate of recombination. This does not fit well with the Perrin's ‘fountain of youth’ hypothesis. However, a new hypothesis can be proposed in the case of salmonids as a jumping sex locus like the sdY locus by continuously reshuffling the sex chromosomes would be also sufficient to prevent their decay. Interestingly this hypothesis shares with the high-turnover model the idea of a regular replacement of the sex chromosomes and keeps the ‘old-wine in-a-new-bottle’ Perrin's idea, that young sex chromosomes might actually harbor old sex-determining genes.
However, the male-specific sex linkage of sdY cannot be extended to all salmonids because we found at least two exceptions in the Coregoninae subfamily, with sdY being detected in both male and female genomes in C. lavaretus and C. clupeaformis. This absence of a sex-specific linkage of sdY in these Coregoninae may be the consequence of the existence of multiple sex chromosomes, as previously described by cytogenetic approaches in two other Coregoninae species, C. albula and C. sardinella (Frolov 1990; Jankun et al. 1991). In such cases, sdY may trigger testicular differentiation through a dosage mechanism, similarly to the dmrt1 gene dosage hypothesis in chicken (Smith et al. 2009). However, other hypotheses may be proposed such like that females have a sex-specific inactive copy of sdY, similarly to the single sex-linked mutation reported in the amhr2 gene in Takifugu (Kamiya et al. 2012) or that sdY is no longer the master sex determinant in these species. The latter hypothesis is supported by the fact that transition between different master sex-determining genes can arise within short evolutionary time periods, as observed in several medaka species (Matsuda et al. 2003; Volff et al. 2003).
Despite many attempts, we have been unable to clone any sdY-related sequence from Northern pike, E. lucius, which belongs to the Esocidae, a family that is considered to be the closest salmonid sister group (Lopez et al. 2004). This result is in agreement with a previous report (Yano et al. 2012) that showed that sdY-related sequences were absent from the teleost sequence databases (genome and expressed sequence tags databases), supporting the idea that sdY is a salmonid innovation. The birth of that master sex determining gene would then coincide with the age of the salmonid family (Fig. 5) that is dated around 60 million years old (Crête-Lafreniere et al. 2012).
Easy and reliable early identification of sex remains an important unsolved challenge in ecology and population genetics. From a practical point of view, molecular tools allowing accurate estimations of sex ratios would represent a considerable progress in the prediction of demographic evolution of stocks, which is an important concern in many salmonid species. From an academic point of view, the availability of such a technique would allow us to renew or greatly facilitate studies centered on or involving sex-ratio fluctuation, evolution or putative environmentally controlled sex reversion in natural populations. In population genetics, this ability offers the possibility of studying male-mediated versus female-mediated dispersion and introgression in natural populations following secondary contact (i.e., stocking or escapement from fish farms). Such a molecular sexing technique would also help to evaluate the impact of endocrine disruptors on complete or partial sex inversion in wild salmonid populations and enable a better management of sex control in salmonid aquaculture species, which is often an important issue for the production of all-female populations (Donaldson and Hunter 1982; Hunter and Donaldson 1983). As we demonstrated that sdY is both conserved in terms of sequence identity and also as a male-specific gene in many salmonids, this information can now be used to develop a simple and efficient molecular sexing technique that could be applied to many salmonid species. The main advantage of this sex-specific marker is its strong sequence conservation, which allows for the development of a unique test that can be used in multiple salmonid species. We provide here preliminary results indicating that this test could be realized quite easily, as we were able to accurately sex genotype different salmonid species from different subfamilies using a simple multiplex PCR approach that was initially designed for rainbow trout (Yano et al. 2012). However, caution should be taken before making any generalizations on applying these results to other species. In addition to sdY being present both in male and female genomes in several Coregoninae, we also found a single sdY-positive female in Chinook salmon (O. tshawytscha) and a single sdY-negative male in lake trout (S. namaycush). These exceptions to the rule may be due to human error in the assessment of the phenotypic sex of these animals, but these apparent discrepancies may also be biologically relevant; for example, the occurrence of ‘apparent’ XY females has been well-documented in Chinook salmon (Nagler et al. 2001; Williamson and May 2005; Williamson et al. 2008). In addition, the occurrence of potential XX males has also been reported in several salmonid species (Quillet et al. 2002; Metcalf and Gemmell 2006), and masculinization of genetic females by environmental factors or pollutants has also been demonstrated in other species (Craig et al. 1996; Azuma et al. 2004).
Taken together, our findings open new avenues for studying the molecular evolution of sex-determination and sex chromosomes in salmonids, including the simple assessment of male heterogamety in additional salmonid species, as shown in this study for the huchen (H. hucho), the Sakhalin taimen (P. perryi), the dolly Varden (S. malma malma), the European grayling (T. thymallus) and the Inconnu (S. leucichthys). These results also provide a biological basis for the development of new molecular sexing approaches that will be very important for ecology and ecotoxicology research and for better control of sex-determination in aquaculture in this economically and environmentally very important fish family.
Acknowledgments
We thank Guillaume Evanno, Martin Gerber, Alexis Champigneulle, Cyrille Chataignier, Vance Trudeau, Jeremy Guiguen, Tomasz Mikolajczyk, Goro Yoshizaki, Masaru Yagisawa, Thibaut Glasser, Julien Periz, Ora Schlei and the PEIMA experimental facility staff for their help in collecting the salmonid samples. This study was supported by funds from the Institut National de la Recherche Agronomique (INRA), the Agence Nationale de la Recherche (ANR Blanc SVSE 7 2011, project SDS) and the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement No. 222719–LIFECYCLE. A.Y. and B.N were supported by a post-doctoral fellowship from INRA and ANR.
Data archiving statement
Sequence information for this study is available at GenBank under the following accession numbers: T. thymallus SdY, JF826018; S. trutta SdY, JF826019; S. salar SdY, JF826020; S. fontinalis SdY, JF826021; S. alpinus, JF826022; O. masou SdY, JF826023; P. perryi SdY, JF826024; H. hucho SdY, JF951962; C. lavaretus SdY, JF826025; S. leucichthys SdY, JX196650.
Literature cited
- Azuma T, Takeda K, Doi T, Muto K, Akutsu M, Sawada M, Adachi S. The influence of temperature on sex-determination in sockeye salmon Oncorhynchus nerka. Aquaculture. 2004;234(1–4):461–473. [Google Scholar]
- Bewick AJ, Anderson DW, Evans BJ. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus. Evolution. 2011;65(3):698–712. doi: 10.1111/j.1558-5646.2010.01163.x. [DOI] [PubMed] [Google Scholar]
- Brunelli JP, Wertzler KJ, Sundin K, Thorgaard GH. Y-specific sequences and polymorphisms in rainbow trout and Chinook salmon. Genome. 2008;51(9):739–748. doi: 10.1139/G08-060. [DOI] [PubMed] [Google Scholar]
- Craig JK, Foote CJ, Wood CC. Evidence for temperature-dependent sex-determination in sockeye salmon (Oncorhynchus nerka. Canadian Journal of Fisheries and Aquatic Sciences. 1996;53(1):141–147. [Google Scholar]
- Crête-Lafreniere A, Weir LK, Bernatchez L. Framing the salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS ONE. 2012;7:e46662. doi: 10.1371/journal.pone.0046662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson WS, Huang TK, Fujiki K, Koop KR, von, Schalburg BF. The sex-determining loci and sex chromosomes in the family salmonidae. Sexual Development. 2009;3(2–3):78–87. doi: 10.1159/000223073. [DOI] [PubMed] [Google Scholar]
- Donaldson EM, Hunter GA. Sex Control in Fish with Particular Reference to Salmonids. Canadian Journal of Fisheries and Aquatic Sciences. 1982;39(1):99–110. [Google Scholar]
- Emonet SF, Grard G, Brisbarre NM, Moureau GN, Temmam S, Charrel RN, de Lamballerie X. Long PCR Product Sequencing (LoPPS): a shotgun-based approach to sequence long PCR products. Nature Protocols. 2007;2(2):340–346. doi: 10.1038/nprot.2006.453. [DOI] [PubMed] [Google Scholar]
- Frolov SV. Differentiation of sex chromosomes in the Salmonidae. III. Multiple sex chromosomes in Coregonus sardinella. Tsitologiya. 1990;32(6):659–663. [Google Scholar]
- Gharbi K, Gautier A, Danzmann RG, Gharbi S, Sakamoto T, Hoyheim B, Taggart JB, et al. A linkage map for brown trout (Salmo trutta): Chromosome homeologies and comparative genome organization with other salmonid fish. Genetics. 2006;172(4):2405–2419. doi: 10.1534/genetics.105.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govoroun M, Mcmeel OM, D'Cotta H, Ricordel MJ, Smith T, Fostier A, Guiguen Y. Steroid enzyme gene expressions during natural and androgen-induced gonadal differentiation in the rainbow trout, Oncorhynchus mykiss. Journal of Experimental Zoology. 2001;290(6):558–566. doi: 10.1002/jez.1106. [DOI] [PubMed] [Google Scholar]
- Guyomard R, Mauger S, Tabet-Canale K, Martineau S, Genet C, Krieg F, Quillet E. A Type I and Type II microsatellite linkage map of Rainbow trout (Oncorhynchus mykiss) with presumptive coverage of all chromosome arms. BMC Genomics. 2006;7:302. doi: 10.1186/1471-2164-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, Sakamoto T, Fernandino JI, et al. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex-determination. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(8):2955–2959. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Schartl M. Sex-determination: switch and suppress. Current Biology. 2011;21(17):R656–R659. doi: 10.1016/j.cub.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hunter G, Donaldson E. Hormonal sex control and its application to fish culture. In: Hoar WS, Randall DJ, Donaldson EM, editors. Fish Physiology. IX. Academic Press; 1983. pp. 223–303. Reproduction. Part B. Behavior and fertility control. [Google Scholar]
- Jankun M, Rab P, Vuorinen J. A karyotype study of vendace, Coregonus albula (Pisces, Coregoninae) Hereditas. 1991;115(3):291–294. doi: 10.1111/j.1601-5223.1992.tb00572.x. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, Fujita M, et al. A Trans-Species Missense SNP in Amhr2 Is Associated with Sex-determination in the Tiger Pufferfish, Takifugu rubripes (Fugu) PLoS Genetics. 2012;8(7):e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Phillips RB, Harwood AS, Koop BF, Davidson WS. Identification of the sex chromosomes of brown trout (Salmo trutta) and their comparison with the corresponding chromosomes in Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss. Cytogenetic and Genome Research. 2011;133(1):25–33. doi: 10.1159/000323410. [DOI] [PubMed] [Google Scholar]
- Lopez JA, Chen WJ, Orti G. Esociform phylogeny. Copeia. 2004;3:449–464. [Google Scholar]
- Mank JE, Avise JC. Evolutionary diversity and turn-over of sex-determination in teleost fishes. Sexual Development. 2009;3(2–3):60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- Marshall Graves JA, Peichel CL. Are homologies in vertebrate sex-determination due to shared ancestry or to limited options? Genome Biology. 2010;11(4):205. doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417(6888):559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Sato T, Toyazaki Y, Nagahama Y, Hamaguchi S, Sakaizumi M. Oryzias curvinotus has DMY, a gene that is required for male development in the medaka, O. latipes. Zoological Sciences. 2003;20(2):159–161. doi: 10.2108/zsj.20.159. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, Paul-Prasanth B, Lau EL, et al. DMY gene induces male development in genetically female (XX) medaka fish. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(10):3865–3870. doi: 10.1073/pnas.0611707104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawaribuchi S, Yoshimoto S, Ohashi S, Takamatsu N, Ito M. Molecular evolution of vertebrate sex-determining genes. Chromosome Research. 2012;20(1):139–151. doi: 10.1007/s10577-011-9265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf VJ, Gemmell NJ. Sexual genotype markers absent from small numbers of male New Zealand Oncorhynchus tshawytscha. Journal of Fish Biology. 2006;68:136–143. [Google Scholar]
- Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, Naruse K, et al. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics. 2012;191(1):163–170. doi: 10.1534/genetics.111.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagler JJ, Bouma J, Thorgaard GH, Dauble DD. High incidence of a male-specific genetic marker in phenotypic female Chinook salmon from the Columbia River. Environmental Health Perspectives. 2001;109(1):67–69. doi: 10.1289/ehp.0110967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(34):13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin N. Sex reversal: a fountain of youth for sex chromosomes? Evolution. 2009;63(12):3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- Quillet E, Aubard G, Queau I. Mutation in a sex-determining gene in rainbow trout: detection and genetic analysis. Journal of Heredity. 2002;93(2):91–99. doi: 10.1093/jhered/93.2.91. [DOI] [PubMed] [Google Scholar]
- Schartl M. Sex chromosome evolution in non-mammalian vertebrates. Current Opinion in Genetics and Development. 2004;14(6):634–641. doi: 10.1016/j.gde.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Shetty S, Kirby P, Zarkower D, Graves JA. DMRT1 in a ratite bird: evidence for a role in sex-determination and discovery of a putative regulatory element. Cytogenetic and Genome Research. 2002;99(1–4):245–251. doi: 10.1159/000071600. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346(6281):240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex-determination in the chicken. Nature. 2009;461(7261):267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Stiglec R, Ezaz T, Graves JA. A new look at the evolution of avian sex chromosomes. Cytogenetic and Genome Research. 2007;117(1–4):103–109. doi: 10.1159/000103170. [DOI] [PubMed] [Google Scholar]
- Takehana Y, Naruse K, Hamaguchi S, Sakaizumi M. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O-dancena. Chromosoma. 2007;116(5):463–470. doi: 10.1007/s00412-007-0110-z. [DOI] [PubMed] [Google Scholar]
- Volff JN, Kondo M, Schartl M. Medaka dmY/dmrt1Y is not the universal primary sex-determining gene in fish. Trends in Genetics. 2003;19(4):196–199. doi: 10.1016/S0168-9525(03)00051-9. [DOI] [PubMed] [Google Scholar]
- Volff JN, Nanda I, Schmid M, Schartl M. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sexual Development. 2007;1:85–99. doi: 10.1159/000100030. [DOI] [PubMed] [Google Scholar]
- Williamson KS, May B. Inheritance studies implicate a genetic mechanism for apparent sex reversal in Chinook salmon. Transactions of the American Fisheries Society. 2005;134(5):1253–1261. [Google Scholar]
- Williamson KS, Phillips R, May B. Characterization of a chromosomal rearrangement responsible for producing “apparent” XY-female fall-run Chinook salmon in California. Journal of Heredity. 2008;99(5):483–490. [Google Scholar]
- Woram RA, Gharbi K, Sakamoto T, Hoyheim B, Holm LE, Naish K, McGowan C, et al. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Research. 2003;13(2):272–280. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A, Guyomard R, Nicol B, Jouanno E, Quillet E, Klopp C, Cabau C, et al. An Immune-Related Gene Evolved into the Master Sex-Determining Gene in Rainbow Trout, Oncorhynchus mykiss. Current Biology. 2012;22:1423–1428. doi: 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Ito M. A ZZ/ZW-type sex-determination in Xenopus laevis. FEBS Journal. 2011;278(7):1020–1026. doi: 10.1111/j.1742-4658.2011.08031.x. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]


