Abstract
A series of neomycin dimers have been synthesized using “Click chemistry” with varying functionality and length in the linker region to target the HIV-1 TAR RNA region of HIV virus. TAR (Transactivation Response) RNA region, a 59 base pair stem loop structure located at 5′-end of all nascent viral transcripts interacts with its target, a key regulatory protein, Tat, and necessitates the replication of HIV-1 virus. Neomycin, an aminosugar, has been shown to exhibit multiple binding sites on TAR RNA. This observation prompted us to design and synthesize a library of triazole linked neomycin dimers using click chemistry. The binding between neomycin dimers and TAR RNA was characterized using spectroscopic techniques including FID (Fluorescent Intercalator displacement), FRET (fluorescence resonance energy transfer) competitive assay, circular dichroism (CD) and UV-thermal denaturation. UV thermal denaturation studies demonstrate that neomycin dimers binding increase the melting temperature (Tm) of the HIV TAR RNA up to 10 °C. (Ethidium bromide) displacement (FID) and FRET competition assay revealed nanomolar binding affinity between neomycin dimers and HIV TAR RNA while in case of neomycin, only a weak binding was detected. More importantly, most of the dimers showed lower IC50s towards HIV TAR RNA, when compared to the fluorescent Tat peptide and show increased selectivity over mutant TAR RNA. Cytopathic effects investigated using MT-2 cells indicate a number of the dimers with high affinity towards TAR show promising anti HIV activity.
Ribonucleic acid-protein interactions are essential for regulation of many important biological processes such as translation, RNA splicing, and transcription.1-3 An important example of such an interaction is involved in the regulation of human immunodeficiency virus type 1 (HIV-1). TAR RNA (trans activation responsive region), a 59 base stem-loop structure located at the 5′-end of the nascent viral transcripts, interacts with Tat protein, (an 86 amino acid protein) and regulates the transcription level of HIV.4, 5 The cooperative interaction of Tat protein along with its cellular cofactor, transactivating elongation factor-b (TEFb) with TAR RNA recruits and activates the CDK9 kinase which phosphorylates the RNA polymerase II (RNAP II) and significantly enhances the processivity of RNAP II.3, 6, 7
HIV transcription in virus infected cells is strongly triggered by the interaction between Tat protein and its cognate TAR RNA. TAR RNA structure is comprised of two stems (upper and lower), a three nucleotide bulge region, and a hairpin. An arginine rich domain of TAT protein interacts with the tri-nucleotide bulge (U23, C24, and U25) of TAR RNA.1, 8, 9 and causes a substantial enhancement in the transcript level (~100 fold).2 NMR studies show that the complexation takes place specifically between arginine residue of TAT protein and a guanine base in the major groove of TAR RNA.10 Disruption of TAR RNA-Tat interaction therefore represents an attractive strategy to inhibit viral replication. A number of molecules have been investigated with this strategy in mind.11, 12 These include, intercalators,12 (ethidium bromide13 and proflavine), DNA minor groove binders14 (Hoechst 33258, and DAPI), phenothiazine,15 argininamide,16 peptides,17 peptidomimetics,18 aminoglycosides,19 and cyclic polypeptides.20 Aminoglycosides are naturally-occurring aminosugars which bind to a wide variety of RNA structures.21 In the past few years, a number of aminoglycoside conjugates have been synthesized to achieve higher binding affinity and specificity towards RNA 21-26 and DNA based targets27-44 such as duplex,45 triplex46-48 and quadruplex structures.29, 30 In an attempt to achieve higher binding affinity and explore multiple binding sites on RNA targets, the homo and hetero dimeric units of aminoglycosides49, 50 (tobramycin, neamine, neomycin B, and kanamycin A) have been synthesized with various linker length and functionalities through disulfide bond formation. These aminoglycosides exhibit higher binding affinity towards dimerized A-site 16S construct, RRE RNA than their corresponding monomeric aminoglycoside units. Also, aminoglycoside dimers exhibit a 20- to 12000- fold higher inhibitory effects towards the catalytic function of Tetrahymena ribozyme than the monomeric units.50 Neamine dimers have been shown to exhibit remarkable antibiotic effects and resistance to aminoglycoside-modifying enzymes.51
Among all the aminoglycosides targeted towards TAR binding, neomycin has shown the highest inhibitory effect (less than 1 μM).19 ESI MS52 and ribonuclease protection experiments22 have suggested that the binding site of neomycin is the stem region just below the tri-nucleotide bulge in TAR RNA. Further ESI MS experiments and gel shift assays have revealed the existence of three neomycin binding sites on HIV TAR RNA.52 These sites do not overlap with the Tat binding site and thus neomycin shows a weak ability to allosterically compete with protein binding leading to weak HIV inhibition. In order to achieve improved binding and specificity profiles, we have explored neomycin’s multiple binding sites on HIV TAR RNA and designed a series of neomycin dimers using click chemistry. Even though these dimers are not expected to directly compete with Tat binding, their binding is expected to lock the conformation of RNA such that Tat-TAR binding is weakened through an allosteric mechanism. We synthesized neomycin dimers using click chemistry with various linker lengths and functionalities to optimize the RNA binding affinity. Our results show that neomycin dimers display nanomolar affinity towards HIV TAR RNA. Spectroscopic techniques, UV thermal denaturation, FID assay, and FRET (Fluorescence Resonance Energy Transfer) assay were utilized to study the binding between neomycin dimers and TAR RNA. In this report, we present our work detailing a simple and efficient route towards the synthesis of triazole linked aminosugar dimers, their binding towards TAR RNA, their ability to effectively inhibit the tat-TAR interaction and their cytopathic effects on HIV in MT-2 cells.
Experimental Procedures
Materials
All of the chemicals were purchased from commercial suppliers and used without further purification. Neomycin B tri-sulfate was purchased from MP Biomedicals (Solon, Ohio, USA). Di-tert-butyl dicarbonate (Boc anhydride) was purchased from Advanced ChemTech (Louisville, KY, USA). SC (Sodium Cacodylate), EDTA (Ethylenediamine Tetraacetic Acid), KCl, sodium phosphate (mono- and di-) salts were purchased from Fisher Scientific. TPS-Cl (2,4,6-triisopropylbenzenesulfonyl chloride) and 4M HCl/dioxane were purchased from Sigma Aldrich. Silica gel for flash column chromatography was purchased from Sorbent Technologies (Atlanta, GA, USA) as silica gel standard grade (particle size = 40-63 μm). All solvents were purchased from VWR. Reaction solvents were distilled over calcium hydride [toluene, pyridine, DCM (dichloromethane)]. EtOH (Ethanol) was first distilled with sodium metal and then redistilled over magnesium turnings. Reactions were carried out under N2 using dry solvent, unless otherwise noted.
Instrumentation
1H NMR spectra were collected on a Bruker 500 MHz FT-NMR spectrometer. MS (MALDI-TOF) spectra were collected using a Bruker Omniflex MALDI-TOF mass spectrometer. All UV spectra were recorded on a Cary 100 Bio UV/Vis spectrophotometer equipped with a thermoelectrically controlled 12-cell holder. Quartz cells with a 1 cm pathlength were used for all the absorbance studies. Spectrophotometer stability and λ alignment were checked prior to initiation of each melting point experiment. Fluorescence spectra were measured on a Photon Technology International instrument (Lawrenceville, New Jersey, USA). The fluorescence measurements in 96-well plates were carried out on a Genios Multi-Detection Microplate Reader, TECAN with Magellan software.
UV thermal denaturation experiment
The UV thermal denaturation samples (1 mL) of TAR-RNA (1 μM/strand) were mixed with ligand (neomycin dimers) with an rdr value of 1 and 2 (rdr = ratio of drug to HIV TAR RNA concentration) in 100 mM KCl, , 10 mM SC, 0.5 mM EDTA, pH 6.8 and incubated for 4 h at 4 °C before starting the experiment. The UV thermal denaturation spectra of the samples in 1 cm path length quartz cuvettes were recorded at 260 nm as a function of temperature (10-95 °C, heating rate: 0.2 °C/min). First derivative plots were used to determine the denaturation temperature.
TAR RNA synthesis
HIV TAR RNA was purchased from Amersham Biosciences (now GE Healthcare Life Sciences, Piscataway, NJ). Before used, HIV TAR RNA (5′-GGC AGA UCU GAG CCU GGG AGC UCU CUG CC -3′) in 10 μM batch in sterile water was heated to 95 °C for 4.5 min, then cooled rapidly in ice bath for 5 min. This snap-cooling causes the RNA to adopt the kinetically favored hairpin rather than the thermodynamically favored duplexes.
FRET mediated competition binding assay
The relative affinity of each dimer for HIV-1 TAR RNA was determined using Fluorescence Resonance Energy Transfer (FRET)-based competitive binding assay with a fluorescein-labeled HIV-1 Tat peptide as described in the literature53. The fluorescence experiments were performed with a Spectra Max Fluorimeter (Molecular Devices) at 25 °C, with excitation and emission wavelengths of 495 and 570 nm, respectively. All samples were prepared in 96 well plates in 1× TK buffer (50 mM Tris, 20 mM KCl, pH = 7.4) with 0.1% Triton-X100 (Sigma). The binding affinity (IC50) values reported for each dimer are the averages of 3~5 individual measurements, and were determined by fitting the experimental data to a sigmoidal dose-response nonlinear regression model on GraphPad Prism 4.0. Prior to the competition experiments, the affinity of fluorescein-labeled Tat peptide for HIV-1 TAR RNA was determined by monitoring fluorescence intensity changes of the fluorescent probe upon addition of HIV-1 TAR RNA. Addition of an increasing concentration (0 nM to 1000 nM) of HIV-1 TAR RNA to a 100 nM solution of fluorescein-labeled Tat peptide in TK buffer at 25 °C afforded a saturation binding curve. The IC50 value obtained from this binding curve was 86 nM.
Competition FRET Assay
To a solution of 100 nM HIV-1 TAR RNA and 100 nM fluorescein-labeled HIV-1 Tat peptide, appropriate concentrations (0 nM to 100 μM) of the dimer antagonists were added at 25 °C; total volume of the incubation solution was 80 μL. After 60 min, fluorescence changes of the sample solution were determined by the Spectra Max Fluorimeter Detector. The experimental dose-response data for a given polyamine were fit to a sigmoidal dose-response nonlinear regression model on GraphPad Prism 4.0 to afford the IC50 values for each dimer.
Ethidium bromide displacement titration
A solution of ethidium bromide (1.25 μM, 1800 μL) was excited at 545 nm, and its fluorescence emission was monitored from 560 nm to 607 nm before and after the addition of HIV TAR RNA. The concentration of HIV TAR RNA was 50 nM/strand. A small fraction of ethidium bromide is bound (less than 20%) under these conditions. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8.
Ethidium bromide displacement titration to determine the binding constant via Scatchard analysis
A solution of ethidium bromide (5.00 μM, 1800 μL) was excited at 545 nm, and its fluorescence emission was monitored from 560 nm to 607 nm before and after the addition of HIV TAR RNA. The concentration of HIV TAR RNA was 200 nM/strand. A small fraction of ethidium bromide is bound (less than 20%) under these conditions. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8.
Assay for inhibition activity towards HIV antigen synthesis in treated cells
500,000 cells were pre-treated with each compound for 1 h. Next, cells were inoculated with approximately 50,000 infectious particles of NL4-3 (an infectious molecular clone of HIV). Infections were performed in triplicate. Every 2 days cells were pooled and stained for HIV antigen synthesis using antibody to all HIV antigens (HIV immune globulin).
In vitro cell culture to determine the amount of reverse transcriptase (RT) activity release
Culture supernatants were precipitated for RT release. In all assays MT-2 cells were used, which are highly susceptible to HIV and are completely lysed by HIV. Anti-HIV activities can be routinely checked by measuring the percentage of cells positive for HIV antigens using immunofluorescence assay (IFA) and for the release of pelletable reverse transcriptase (RT) into the supernatant. For these assays, 500,000 MT-2 cells in 1 mL of media were added to the wells of a 24 well tissue culture plate. Next, 0.5 mL of 4x concentrated compound was added to triplicate wells of the plate. The cells and compounds were incubated for 1 h at 37 °C. Finally, 0.5 mL of HIVNL4-3, produced in H9 cells (a CD4+ lymphoblastoid cell line), 100,000 cpm of RT activity per well, was added to each well. Virus control wells contained no compounds. This inoculum is at a multiplicity of infection less than 1. On days 2, 4, and 6, 0.75 mL of supernatants were removed and placed into individual microfuge tubes for RT assay. The supernatants from each well were precipitated at 4 °C overnight in a solution including 30% polyethylene glycol. After precipitation, precipitate was lysed in a solution containing Triton X100, Tris buffer, and DTT. RT activity of each aliquot was determined as incorporation of 3H-dTTP into a poly rA-oligo dT template. After a one hour assay at 37 °C, the incorporated dTTP was precipitated onto a ZetaProbe (BioRad) membrane. Slots were excised and placed into liquid scintillation cocktail. After overnight incubation, counts per minute for each sample were determined on a beta counter. Results were calculated as cpm/ml of original culture supernatant. Next, cells were resuspended in the media and 0.5 mL of the total remaining volume was removed for IFA. Cells were combined from their triplicate infections, pelleted, and the enriched cells were air-dried onto glass slides. The dried cells were fixed in acetone:methanol (50:50). After fixing, cells were stained with HIV immunoglobulin, washed in PBS, and counter-stained with FITC-conjugated goat and human IgG. Slides were washed in PBS and the percentage of HIV-positive cells was determined by epifluorescence. To the remaining cells in culture, 1.25 mL of media were added and the cultures maintained at 37 °C.
Synthesis and characterization of dimers DPA51-DPA65
General procedure for synthesis of N-Boc DPA51-DPA65
To a solution of neomycin-boc-5”-azide (0.05 mmol) in dry toluene (5 mL), dialkyne linker (0.025 mmol, 0.50 eq) was added followed by the addition of CuI (4.76 mg, 0.025 mmol) and DIPEA (6.46 mg, 0.05 mmol). The reaction mixture was stirred at 90 °C for 18 h in an atmosphere of argon. The progress of the reaction was monitored by TLC. The volatiles were rotoevaporated in vacuo. Purification by flash column chromatography (Rf =0.38-0.44, 0 to 10 % ethanol in CH2Cl2) afforded the desired product(s) as a white solid (% yields are reported for individual compounds in Table 1). [Rf 0.34-0.42, 10 % ethanol in CH2Cl2 (v/v)].
Table 1.
Linker structures and % yield (for two steps) for the synthesis of dimeric neomycins.
| Neomycin dimer | Linker length | Structure of the linker (-x-, from Scheme 1 and Scheme 2) |
% yield |
|---|---|---|---|
| DPA51 | 7 |
|
89.5 |
| DPA52 | 7 |
|
88.2 |
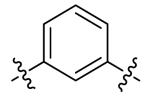
| DPA65 | 7 |
|
83.0 |
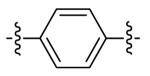
| DPA53 | 8 |
|
87.0 |
| DPA54 | 8 |
|
88.5 |
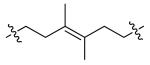
| DPA55 | 14 |
|
82.0 |
| DPA56 | 14 |
|
88.8 |
| DPA58 | 20 |
|
83.5 |
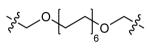
| DPA60 | 24 |
|
82.8 |
General procedure for deprotection of N-Boc DPA51-DPA65
To a solution of neomycin dimer (0.012 mmol) in dioxane (3 mL), 4 M HCl/dioxane (1 mL) was added and the reaction stirred at room temperature. There was a formation of white precipitate after 15 min. The reaction mixture was centrifuged and the solid was collected. The solid was washed with a solution of diethyl ether/hexane [3 × 5 mL, 1:1 (v/v)]. The solid was dissolved in water and lyophilized to afford the desired product(s) as a powder (% yields are reported for individual compounds in Table 1).
DPA51
IR (neat, cm−1) 3421 (br, OH), 1686, 1524, 1366; 1H NMR (500 MHz, D2O) δ 8.02 (s, 2 H, triazole), 5.92 (d, J = 3.94 Hz, 2 H, H1II), 5.28 (d, J = 3.16 Hz, 2 H, H1III), 5.17 (s, 2 H, H1IV), 4.41 (m, 2 H, H4III), 4.34 (d, J = 5.20 Hz, 2 H, H2III), 4.18 (t, J = 4.73 Hz, 2 H, H4IV), 4.10 (d, J = 2.84 Hz, 2 H, H4I), 3.96 (t, J = 9.62 Hz, 2 H, H6II), 3.90 (t, J = 9.93 Hz, 2 H, H5I), 3.86-3.76 (m, 8 H, H2IV, H4II, H5IV, and H3III), 3.70 (d, J = 1.89 Hz, 2 H, H6I), 3.54 (t, J = 9.61 Hz, 2 H, H3II), 3.49-3.45 (m, 4 H, H3I, H5II), 3.44-3.32 (m, 2 H, H1I), 3.30-3.20 (m, 8 H, H5III and propargyl ether protons), 2.39-2.32 (dt, J1 = 3.94 Hz, J2 = 4.25 Hz, 2 H, H2Ieq.), 1.72-1.82 (q, J = 12.45 Hz, 2 H, H2Iax.); 13C NMR (500 MHz, D2O) δ 162-162 (q, CF3COOH), 127, 125, 115, 110, 95, 85, 79, 77, 75, 73, 72, 70, 69, 68, 67, 63, 62, 53, 52, 50, 49, 48, 40.5, 40.1, 28; MS (MALDI-TOF) m/z calcd for C52H96N18O25 1391.44, found 1392.66 [M+H2O]+.
DPA52
IR (neat, cm−1) 3375 (br, OH), 2979, 2932, 2108, 1727, 1686, 1522, 1457; 1H NMR (500 MHz, D2O) δ 8.06 (s, 2 H, triazole), 5.95 (d, J = 3.94 Hz, 2 H, H1II), 5.32 (d, J = 3.15 Hz, 2 H, H1III), 5.17 (d, J = 1.26 Hz, 2 H, H1IV), 4.47 (d, J = 1.26 Hz, 2 H), 4.48-4.42 (m, 2 H, H4III), 4.40-4.36 (t, J = 5.20 Hz, 2 H, H2III), 4.24-4.20 (t, J = 4.41 Hz, 2 H, H4IV), 4.14-4.11 (t, J = 2.99 Hz, 2 H, H4I), 4.03-3.98 (t, J = 9.62 Hz, 2 H, H6II), 3.96-3.92 (m, 2 H, H5I), 3.92-3.80 (m, 8 H, H2IV, H4II, H5IV, and H3III), 3.74-3.72 (m, 2 H, H6I), 3.61-3.55 (t, J = 9.14 Hz, 2 H, H3II), 3.54-3.48 (m, 4 H, H3I, H5II), 3.48-3.40 (m,4 H), 3.38-3.20 (m, 8 H), 2.42-2.35 (dt, J1 = 3.47 Hz, J2 = 4.25 Hz, 2 H, H2Ieq.), 1.85-1.75 (q, J = 12.61 Hz, 2 H, H2Iax.); MS(MALDI-TOF) m/z calcd for C53H98N18O24 1389.45, found 1389.66 [M+H2O]+; Anal. Calcd for C53H110N18O24Cl12: C, 35.19; H, 6.13; Cl, 23.52; N, 13.94; O, 21.23. Found: C, 34.89; H, 6.21; N, 13.71.
DPA53
IR (neat, cm−1) 3368 (br, OH), 2090, 1642; 1H NMR (500 MHz, D2O) δ 8.51 (s, 2 H, triazole), 7.91 (s, 4 H, Ar), 6.02 (d, J = 3.60 Hz, 2 H, H1II), 5.39 (d, J = 3.00 Hz, 2 H, H1III), 5.24 (s, 2 H, H1IV), 4.27 (t, J = 4.70 Hz, 2 H),4.20- 4.05 (m, 6 H, H5I, H6II), 4.00-3.80-4.00 (m, 6 H), 3.73 (m, 6 H), 3.65 (m, 12 H), 3.55 (m, 8 H), 3.45-3.20 (m, 10 H), 3.18-3.10 (m, 2 H), 2.43-2.35 (dt, J1 = 4.15 Hz, J2 = 4.28 Hz, 2 H, H2Ieq.), 1.82-1.72 (q, J = 12.45 Hz, 2 H, H2Iax.); MS (MALDI-TOF) m/z calcd for C56H96N18O24 1423.47, found 1424.41[M+H2O]+; Anal. Calcd for C56H108N18O24Cl12 C, 36.49; H, 5.91; Cl, 23.08; N, 13.68; O, 20.83. Found: C, 36.04; H, 5.79; N, 13.31; UV (water) λmax = 275 nm.
DPA54
1H NMR (500 MHz, D2O) δ 7.86 (s, 2 H, triazole), 5.79 (s, 2 H, H1II), 5.30 (s, 2 H, H1III), 5.20 (s, 2 H, H1IV), 4.47 (d, J = 1.26 Hz, 2 H), 4.48-4.42 (m, 2 H, H4III), 4.38 (t, J = 5.20 Hz, 2 H, H2III), 4.46 (s, 8 H), 4.25 (s, 4 H), 4.22 (s, 4 H), 4.12 (m, 4 H), 3.97-3.90 (m, 6 H), 3.75-3.65 (m, 10 H), 3.55-3.45 (m, 6 H), 3.40-3.20 (m, 12 H), 3.40-3.23 (m,12 H), 3.20-3.05 (m, 8 H), 2.38 (d, J = 12.14 Hz, 2 H, H2Ieq.), 1.70-1.50 (m, 2 H, H2Iax.); MS (MALDI-TOF) m/z calcd. for C54H100N18O24 [M+H2O]+, calcd 1405.48, found 1404.64; UV (water) λmax = 225 nm.
DPA55
1H NMR (500 MHz, D2O) δ 7.82 (s, 2 H, triazole), 5.95 (s, 2 H, H1II), 5.33 (s, 2 H, H1III), 5.22 (s, 2 H, H1IV), 4.55-4.43 (br, 4 H, H4III), 4.24 (m, 2 H), 4.13 (s, 2H), 3.99 (m, 2H), 3.95-3.80 (m, 8H), 3.78-3.70 (4 H), 3.66-3.58 (m, 4 H), 3.56 (m, 2 H), 3.54 (s, 2H), 3.51 (s, 2 H), 3.45-3.35 (m, 6H), 3.34-3.30 (s, 2 H), 3.30-3.19 (m, 6 H), 2.40-2.31 (m, 6 H), 2.26 (m, 2 H, H2Ieq), 1.81-1.72 (m, 2 H, H2Iax.); MS (MALDI-TOF) m/z calcd. for C58H106N18O24 [M+H2O]+, found 1457.90; UV (water) λmax = 227 nm.
DPA56
[Rf 0.42 in 10% EtOH in DCM (v/v)]; 1H NMR (500 MHz, D2O): δ 7.80 (s, 2 H, triazole), 6.00 (d, J = 3.63 Hz, 2 H, H1II), 5.36 (d, J = 2.84 Hz, 2 H, H1III), 5.21 (s, 2 H, H1IV), 4.53-4.47 (m, 4 H, H4III), 4.45 (t, J = 5.05 Hz, 2 H), 4.23 (t, J = 5.36 Hz, 4 H), 4.17-4.11 (m, 6 H), 3.97 (t, J = 9.77 Hz, 4 H, H5I, H6II), 3.93-3.83 (m, 8 H, H2IV, H4II, H5IV, and H3III), 3.75-3.70 (m, 6 H), 3.69-3.59 (m, 10 H), 3.55-3.59 (m, 2 H), 3.54 (s, 4 H), 3.50-3.53 (m, 4 H), 3.43-3.34 (m, 8 H), 3.34-3.24 (m, 8 H), 3.22-3.15 (m, 8 H), 2.64-2.58 (m, 4 H), 2.40-2.33 (m, 2 H, H2Ieq), 1.96 (m, 6 H, H2Iax. and protons from linker), 1.55 (m, 4 H, protons from linker); MS (MALDI-TOF) m/z calcd.for C56H104N18O24 [M+Na]+ 1435.53, found 1434.83; Anal. Calcd for C56H116N18O24Cl12: C, 36.34; H, 6.32; Cl, 22.98; N, 13.62; O, 20.74, found C, 36.24; H, 6.11; N, 13.41; UV (water) λmax = 227 nm.
DPA58
1H NMR (500 MHz, D2O) δ 8.06 (s, 2 H, triazole), 5.97 (d, J = 3.47 Hz, 2 H, H1II), 5.33 (d, J = 3.00 Hz, 2 H, H1III), 5.22 (s, 2 H, H1IV), 4.80-4.74 (m, 2 H), 4.51-4.43 (m, 4 H, H4III), 4.24 (t, J = 4.42 Hz, 2 H), 4.13 (t, J = 2.37 Hz, 2 H), 3.97 (t, J = 9.61 Hz, 2 H, H6II), 3.93 (t, J = 10.40 Hz, 2 H), 3.90-3.78 (m, 6 H), 3.76-3.70 (m, 4 H), 3.69-3.58 (m, 8 H), 3.57-3.53 (m, 6 H), 3.52-3.47 (m, 6 H), 3.44 (t, J = 9.14 Hz, 4 H), 3.41-3.34 (m, 4 H), 3.32 (t, J = 6.14 Hz, 2 H), 3.30-3.16 (m, 6 H), 2.41-2.33 (dd, J1 = 3.94 Hz, J2 = 3.62 Hz, 2 H), 1.84-1.72 (q, J = 13.24 Hz, 2 H, H2Iax.), 1.53-1.43 (m, 4 H, linker protons), 1.25-1.14 (m, 8 H, linker protons), 1.13-1.04 (t, J = 6.62 Hz, 4 H, linker protons); MS (MALDI-TOF) m/z calcd.for C60H112N18O26 [M+2H2O]+ 1537.64, found 1538.11; UV (water) λmax = 229 nm.
DPA60
1H NMR (500 MHz, D2O): δ 8.05 (s, 2 H, triazole), 5.97 (d, J = 3.78 Hz, 2 H, H1II), 5.34 (d, J = 3.31 Hz, 2 H, H1III), 5.22 (s, 2 H, H1IV), 4.55 (s, 4 H), 4.50-4.46 (m, 2 H, H4III), 4.43 (t, J = 4.57 Hz, 2 H), 4.23 (t, J = 4.42 Hz, 2 H), 4.19-4.15 (m, 2 H), 3.94 (t, J = 2.53 Hz, 2 H), (N 4.05 (t, J = 9.93 Hz, 2 H), 3.96-3.90 (t, J = 10.25 Hz, 2 H), 3.90-3.79 (m, 8 H), 3.76-3.69 (m, 6 H), 3.70-3.58 (m, 12 H), 3.55-3.42 (m, 12 H), 3.42-3.38 (m, 2 H), 3.38-3.35 (m, 2 H), 3.34-3.30 (m, 4 H), 3.30-3.22 (m, 6 H), 2.38 (dd, J1 = 4.10 Hz, J2 = 4.73 Hz, 2 H), 1.85-1.75 (q, J = 11.98 Hz, 2 H, H2Iax.), 1.54-1.41 (m, 4 H, linker protons), 1.24-1.10 (m, 18 H, linker protons); MS (MALDI-TOF) m/z calcd.for C64H120N18O26 [M+H2O]+, found 1576.11; UV (water) λmax = 228 nm.
DPA65
IR (KBr, cm−1) 3434 (br, OH), 2086 (weak), 1637; 1H NMR (500 MHz, D2O) δ 8.52 (s, 2 H, triazole), 8.24 (s, 1 H, Ar), 7.80 (d, J = 7.72 Hz, 2 H, Ar), 7.58 (t, J = 7.88 Hz, 1 H, Ar), 5.94 (d, J = 3.62 Hz, 2 H, H1II), 5.38 (d, J = 3.00 Hz, 2 H, H1III), 5.25 (d, J = 1.10 Hz, 2 H, H1IV), 4.90-4.83 (m, 2 H, H4III), 4.81-4.76 (m, 2 H), 4.64-4.56 (m, 6 H), 4.24 (t, J = 4.73 Hz, 2 H), 4.16 (t, J = 4.10 Hz, 2 H), 4.13 (t, J = 2.99 Hz, 2 H), 4.05 (t, J = 9.77 Hz, 4 H, H5I, H6II), 3.95-3.85 (m, 6 H), 3.85-3.79 (m, 2 H), 3.74-3.71 (m, 2 H), 3.67-3.59 (m, 4 H), 3.58-3.49 (m, 8 H), 3.49-3.38 (m, 4 H), 3.47-3.34 (m, 2 H), 3.47-3.31 (m, 4 H), 3.31-3.29 (m, 4 H), 3.29-3.22 (m, 6 H), 3.07-3.01 (m, 2 H), 2.37 (dd, J1 = 4.10 Hz, J2 = 4.25 Hz, 2 H, H2Ieq.), 1.82-1.72 (q, 2 H, H2Iax); (M+Na+)for C56H96N18O24, calcd 1423.47, found 1424.41[M+H2O]; Anal. Calcd for C56H108N18O24Cl12: C, 36.49; H, 5.91; Cl, 23.08; N, 13.68; O, 20.83. Found: C, 36.22; H, 5.81; N, 13.48; UV (water) λmax = 236 nm.
Results and Discussion
Design and synthesis of triazole linked neomycin dimers
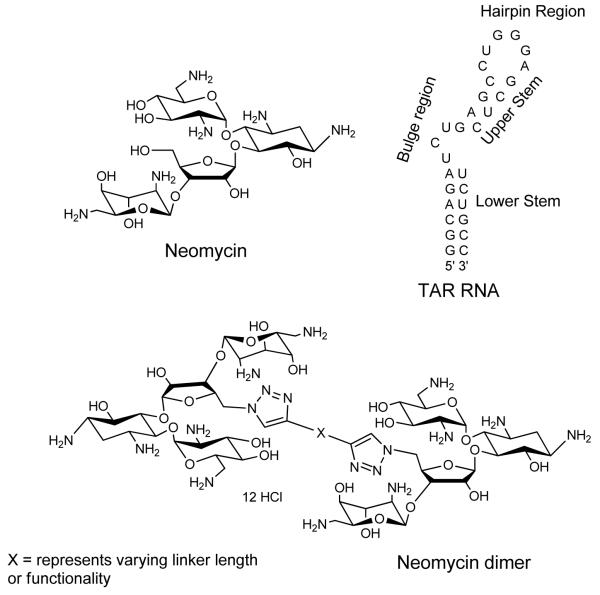
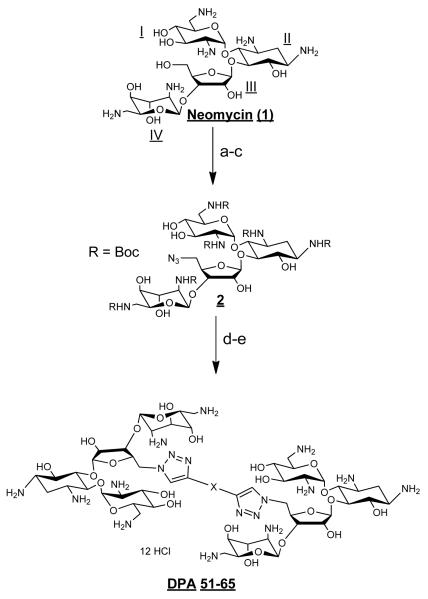
Previous MS- ESI data has shown that neomycin has at least two binding sites on HIV TAR RNA.52 One binding site has been reported to be below the tri-nucleotide bulge region (see Scheme 1 for structures), and the second binding site lies at the upper stem or hairpin region. To take advantage of these multiple sites, and to improve ligand affinity and selectivity, a series of neomycin dimers was synthesized. The neomycin dimers were synthesized using a high yielding, inert, and robust synthetic “click chemistry” approach as shown in Scheme 2).54-56 The length and functionality of the linker, that tethers the two neomycin units, has been varied to investigate the optimal binding interaction between neomycin dimers and HIV TAR RNA. Azide 2 was synthesized in three steps from commercially available neomycin 157 and then reacted with terminal dialkynes in a 2:1 ratio to yield the traizole linked dimers (Scheme 2). Deprotection of tert-Butoxycarbonyl (Boc) groups using 4 N HCl yielded the dimer hydrochloride salts58-61 DPA52-DPA65 in excellent yields.
Scheme 1.
Structures of small molecule ligands (neomycin and neomycin dimer) and TAR RNA used in the study.
Scheme 2.
Reagents and conditions: (a) (Boc)2O, DMF, H2O, Et3N, 60°C, 5 h, 60%. (b) TPS-Cl, pyridine, r.t., 40 h, 50%. (c) NaN3, DMF/H2O (10/1), 12 h, 90 °C, 90%. (d) Toluene, CuI, DIPEA, r.t., 90 °C. (e) 4 M HCl/dioxane, r.t., 5 min. Yield for steps d-e=82-90%.
Neomycin dimers significantly enhance the thermal stability of HIV TAR RNA
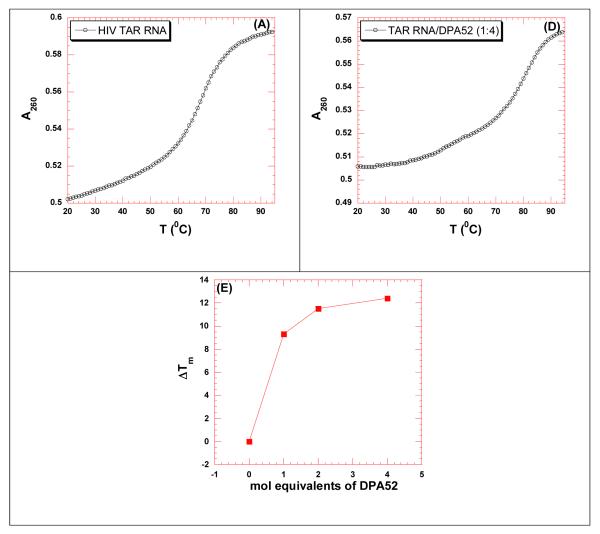
UV thermal denaturation studies were carried out between the dimers and HIV 1 TAR RNA (Figure 1 and Table 2, see section S3 in Supplementary Information for UV denaturation plots). Neomycin dimers significantly enhance the thermal stability of HIV TAR RNA (ΔTm = 3.3-10.2 °C, Table 2). The thermal stabilization of HIV TAR RNA by neomycin dimers decreases with increasing linker length between the two neomycin units. The UV thermal denaturation data indicates that the probable binding sites of two neomycin units are in close proximity. Additionally, neomycin displayed no thermal stabilization of TAR RNA, as opposed to the neomycin dimers. There was very little thermal stabilization observed (ΔTm = 0.5 °C) of HIV TAR RNA even when the concentration of neomycin was increased above a 1:1 ratio.
Figure 1.
Concentration dependent UV thermal denaturation profile of HIV TAR RNA with neomycin dimer DPA52. UV thermal denaturation data of HIV-1 TAR RNA at (A) 0, (B) 4 μM (4 mol. eq.) of DPA52. (E) The plot shows the change in thermal stabilization (ΔTm) as a function of molar equivalents of DPA52. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. [HIV TAR RNA] = 1 μM/strand. The heating rate was 0.3 °C.
Table 2.
UV thermal denaturation data of HIV TAR RNA in the presence of increasing conc. of neomycin dimer (DPA52). Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. [HIV TAR RNA] = 1 μM/strand. The heating rate was 0.3 °C.
| Ratio (DPA52/TAR RNA) | Tm (°C) | ΔTm (°C) |
|---|---|---|
| TAR RNA | 68.9 | - |
| 1 | 78.2 | 9.3 |
| 2 | 80.4 | 11.5 |
| 4 | 81.3 | 12.4 |
The UV denaturation data suggests that the thermal stabilization of HIV TAR RNA by neomycin dimers is a result of specific interaction as opposed to a non-specific electrostatic cation (neomycin dimers) –anion (HIV TAR RNA) interaction. To further validate our assumption, a concentration dependent thermal denaturation study was performed where the concentration of the dimer was varied at a fixed RNA concentration. UV thermal denaturation of HIV TAR RNA was monitored at 2 mole equivalent of neomycin dimers and the results are summarized in Table 3. Additionally, a concentration dependent UV thermal denaturation study was carried out between HIV TAR RNA and DPA52 (up to 4 mole equivalent of DPA52, Figure 1) and the results are also summarized in Table 2.
Table 3.
Representative table of UV thermal denaturation data of ligand / HIV TAR RNA complexes at rdr = 1 and 2 (rdr = ratio of neomycin dimer to HIV TAR RNA conc.). Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. [HIV TAR RNA] = 1 μM/strand. [Neomycin dimer] = 1 and 2 μM. [Neomycin] = 1 and 2 μM.
| Neomycin dimer | Tm | ΔTm (at rdr = 1) | ΔΔTm (rdr = 2-rdr=1) |
|---|---|---|---|
| HIV 1 TAR RNA | 68.9 | NA | NA |
| DPA51 | 79.1 | 10.2 | 2.8 |
| DPA52 | 78.2 | 9.3 | 2.5 |
| DPA65 | 78.2 | 9.3 | 2.1 |
| DPA53 | 78.5 | 9.6 | 2.9 |
| DPA54 | 77.1 | 8.2 | 2.2 |
| DPA55 | 76.4 | 7.6 | 2.8 |
| DPA56 | 74.9 | 6.1 | 3.2 |
| DPA58 | 74.3 | 5.4 | 2.9 |
| DPA60 | 72.1 | 3.3 | 3.0 |
| Neomycin | 69.1 | 0.2 | 0.4 |
There are few important points to be noted. (1) There was only 2-3 °C additional thermal stabilization observed in going from one to two mole equivalent of neomycin dimers. This is in contrast to the UV-thermal stabilization of HIV TAR RNA of ~11 °C, when going from 0:1 to a 1:1 (neomycin dimer:HIV TAR RNA). (2) The UV thermal denaturation of HIV TAR RNA upon increase of all the neomycin dimers from one to two mole equivalent shows the same change in Tm (2-3 °C). (3) There was a thermal stabilization of 12.4 °C observed for HIV TAR RNA in the presence of 4 mole equivalent of DPA52 in comparison to 9.3 °C at one mole equivalent. The thermal denaturation data suggests that in all likelihood: (1) A specific binding interaction takes place between neomycin dimers and HIV TAR RNA at a binding stoichiometry of 1:1 (neomycin dimer:HIV TAR RNA). The concentration dependent study suggests that neomycin dimer binding with HIV TAR RNA is not simply a result of charge interactions, but involves potential and shape complementarity to the RNA target. (2) Higher molar ratio of neomycin dimer does not enhance the UV thermal stability significantly. It is indicative of 1:1 binding ratio between neomycin dimer and HIV TAR RNA. (3) Addition of large excess of neomycin to TAR does not have any effect on the thermal stability of HIV TAR RNA, suggesting that sum of the monomer parts clearly leads to a much improved ligand with enhanced RNA affinity.
CD spectroscopy studies to characterize the binding between neomycin dimer and HIV TAR RNA
CD spectroscopy is a powerful tool to monitor and characterize the binding interaction between macromolecules (nucleic acids and proteins) and ligands.32, 33 CD spectroscopy can provide useful insight into the change in the conformation of the macromolecule during the binding reaction.32 CD spectroscopy was used to investigate the binding interaction between neomycin dimer and HIV TAR RNA. CD spectrum of HIV TAR RNA consists of a strong positive peak at 265 nm and an equally strong peak with a negative magnitude at 211 nm. Additionally, there is a negative peak observed at ~240 nm. This CD spectrum is indicative of an A-form conformation. Earlier CD studies of Tat/HIV TAR RNA complex revealed a substantial change in the structure of HIV TAR RNA induced by TAT protein. During the Tat-HIV TAR RNA binding interaction there is a slight red shift observed at 265 nm. Additionally, the CD intensity of HIV TAR RNA at 265 nm decreases by 15%.62 The CD peak at 265 nm is a signature peak for tri-nucleotide bulge region (UCU) and has been attributed to the arrangement of base stacking in the bulge region.22 During the Tat-HIV TAR RNA interaction, the Tat protein binds at its primary binding site which is the tri-nucleotide region and modifies the arrangement of base stacking in the bulge region. The binding interaction was monitored by a decrease in the CD intensity at 265 nm. There is a marginal red shift observed at 265 nm, indicative of the small distortion of A-form structure.
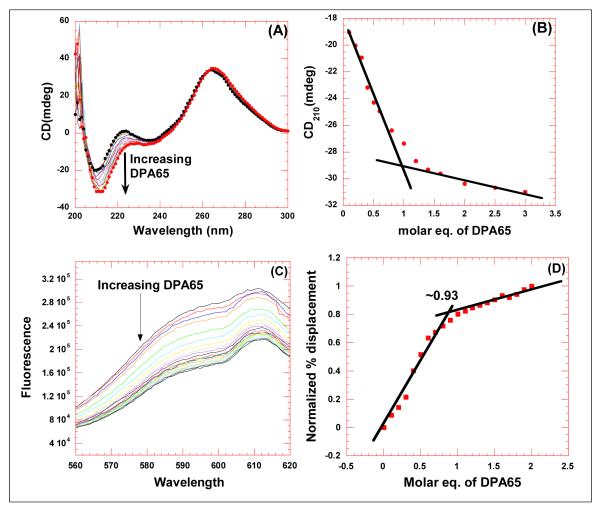
A CD titration was performed between neomycin dimer and HIV TAR RNA by continuous addition of the dimer to an HIV TAR RNA solution (Figure 2A). The overall shape of the CD spectra, in the absence as well as presence of the dimer are similar, suggesting that the overall conformation of HIV TAR RNA is conserved. There are some changes observed in the spectrum of HIV TAR RNA upon the addition of the dimer. The addition of neomycin dimer causes a gradual decrease in the peak at 210 nm and at 240 nm. Contrary to TAT protein binding, there was no change in the CD intensity of HIV TAR RNA/dimer complex at 265 nm, however a red shift was observed. This observation suggests that the dimer does not interact with the tri-nucleotide bulge region (Figure 2 A). However, the interaction of neomycin dimer causes a slight distortion of the A-form conformation of HIV TAR RNA and pushes it towards B-form. All these observations collectively give insight into the probable binding site of neomycin dimer on HIV TAR RNA. The primary binding site of neomycin is just below the tri-nucleotide bulge region (see Scheme 1), hence one unit of neomycin likely binds to the lower stem region of HIV TAR RNA, and the second neomycin unit binds to the upper stem or the hairpin region. Further structural studies are ongoing to evaluate the effect of dimer binding on RNA conformation and will be reported in due course.
Figure 2.
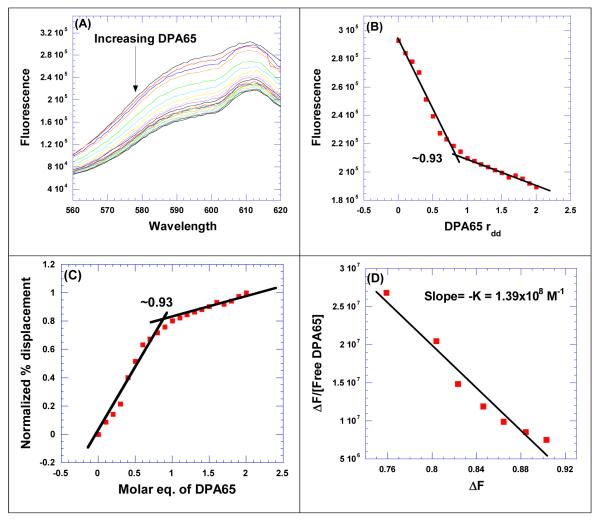
Comparison of CD titration and FID titration for determining the binding stoichiometry between HIV TAR RNA and DPA65. (A) CD titration of HIV TAR RNA with increasing concentration of neomycin dimer. The figure represents molar ellipticity for CD titration of HIV TAR RNA with neomycin dimer. The continuous changes in the CD spectra correspond to the incremental amount of neomycin dimer ranging from an rdr of 0 to 3. (B) A plot of normalized molar ellipticity versus rdr for CD titration of HIV TAR RNA with neomycin dimer. The continuous lines in the plot reflect the linear least squares fit of each apparent linear domain of the experimental data (filled circles) before and after the apparent inflection point. (C) Raw fluorescence emission spectra in the presence of increasing concentration of DPA65. (D) The plot between normalized fluorescence intensity (at 605 nm) of HIV TAR RNA/EtBr complex as a function of concentration of DPA65 results in a saturating binding plot. For CD, Molar ellipticity is per molecule of HIV TAR RNA, and rdr = drug/ HIV TAR RNA. [HIV TAR RNA] = 4 μM/strand. For FID titration, [HIV TAR RNA] = 100 nM/strand.
CD spectroscopy is useful in identifying global changes in the macromolecule structure but it also can provide valuable information about the binding stoichiometry of ligands involved in the study.33 The change in CD intensity was plotted as a function of molar ratio of neomycin dimer/HIV TAR RNA, and the plot results in an inflexion point (Figure 2 B) suggesting that the binding stoichiometry is 1:1 (neomycin dimer: HIV TAR RNA). To verify the CD spectroscopy results, an FID (fluorescent Intercalator Displacement) titration was also performed63 (ethidium bromide as an intercalator) (Figure 2 C). The change in fluorescence was plotted as a function of molar equivalents of neomycin dimer and the inflexion point allows us to also estimate the binding stoichiometry of neomycin dimer to HIV TAR RNA. Both CD and FID titration data reveal a binding stoichiometry of 1:1 between neomycin dimer and HIV TAR RNA.
Ethidium bromide displacement and FRET competition binding assay for characterizing the binding of neomycin dimers with HIV TAR RNA
To further investigate the binding between HIV TAR RNA and neomycin dimers, ethidium bromide displacements63 were performed and the results are summarized in Table 4 (see section S4 in Supplementary Information for FID assay plots). The affinities of neomycin dimers are directly proportional to the amount of ethidium bromide displaced from HIV TAR RNA and reflected in the IC50 (the concentration of ligand required to displace 50% ethidium bromide from HIV TAR RNA) values. Ethidium bromide displacement experiments indicate that in general, neomycin dimers with shorter linker lengths have a higher affinity towards HIV TAR RNA than neomycin dimers with a longer linker. The IC50 values for neomycin dimers fall within a range of 36-99 nM. In comparison to neomycin dimers, neomycin shows very low affinity towards HIV TAR RNA. The IC50 value of neomycin towards HIV TAR RNA is 417±115 nM. The trend of linker length versus IC50 values observed here is the same as observed from the linker length versus δTm values from UV thermal denaturation experiments. These results also indicate that the two neomycin binding sites on HIV TAR RNA are very close to each other. Hence, the neomycin dimers with shorter linkers bind tightly, as reflected in the lower IC50 and higher thermal stabilization towards HIV TAR RNA.
Table 4.
Representative table of ethidium bromide displacement assay. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. HIV TAR RNA = 50 nM/strand.
| Ligand | Linker length | IC50 (nM) |
|---|---|---|
| DPA51 | 7 | 56 |
| DPA52 | 7 | 52 |
| DPA65 | 7 | 36 |
| DPA53 | 8 | 67 |
| DPA54 | 8 | 81 |
| DPA55 | 10 | 99 |
| DPA56 | 10 | 97 |
| DPA58 | 16 | 67 |
| DPA60 | 20 | 74 |
| Neomycin | NA | 417 |
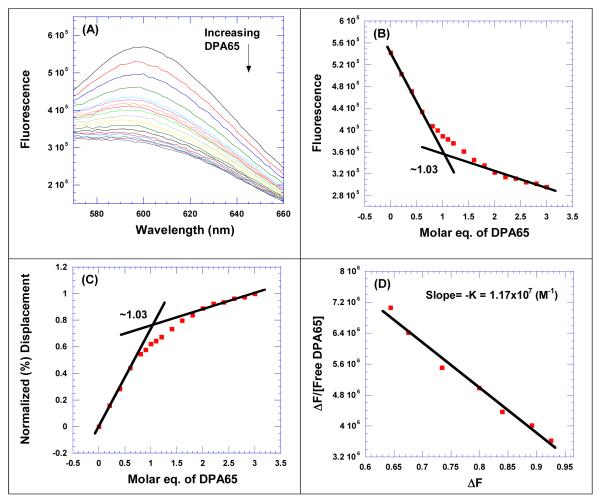
The CD titration and UV denaturation, data show that the dimers bind with a stoichiometry of 1:1 with TAR RNA. We therefore performed ethidium bromide displacement titrations between HIV TAR RNA and neomycin dimer to determine the association constant using Scatchard analysis63 (Figure 3A-D). The plot between the change in fluorescence versus molar equivalents of neomycin dimer(s) to TAR RNA was used (as described above) to determine the binding stoichiometry of neomycin dimer towards HIV TAR RNA (Figure 3B). The data from ethidium bromide displacement titrations was analyzed and transformed using Scatchard analysis (Figure 3C). The slope of the plot allows us to determine the association constant of neomycin dimer(s) with HIV TAR RNA (Figure 3D).
Figure 3.
FID titration of DPA65 with HIV TAR RNA. (A) Raw fluorescence emission spectra in the presence of increasing concentration of DPA65. (B) The decrease of fluorescence intensity (at 605 nm) of HIV TAR RNA/EtBr complex with increasing concentration of DPA65 results in a saturating binding plot. (C) The plot between normalized fluorescence intensity (at 605 nm) of HIV TAR RNA-EtBr complex as a function of concentration of DPA65 results in a saturating binding plot. (D) The Scatchard plot analysis of DPA65 with HIV TAR RNA. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. HIV TAR RNA = 200 nM/strand. [EtBr] = 5 μM.
FID titrations were performed with all the neomycin dimers and monomer (neomycin) (see section S6 in Supplementary Information for FID titrations) with TAR RNA and the results are summarized in Table 5. The binding constant of neomycin with TAR RNA could not be determined using this analysis, due to the weak affinity of neomycin towards HIV TAR RNA (as indicated by small displacement of EtBr). This is consistent with the earlier observations (UV melt, FID assay) which show a weak affinity of neomycin towards HIV TAR RNA.
Table 5.
Representative table of binding constants derived from Scatchard analysis of the FID assay (ethidium bromide as in intercalator) between neomycin dimer and HIV TAR RNA. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. HIV TAR RNA = 200 nM/strand. [EtBr] = 5 μM.
| Neomycin Dimer | Linker Length | K (M−1) |
|---|---|---|
| DPA51 | 7 | 1.17×108 |
| DPA52 | 7 | 7.08×107 |
| DPA65 | 7 | 1.39×108 |
| DPA53 | 8 | 1.46×108 |
| DPA54 | 8 | 2.61×107 |
| DPA55 | 10 | 1.06×107 |
| DPA56 | 10 | 6.60×107 |
| DPA58 | 16 | 7.58×106 |
| DPA60 | 20 | 2.53×107 |
| Neomycin | NA | - |
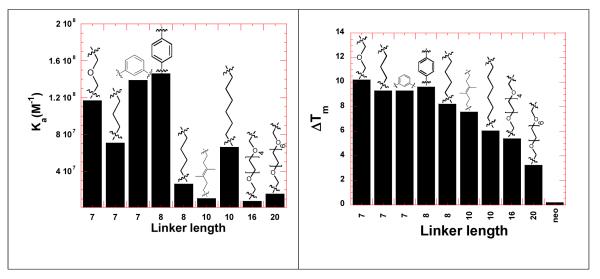
A comparison was then made between the linker lengths of neomycin dimers and the binding constants derived from Scatchard analysis (Figure 4). The binding constant decreases with an increase in the length of the linker. Neomycin dimer (DPA56, linker length 10) was an exception, as it showed a higher affinity than DPA54, linker length 8, but still much lower affinity than linker lengths 7-8. The trend in binding constant versus Ka was compared with the data from the UV thermal denaturation experiment (Figure 4). The trends are similar if not identical. The binding constant decreases with increase in the linker length; similarly the UV thermal stabilization of HIV TAR decreases with increase in the linker length of neomycin dimers.
Figure 4.
Bar graph showing the comparison of binding constants derived from Scatchard analysis of the FID titrations (ethidium bromide as in intercalator) (left) and UV thermal denaturation profile of neomycin dimers with HIV TAR RNA (right). Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. For FID titration, [HIV TAR RNA] = 200 nM/strand. For UV thermal denaturation experiment, [HIV TAR RNA] = 1 μM/strand. [Neomycin dimer] = 1 μM. [Neomycin] = 1 μM.
Salt dependent studies of neomycin dimer with TAR RNA
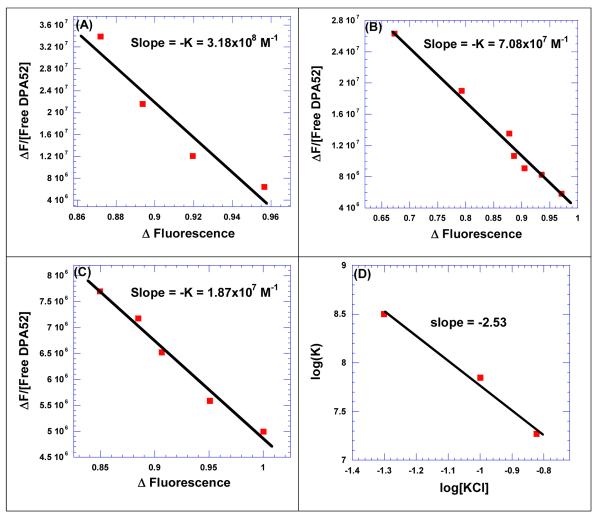
We then studied the effect of salt on the binding between neomycin dimer and HIV TAR RNA. FID titrations were carried out at three different salt concentrations (Figure 5) and the data were analyzed and fitted using Scatchard analysis to determine the binding constants. The binding constants at three different salt concentrations are summarized in Table 6.
Figure 5.
Salt dependent studies between neomycin dimer (DPA52) and HIV TAR RNA. The binding constants were determined from FID titration using Scatchard plot analysis at (A) 50 mM KCl, (B) 100 mM KCl, and (C) 150 mM KCl. (D) A plot between log (K) as a function of log(KCl) was fit with linear regression and the solid black line reflects the linear fit which results in a slope that reflects the number of ion pairs forming between DPA52 and HIV TAR RNA. Buffer conditions: 10 mM SC, 0.5 mM EDTA, pH 6.8. [HIV TAR RNA] = 200 nM/strand. [EtBr] = 5 μM.
Table 6.
Representative table of binding constants derived from Scatchard analysis showing the salt dependence of binding between DPA52 and HIV TAR RNA. Buffer conditions: 10 mM SC, 0.5 mM EDTA, pH 6.8. [HIV TAR RNA] = 200 nM/strand. [EtBr] = 5 μM.
| [KCl] in mM | K (M−1) |
|---|---|
| 50 | 3.18×108 |
| 100 | 7.08×107 |
| 150 | 1.87×107 |
The binding affinity of neomycin dimer (DPA52) towards HIV TAR RNA continuously decreases with an increase in the salt concentration from 50 to 150 mM KCl. The salt dependent binding parameters were used to calculate the number of ion pairs formed during the association of neomycin dimer and HIV TAR RNA, as described by Record.64 In case of HIV TAR RNA and neomycin dimer interaction, there are 2-3 ion pairs between ligand and RNA. The free energy of electrolytic contribution was then calculated using equation 1.64, 65
| 1 |
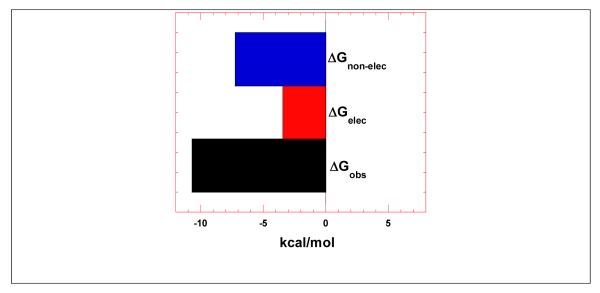
In equation 1, Z denotes the charge on the ligand, φ is the number of the counter-ions associate with each phosphate group on nucleic acid, and the value normally varies for nucleic acids. The value of φ for A-form RNA duplex [poly(rA).poly(rU)] is 0.89, while that for single-stranded RNA [poly(rA)] is 0.78. HIV TAR RNA contains hairpin, bulge, and duplex region; therefore it is likely that the value of φ fall within the range of these two values, hence the average of these two values was used in the analysis. The free energy of electrolytic contribution was calculated by inserting the experimental values in equation 1. The electrolytic contribution to the free energy (ΔGpe) obtained is −3.45 kcal/mole at 100 mM KCl and the total free energy of interaction (ΔGobs) is −10.69 kcal/mole. The free energy contribution of non-electrolytic energy was calculated using equation 2 as −7.24 kcal/mole (Figure 6).
Figure 6.
Bar graph showing the dissection of the free energy into its components including electrolytic and non-electrolytic contributions.
| 2 |
Figure 6 shows that the major contribution to the binding event between neomycin dimer and HIV TAR RNA is non-electrolytic. This observation gives further insight into the mode of binding between neomycin dimer and HIV TAR RNA. Though the dimer is a highly positively charged ligand (12 aliphatic amines, 10 of which are expected to be protonated at physiological pH), the free energy of electrolytic contribution does not play a major part in the binding to RNA. The observation suggests that the shape complementarity of neomycin dimer to TAR RNA conformation is largely responsible for the high binding affinity of this highly positively charged pharmacophore towards HIV TAR RNA.
FRET (Fluorescence Resonance Energy Transfer) mediated competition binding assay
A FRET mediated fluoremetric competition binding assay was performed to characterize the binding interaction of neomycin dimers with HIV TAR RNA. The assay developed by Hamashaki and coworkers53 is very useful in defining the binding interaction of ligands with HIV TAR RNA. This particular assay is different than ethidium bromide displacement assay. The assay gives a direct competition between an analog of TAT protein, a fluorescent labeled TAT peptide and the ligands used in the study. In this assay, TAT49-57 peptide was functionalized using fluorescein dye on its N-terminus and tetramethylrhodamine dye on C-terminus to collectively form a FRET system.53
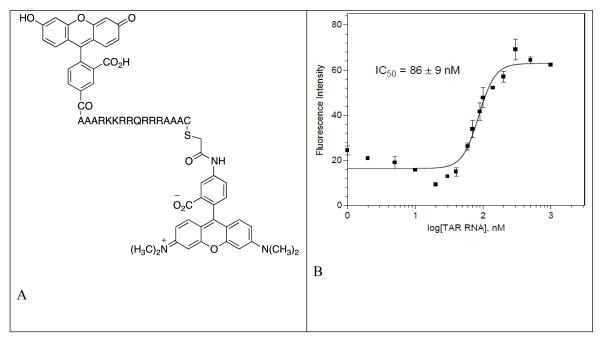
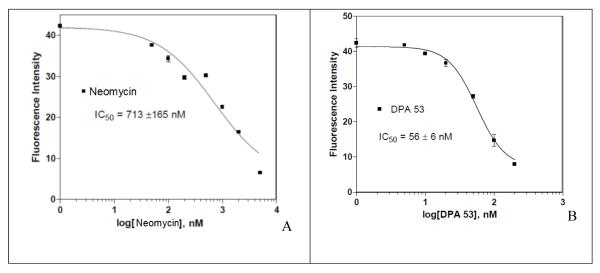
The fluorescein labeled TAT49-57 peptide was first titrated against HIV TAR RNA to determine its affinity (Figure 7). Fluorescence increase is seen with the increase in the concentration of HIV TAR RNA in fluorescein labeled TAT49-57 peptide solution and reaches a constant value at a saturating concentration of HIV TAR RNA. Dimers were then titrated against the fluorescein labeled TAT49-57 peptide + HIV TAR RNA solution (Figure 8, see section S5 in Supplementary Information for FRET assays). The binding affinity of ligands is directly related to the decrease in the fluorescence. The FRET assay was performed with neomycin dimers and neomycin and the results are summarized in Table 7. The IC50 value of neomycin is almost an order of magnitude higher than the neomycin dimers, suggesting much higher affinities for the dimers towards TAR RNA than the monomer neomycin. The IC50 value of Fluorescein labeled TAT49-57 peptide is 86 ± 9 nM. Neomycin dimers show IC50 values within the range of 47-80 nM. In general, neomycin dimers exhibit higher binding affinity in comparison to neomycin or TAT49-57 peptide towards HIV TAR RNA. The FRET binding results of dimer-TAR RNA are consistent with the data obtained from the aforementioned techniques such as UV thermal denaturation and Ethidium bromide displacement.
Figure 7.
Fluorescein labeled TAT49-57 peptide used for the FRET mediated fluoremetric competition binding assay. (B) Saturating binding curve of Fluorescein labeled TAT49-57 peptide with HIV TAR RNA at 25 °C. The binding affinity (IC50) value reported is the average of 3~5 individual measurements, and was determined by fitting the experimental data to a sigmoidal dose-response nonlinear regression model on GraphPad Prism 4.0
Figure 8.
Determination of IC50 values using FRET competition binding assay. Titration curves are shown for determining the IC50 values of (A) Neomycin and (B) DPA53. The binding affinity (IC50) values reported for each ligand are the averages of 3~5 individual measurements, and were determined by fitting the experimental data to a sigmoidal dose-response nonlinear regression model on GraphPad Prism 4.0.
Table 7.
The IC50 values of ligand determined from FRET mediated competition binding assay.
| Ligands | Linker length | Structure of the linker (-x-, from Scheme 1 and Scheme 2) |
IC50(nM) |
|---|---|---|---|
| Fluorescin-labeled- TAT peptide |
NA | NA | 86 ± 9 |
| DPA51 | 7 |
|
77 ± 27 |
| DPA52 | 7 |
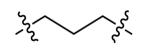
|
60 ± 8 |
| DPA65 | 7 |
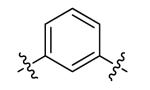
|
47 ± 6 |
| DPA53 | 8 |
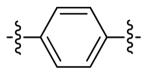
|
56 ± 6 |
| DPA54 | 8 |
|
58 ± 6 |
| DPA55 | 10 |
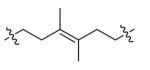
|
80 ± 9 |
| DPA56 | 10 |
|
59 ± 11 |
| DPA58 | 16 |
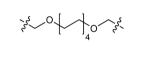
|
61 ± 13 |
| DPA60 | 20 |
|
67 ± 9 |
| Neomycin | NA | NA | 713 ± 165 |
The results from FRET mediated assay showed that neomycin dimers exhibit nanomolar affinity towards HIV TAR RNA. A comparison of binding affinity can be made with the earlier reported neamine dimers. Previously reported neamine dimers showed IC50 in the range of 150-400 nM (pH =7.4, 50 mM Tris-HCl, 20 mM KCl) using FRET mediated competition binding assay, and have a much lower affinity when compared to neomycin dimers reported here (47-128 nM at pH =7.4, 50 mM Tris-HCl, 20 mM KCl). The results clearly show a 3-4 fold preference of neomycin dimers than neamine dimers towards HIV TAR RNA.51 These observations acknowledge the contribution of rings 3 and 4 in the binding event of neomycin and TAR RNA.
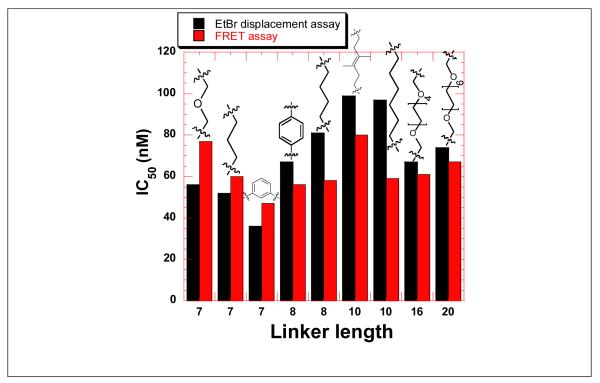
The affinity of neomycin dimers with HIV TAR RNA was compared using FRET and ethidium bromide FID assay (Figure 10). The results from both the assays were comparable as observed from the trends in IC50 values as a function of linker length. The binding affinity of shorter and rigid linkers is higher and reflected in the lower IC50 value, as compared to the higher IC50 obtained with longer linkers. The lowest IC50 was displayed by DPA53 and DPA65 with HIV TAR RNA. Dimers DPA65 and DPA53 are the most rigid structures in the triazole library reported here. They have a phenyl ring between two neomycin units in addition to the two triazole rings leading to an extended aromatic system which enhances the linker rigidity of these dimers in comparison to other more flexible dimers such as DPA 52 and DPA 54.
Figure 10.
Bar graph showing the comparison of IC50 values of neomycin dimers with HIV TAR RNA using FRET-mediated competition binding assay and ethidium bromide displacement assay.
To investigate the specificity of the tight binders, we then studies the binding of DPA65 with a TAR mutant (tetraloop TAR), as shown in Figure 11. FID titrations were conducted between DPA65 and mutant TAR (Figure 12) and the Scatchard analysis of the data yielded a Ka~107M−1. When compared to its affinity for wild type TAR, DPA65 binds to mutant TAR by an order of magnitude lower affinity, suggesting that linker length variations can lead to RNA binders with higher affinities and selectivity than monomeric aminoglycosides.
Figure 11.
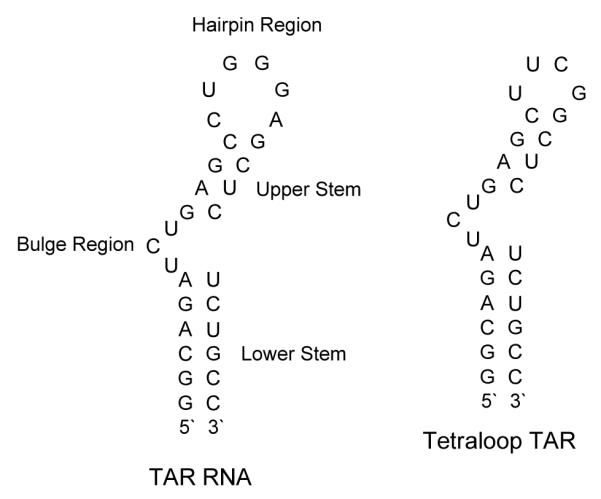
Structure of TAR RNA and mutant tetraloop TAR used in the study.
Figure 12.
FID titration of DPA65 with tetraloop TAR RNA Mutant. (A) Raw fluorescence emission spectra in the presence of increasing concentration of DPA65. (B) The decrease of fluorescence intensity (at 610 nm) of tetraloop TAR RNA/EtBr complex with increasing [DPA65]. (C) The plot between normalized fluorescence intensity (at 610 nm) of tetraloop TAR RNA-EtBr complex as a function of [DPA65]. (D) Scatchard analysis of DPA65 with tetraloop TAR RNA. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. Tetraloop TAR RNA = 200 nM/strand. [EtBr] = 5 μM.
In vitro assay to study the effect of neomycin dimers in MT-2 cells
An important step in the life cycle of HIV is the death of the infected cell population. The HIV envelope uses CD4 cell as a receptor. The infection caused by the HIV virus through CD4 results in an immense enhancement in the number of the infected cells, including cell-cell fusion and cell death, a phenomenon called cytopathic effect (CPE).66 Neomycin dimers and neomycin were monitored for their ability to protect MT2 cells from the cytopathic effect caused by HIV (Table 9). We first assessed toxicity against MT-2 cells, a CD4+, lymphoblastoid cell line. We utilized the highly restrictive definition of 5% cytotoxicity (CT5), rather than the more commonly reported 50% cytotoxicity (CT50), as HIV is an obligate intracellular parasite. Any toxic effects on the cell may manifest themselves as a perceived anti-HIV effect. The CT5 is a non-toxic concentration where 95% of cells are viable. The maximum concentration of the neomycin conjugates used is listed in Table 9. For all compounds, this is the concentration of ligands where no more than 5% cell death was observed (CT5). For comparison, water was used as control.
Table 9.
The inhibition effect of neomycin dimers on HIV antigen synthesis in treated cells.
| Compound | Concentration (μM) |
% of HIV antigen synthesis | ||
|---|---|---|---|---|
|
| ||||
| Day 2 | Day 4 | Day 6 | ||
|
| ||||
| DPA52 | 25 | 15% | 100% | 100% |
| DPA53 | 9 | 2% | 40% | 100% |
| DPA54 | 17 | 3-5% | 80% | 100% |
| DPA55 | 4 | 3-5% | 30% | 100% |
| DPA56 | 8 | 5-7% | 40% | 100% |
| Virus Control | NA | 15% | 100% | 100% |
We next assessed anti-HIV activities of the compounds. This assay is performed in triplicate wells of a 96 well plate. It utilizes Finter’s neutral red dye, a vital dye, to assess the ability of any given antibody or compound to inhibit HIV-induced cytopathic effect. As indicated in Table 8, neomycin dimers showed much improved protection from cytopathic effect as opposed to neomycin and water (control). Neomycin dimer (DPA52) showed little protection from cytopathic effect, suggesting that minor variations in linker length can significantly affect the pharmacodynamic and/or pharmacokinetic properties of these ligands. Neomycin dimers, DPA53-DPA56, showed 20-33% protection from the cytopathic effect in the concentration range of 4-17 μM. Overall, the compounds showed modest anti-HIV activity in this assay with a maximal 33% suppression of cell death.
Table 8.
5% Toxicity concentrations of neomycin dimers and neomycin showing the maximum protection obtained from HIV cytopathic effects.
| Compound | 5% Toxicity (μM) | Maximum protection (concentration achieved in μM) |
|---|---|---|
| Neomycin | >206 | 9% (206) |
| DPA52 | >138 | 1% (9) |
| DPA53 | 17 | 33% (8) |
| DPA54 | 69 | 31% (17) |
| DPA55 | 8 | 20% (4) |
| DPA56 | 34 | 33% (17) |
| Water | None | 2% |
We then confirmed the anti-HIV activity by two additional assays. These include spread of HIV through culture, as measured by immunofluorescence assay (IFA) and reverse transcriptase (RT) release into supernatant fluids. In this experiment, non-toxic concentrations of each of five neomycin dimers, with some anti-HIV activity (Table 8) were selected. We pre-treated 500,000 cells with each compound for 1 h. Next, cells were inoculated with approximately 50,000 infectious particles of HIVNL4-3. Infections were performed in triplicate. Every 2 days cells were collected and stained for HIV antigen synthesis. Culture supernatants were analyzed for RT release. As shown in Table 9, the compounds had a more robust effect on HIV antigen synthesis than was apparent in the cytopathicity assay. Indeed, several compounds inhibited HIV replication by as much as 70% (day 4).
The neomycin dimers inhibit HIV antigen synthesis effectively up to 4 days post inoculation. Neomycin dimers (DPA55, DPA53, and DPA56) inhibit HIV antigen synthesis from 60-70% up to the 4th day as opposed to other neomycin dimers. The virus control contains the infected cells plus water solvent. Dimer DPA53, is among the highest affinity dimer towards HIV TAR RNA as borne out by FRET mediated competition binding assay, ethidium bromide assay, and UV thermal denaturation experiment.
Neomycin dimers also reduced the amount of reverse transcriptase (RT) released into the cell culture supernatant as opposed to the virus control.67 HIV replication was suppressed by an even greater amount in this assay. For example, DPA56 suppressed RT release by 90% at day 4 and at least 20% at day 6 (Table 10). A couple of important observations need to be noted (1) all the neomycin dimers inhibit the release of RT. (2) Neomycin dimer, DPA53, is the most effective among all the neomycin dimers studied reflected by the maximum inhibition (Table 10) at moderate concentration. (3) As opposed to the HIV antigen inhibition assay, neomycin dimers inhibit the release of RT even after 6 days.
Table 10.
Release of reverse transcriptase into the culture supernatant (cpm/ml). Values are mean of triplicate infections, values in parentheses are one standard deviation.
| Compound | Concentration (μM) |
Day 2 (cpm/mL) | Day 4 (cpm/mL) | Day 6 (cpm/mL) |
|---|---|---|---|---|
|
| ||||
| DPA52 | 25 | 21,485 (4,574) | 347,845 (10,680) | 268,357 (57,519) |
| DPA53 | 9 | 8,880 (1,373) | 45,539 (32,831) | 221,445 (20,678) |
| DPA54 | 17 | 14,805 (1,309) | 165,301 (22,967) | 427,475 (49,583) |
| DPA55 | 4 | 20,072 (3,748) | 107,933 (17,963) | 305,277 (59,913) |
| DPA56 | 8 | 15,989 (2,523) | 105,704 (15,948) | 412,475 (19,612) |
| Virus Control | NA | 46,029 (6,651) | 928,112 (126,547) | 1,078,741 (188,673) |
Cpm/mL: counts per minute/mL
These findings were somewhat surprising, as we do not usually see a significant anti-HIV effect in confirmatory assays that is stronger than the anti-HIV activity seen in the cytopathic effect assay (Table 8). A much more significant anti-HIV effect of neomycin dimers was seen in the confirmatory assays. Since DPA65 was the one of the most potent binding agent, two separate experiments were performed to determine the anti-HIV activities of this compound. As with the other compounds, DPA65 was inactive to weakly active in the 96-well plate screening assay. When the anti-HIV properties of the compound as measured by RT release into the culture media were determined, they were highly reproducible over a period of months and replicates. The compound was tested three separate times, in triplicate each time. On day 2 post inoculation, it protected cells by 40-67%. On day 4, it protected by 68-73% and on day 6 by 24-27%. The lower levels of protection at day 6 were due to lysis of the virus control infections on day 4, and thus little additional release of virus into the culture media from day 4 to day 6. To ascertain whether the compounds were acting at Tat-TAR interactions within cells, we attempted to raise inhibitor-resistant virus by culturing HIV in increasing concentrations of neomycin dimers. We were not able to raise a resistant virus, indicating that the combination of anti-HIV activity with the likelihood of deleterious mutations within Tat or TAR precluded the selection of viable, inhibitor-resistant HIV. Nevertheless, the unusual finding of weaker activity against HIV-induced cytopathic effect but a more potent inhibition of antigen synthesis is consistent with an effect on HIV transcription.
Conclusion
A series of neomycin dimers were synthesized via click chemistry in high yields. The dimers were then studied with HIV TAR RNA using various analytical techniques. UV-thermal denaturation, CD titration, FID assay, FRET mediated competition binding assay and in vitro cellular assays for HIV inhibition. There are a number of conclusions that can be drawn from the studies. (1) UV thermal denaturation studies show that neomycin dimers enhance the melting temperature of HIV TAR RNA up to 10.3 °C. The monomer neomycin does not show any effect on the thermal stabilization of HIV TAR RNA. The neomycin dimers with the shortest linker length displayed the highest thermal stabilization and it decreases with an increase in the linker length. The concentration dependent thermal denaturation studies indicate that neomycin dimer binds with a stoichiometry of 1:1 with HIV TAR RNA. (2) The CD spectra in presence of neomycin dimer suggest that it does not interact with the tri-nucleotide bulge region, which is the binding site of TAT protein. Additionally, the neomycin dimer binding pushes the conformation of HIV TAR RNA away from an A-form. (3) The ethidium bromide displacement titration between neomycin dimers and HIV TAR RNA showed a binding stoichiometry of 1:1, validating the results from CD, and UV thermal denaturation experiments. (4) Scatchard analysis of FID titrations result in a binding constant of 107-108 M−1 between neomycin dimers and HIV TAR RNA depending on the linker length. The trends in binding affinity obtained are similar to the trends in UV thermal denaturation stabilization. The neomycin dimers with shorter linker lengths show the highest affinity towards HIV TAR RNA. (5) Salt dependent studies show that despite the highly charged nature of the dimeric ligands, the major contributor to the free energy of binding comes from non-electrolytic terms. (6) The FRET mediated competition binding assay confirms the high affinity of neomycin dimers towards HIV TAR RNA. The patterns of TAR RNA –dimer binding affinity are similar to those obtained from UV thermal denaturation and FID assay. The binding affinity of neomycin is much weaker than all the neomycin dimers. (7) In vitro assays show that neomycin dimers are effective inhibitors of HIV. Some of the neomycin dimers inhibit the release of reverse transcriptase effectively at low concentration. Our studies outline the growing potential of click chemistry in efficiently generating multivalent ligands to improve ligand binding to therapeutic targets. Further studies are ongoing to explore SAR of modified aminosugars, structural studies of binding, and the details of mechanisms of HIV inhibition by these dimers. These studies will be reported in due course.
Supplementary Material
Figure 9.
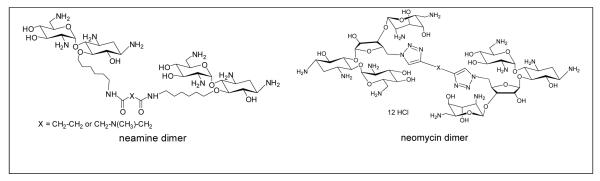
Chemical structure of neomycin and neamine dimers.
Footnotes
Supporting Information.
Characterization of triazole linked neomycin dimers using click chemistry. UV-thermal denaturation profiles, ethidium bromide displacement assay plots, FRET-mediated competition binding assay plots, and ethidium bromide displacement titration to determine the binding constants plots. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Berkhout B, Jeang KT. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J. Virol. 1989;63:5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KA, Peterlin BM. Control of Rna Initiation and Elongation at the Hiv-1 Promoter. Annu. Rev. Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 3.Draper DE. Protein-Rna Recognition. Annu. Rev. Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 4.Rosen CA, Sodroski JG, Haseltine WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 5.Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 6.Aboul-ela F, Karn J, Varani G. The Structure of the Human Immunodeficiency Virus Type-1 TAR RNA Reveals Principles of RNA Recognition by Tat Protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 7.Rana TM, Jeang KT. Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch. Biochem. Biophys. 1999;365:175–185. doi: 10.1006/abbi.1999.1206. [DOI] [PubMed] [Google Scholar]
- 8.Hauber J, Cullen BR. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J. Virol. 1988;62:673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng S, Holland EC. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 10.Puglisi J, Tan R, Calnan B, Frankel A. Williamson Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 11.Thomas JR, Hergenrother PJ. Targeting RNA with Small Molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 12.Bailly C, Colson P, Houssier C, Hamy F. The Binding Mode of Drugs to the TAR RNA of HIV-1 Studied by Electric Linear Dichroism. Nucleic Acids Res. 1996;24:1460–1464. doi: 10.1093/nar/24.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratmeyer LS, Vinayak R, Zon G, Wilson WD. An ethidium analog that binds with high specificity to a base-bulged duplex from the TAR RNA region of the HIV-1 genome. J. Med. Chem. 1992;35:966–968. doi: 10.1021/jm00083a024. [DOI] [PubMed] [Google Scholar]
- 14.Dassonneville L, Hamy F, Colson P, Houssier C, Bailly C. Binding of Hoechst 33258 to the TAR RNA of HIV-1. Recognition of a pyrimidine bulge-dependent structure. Nucleic Acids Res. 1997;25:4487–4492. doi: 10.1093/nar/25.22.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind KE, Du Z, Fujinaga K, Peterlin BM, James TL. Structure-Based Computational Database Screening, In Vitro Assay, and NMR Assessment of Compounds that Target TAR RNA. Chem. Biol. 2002;9:185–193. doi: 10.1016/s1074-5521(02)00106-0. [DOI] [PubMed] [Google Scholar]
- 16.Tao J, Frankel AD. Specific binding of arginine to TAR RNA. Proc. Natl. Acad. Sci. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S, Tamilarasu N, Ryan K, Huq I, Richter S, Still WC, Rana TM. Inhibition of gene expression in human cells through small molecule-RNA interactions. Proc. Natl. Acad. Sci. 1999;96:12997–13002. doi: 10.1073/pnas.96.23.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Structure-Guided Peptidomimetic Design Leads to Nanomolar Î2-Hairpin Inhibitors of the Tatâ^’TAR Interaction of Bovine Immunodeficiency Virusâ€,‡. Biochemistry (N. Y.) 2007;46:741–751. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 19.Mei H, Mack DP, Galan AA, Halim NS, Heldsinger A, Loo JA, Moreland DW, Sannes-Lowery KA, Sharmeen L, Truong HN, Czarnik AW. Discovery of selective, small-molecule inhibitors of RNA complexes—1. The tat protein/TAR RNA complexes required for HIV-1 transcription. Bioorg. Med. Chem. 1997;5:1173–1184. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 20.Raghunathan D, Sánchez-Pedregal VM, Junker J, Schwiegk C, Kalesse M, Kirschning A, Carlomagno T. TAR-RNA recognition by a novel cyclic aminoglycoside analogue. Nucleic Acids Res. 2006;34:3599–3608. doi: 10.1093/nar/gkl494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tor Y. The ribosomal A-site as an inspiration for the design of RNA binders. Biochimie. 2006;88:1045–1051. doi: 10.1016/j.biochi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Huber PW, Cui M, Czarnik AW, Mei H. Binding of Neomycin to the TAR Element of HIV-1 RNA Induces Dissociation of Tat Protein by an Allosteric Mechanismâ€. Biochemistry (N. Y.) 1998;37:5549–5557. doi: 10.1021/bi972808a. [DOI] [PubMed] [Google Scholar]
- 23.Charles I, Xi H, Arya DP. Sequence-specific targeting of RNA with an oligonucleotide-neomycin conjugate. Bioconjug. Chem. 2007;18:160–169. doi: 10.1021/bc060249r. [DOI] [PubMed] [Google Scholar]
- 24.Shaw NN, Xi H, Arya DP. Molecular recognition of a DNA:RNA hybrid: sub-nanomolar binding by a neomycin-methidium conjugate. Bioorg. Med. Chem. Lett. 2008;18:4142–4145. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 25.Shaw NN, Arya DP. Recognition of the unique structure of DNA:RNA hybrids. Biochimie. 2008;90:1026–1039. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Xi H, Gray D, Kumar S, Arya DP. Molecular recognition of single-stranded RNA: neomycin binding to poly(A) FEBS Lett. 2009;583:2269–2275. doi: 10.1016/j.febslet.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Arya DP. New approaches toward recognition of nucleic Acid triple helices. Acc. Chem. Res. 2011;44:134–146. doi: 10.1021/ar100113q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi H, Davis E, Ranjan N, Xue L, Hyde-Volpe D, Arya DP. Thermodynamics of Nucleic Acid “Shape Readout” by an Aminosugar. Biochemistry (N. Y.) 2011 doi: 10.1021/bi201077h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue L, Ranjan N, Arya DP. Synthesis and spectroscopic studies of the aminoglycoside (Neomycin)-perylene conjugate binding to human telomeric DNA. Biochemistry. 2011;50:2838–2849. doi: 10.1021/bi1017304. [DOI] [PubMed] [Google Scholar]
- 30.Ranjan N, Andreasen KF, Kumar S, Hyde-Volpe D, Arya DP. Aminoglycoside Binding to Oxytricha nova Telomeric DNA. Biochemistry. 2010;49:9891–9903. doi: 10.1021/bi101517e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis B, Arya DP. Triple recognition of B-DNA by a neomycin-Hoechst 33258-pyrene conjugate. Biochemistry. 2010;49:452–469. doi: 10.1021/bi9016796. [DOI] [PubMed] [Google Scholar]
- 32.Xi H, Kumar S, Dosen-Micovic L, Arya DP. Calorimetric and spectroscopic studies of aminoglycoside binding to AT-rich DNA triple helices. Biochimie. 2010;92:514–529. doi: 10.1016/j.biochi.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue L, Xi H, Kumar S, Gray D, Davis E, Hamilton P, Skriba M, Arya DP. Probing the recognition surface of a DNA triplex: binding studies with intercalator-neomycin conjugates. Biochemistry. 2010;49:5540–5552. doi: 10.1021/bi100071j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willis B, Arya DP. Triple Recognition of B-DNA. Bioorg. Med. Chem. Lett. 2009;19:4974–4979. doi: 10.1016/j.bmcl.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 35.Willis B, Arya DP. Major groove recognition of DNA by carbohydrates. Curr. Org. Chem. 2006;10:663–673. [Google Scholar]
- 36.Willis B, Arya DP. Recognition of B-DNA by Neomycin-Hoechst 33258 Conjugates. Biochemistry. 2006;45:10217–10232. doi: 10.1021/bi0609265. [DOI] [PubMed] [Google Scholar]
- 37.Willis B, Arya DP. An expanding view of aminoglycoside-nucleic acid recognition. Adv. Carbohydr. Chem. Biochem. 2006;60:251–302. doi: 10.1016/S0065-2318(06)60006-1. [DOI] [PubMed] [Google Scholar]
- 38.Arya DP. Aminoglycoside-Nucleic Acid Interactions: The Case for Neomycin. Top. Curr. Chem. 2005;253:149–178. [Google Scholar]
- 39.Xi H, Arya DP. Recognition of triple helical nucleic acids by aminoglycosides. Curr Med Chem Anticancer Agents. 2005;5:327–338. doi: 10.2174/1568011054222328. [DOI] [PubMed] [Google Scholar]
- 40.Arya DP, Xue L, Willis B. Aminoglycoside (Neomycin) Preference Is for A-Form Nucleic Acids, Not Just RNA: Results from a Competition Dialysis Study. J. Am. Chem. Soc. 2003;125:10148–10149. doi: 10.1021/ja035117c. [DOI] [PubMed] [Google Scholar]
- 41.Arya DP, Micovic L, Charles I, Coffee RL, Jr., Willis B, Xue L. Neomycin Binding to Watson-Hoogsteen (W-H) DNA Triplex Groove: A Model. J. Am. Chem. Soc. 2003;125:3733–3744. doi: 10.1021/ja027765m. [DOI] [PubMed] [Google Scholar]
- 42.Arya DP, Willis B. Reaching into the major groove of B-DNA: synthesis and nucleic acid binding of a neomycin-hoechst 33258 conjugate. J. Am. Chem. Soc. 2003;125:12398–12399. doi: 10.1021/ja036742k. [DOI] [PubMed] [Google Scholar]
- 43.Arya DP, Xue L, Tennant P. Combining the best in triplex recognition: synthesis and nucleic acid binding of a BQQ-neomycin conjugate. J. Am. Chem. Soc. 2003;125:8070–8071. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 44.Xue L, Charles I, Arya DP. Pyrene-neomycin conjugate: dual recognition of a DNA triple helix. Chem. Commun. 2002;1:70–71. doi: 10.1039/b108171c. [DOI] [PubMed] [Google Scholar]
- 45.Hamilton PL, Arya DP. Natural product DNA major groove binders. Nat. Prod. Rep. 2012;29:134–143. doi: 10.1039/c1np00054c. [DOI] [PubMed] [Google Scholar]
- 46.Arya DP, Coffee RL., Jr. DNA Triple Helix Stabilization by Aminoglycoside Antibiotics. Bioorg. Med. Chem. Lett. 2000;10:1897–1899. doi: 10.1016/s0960-894x(00)00372-3. [DOI] [PubMed] [Google Scholar]
- 47.Arya DP, R.Lane Coffee J, Willis B, Abramovitch AI. Aminoglycosides-Nucleic Acid interactions: Remarkable Stabilization of DNA and RNA Triple helices by Neomycin. J. Am. Chem. Soc. 2001;123:5385–5395. doi: 10.1021/ja003052x. [DOI] [PubMed] [Google Scholar]
- 48.Arya DP, Jr., R LC, Charles I. Neomycin-Induced Hybrid Triplex Formation. J. Am. Chem. Soc. 2001;123:11093–11094. doi: 10.1021/ja016481j. [DOI] [PubMed] [Google Scholar]
- 49.Wang H, Tor Y. Dimeric aminoglycosides: design, synthesis and RNA binding. Bioorg. Med. Chem. Lett. 1997;7:1951–1956. [Google Scholar]
- 50.Michael K, Wang H, Tor Y. Enhanced RNA binding of dimerized aminoglycosides. Bioorg. Med. Chem. 1999;7:1361–1371. doi: 10.1016/s0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 51.Riguet E, Desire J, Boden O, Ludwig V, Gobel M, Bailly C, Decout JL. Neamine dimers targeting the HIV-1 TAR RNA. Bioorg. Med. Chem. Lett. 2005;15:4651–4655. doi: 10.1016/j.bmcl.2005.07.082. [DOI] [PubMed] [Google Scholar]
- 52.Sannes-Lowery KA, Mei H, Loo JA. Studying aminoglycoside antibiotic binding to HIV-1 TAR RNA by electrospray ionization mass spectrometry. Intl. J. Mass Spectrom. 1999;193:115–122. [Google Scholar]
- 53.Matsumoto C, Hamasaki K, Mihara H, Ueno A. A high-throughput screening utilizing intramolecular fluorescence resonance energy transfer for the discovery of the molecules that bind HIV-1 TAR RNA specifically. Bioorg. Med. Chem. Lett. 2000;10:1857–1861. doi: 10.1016/s0960-894x(00)00359-0. [DOI] [PubMed] [Google Scholar]
- 54.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov. Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 55.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 56.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective ligation of azides and terminal alkynes. Angew Chem Int Edit Angew Chem Int Edit. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 57.Kumar S, Arya DP. Recognition of HIV TAR RNA by triazole linked neomycin dimers. Bioorg. Med. Chem. Lett. 2011;21:4788–4792. doi: 10.1016/j.bmcl.2011.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S, Xue L, Arya DP. Neomycin-Neomycin Dimer: An All-Carbohydrate Scaffold with High Affinity for AT-Rich DNA Duplexes. J. Am. Chem. Soc. 2011;133:7361–7375. doi: 10.1021/ja108118v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arya DP, Coffee RL, Xue L. From triplex to B-form duplex stabilization: reversal of target selectivity by aminoglycoside dimers. Bioorg. Med. Chem. Lett. 2004;14:4643–4646. doi: 10.1016/j.bmcl.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 60.Charles I, Xue L, Arya DP. Synthesis of aminoglycoside-DNA conjugates. Bioorg. Med. Chem. Lett. 2002;12:1259–1262. doi: 10.1016/s0960-894x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 61.Charles I, Arya DP. Synthesis of neomycin-DNA/peptide nucleic acid conjugates. J. Carbohydr. Chem. 2005;24:145–160. [Google Scholar]
- 62.Suryawanshi H, Sabharwal H, Maiti S. Thermodynamics of Peptide-RNA Recognition: The Binding of a Tat Peptide to TAR RNA. J. Phys. Chem. B. 2010;114:11155–11163. doi: 10.1021/jp1000545. [DOI] [PubMed] [Google Scholar]
- 63.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A Simple, High-Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 64.Record MT, Jr, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 65.Spolar RS, Livingstone JR, Record MT. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry (N. Y.) 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 66.Leonard R, Zagury D, Desportes I, Bernard J, Zagury JF, Gallo RC. Cytopathic effect of human immunodeficiency virus in T4 cells is linked to the last stage of virus infection. Proc. Natl. Acad. Sci. 1988;85:3570–3574. doi: 10.1073/pnas.85.10.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson WE, Montefiori DC, Gillespie DH, Mitchell WM. Complement-Mediated, Antibody-Dependent Enhancement of HIV-1 Infection in Vitro Is Characterized by Increased Protein and RNA Syntheses and Infectious Virus Release. JAIDS J. Acquired Immune Defic. Syndromes. 1989;2 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.