Abstract
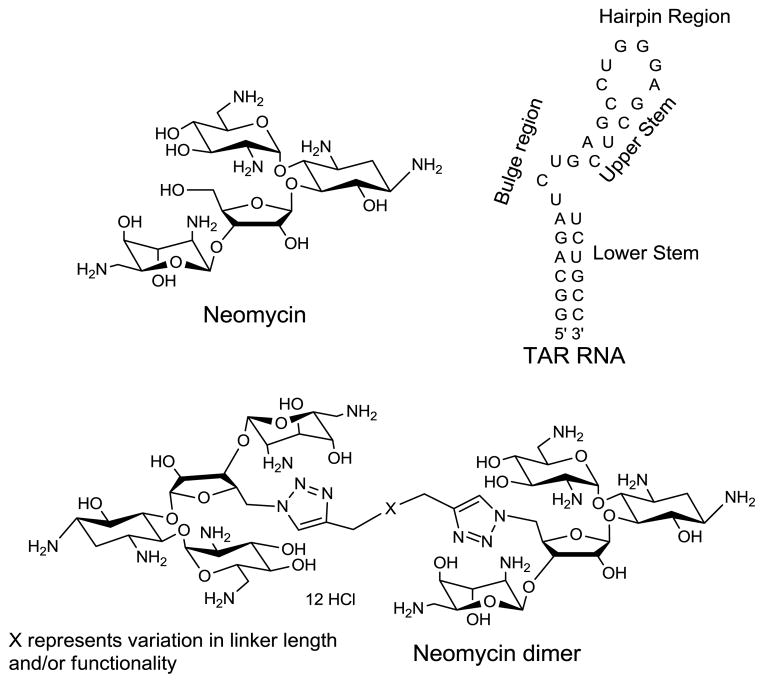
A series of neomycin dimers have been synthesized using “click chemistry” with varying linker functionality and length to target the TAR RNA region of HIV virus. TAR (Trans Activation Response) RNA region, a 59 base pair stem loop structure located at 5′-end of all nascent HIV-1 transcripts interacts with a key regulatory protein, Tat, and necessitates the replication of HIV-1 virus. Neomycin, an aminosugar, has been shown to exhibit more than one binding site with HIV TAR RNA. Multiple TAR binding sites of neomycin prompted us to design and synthesize a small library of neomycin dimers using click chemistry. The binding between neomycin dimers and HIV TAR RNA was characterized using spectroscopic techniques including FID (Fluorescent Intercalator Displacement) titration and UV-thermal denaturation. UV thermal denaturation studies demonstrate that neomycin dimer binding increase the melting temperature (Tm) of the HIV TAR RNA up to 10 °C. Ethidium bromide displacement titrations revealed nanomolar IC50 between neomycin dimers and HIV TAR RNA, whereas with neomycin, a much higher IC50 in the micromolar range is observed.
RNA-protein interactions are essential for regulation of many important biological processes such as translation, RNA splicing, and transcription.1–3 An important example involved in the mechanism of regulatory process of human immunodeficiency virus type 1 (HIV-1). TAR RNA (trans activation responsive region), a 59 base stem-loop structure located at the 5′-end of the nascent viral transcripts,4–7 interacts with Tat protein, (a 86 amino acid protein) and regulates the transcription of HIV virus. TAR RNA-Tat protein interaction is an essential interaction in the replication cycle of HIV virus. TAR, a 29-mer RNA, used in this study is the shorter version of 59-mer. It contains the binding site for TAT protein (the tri-nucleotide bulge), along with the wild type TAR RNA core. HIV TAR structure is comprised of two stems (upper and lower), a three nucleotide bulge region, and a hairpin (for structures, see Scheme 1). An arginine rich domain of Tat protein interacts with the tri-nucleotide bulge (U23, C24, and U25) of TAR RNA3,8,9 and causes a substantial enhancement in the transcript level (~100 fold).2 NMR studies have shown that complexation takes place specifically between arginine residue of Tat protein and a guanine base in the major groove of TAR RNA.10 Disruption of TAR RNA-Tat interaction represents an attractive strategy to inhibit viral replication. There are several molecules investigated so far11 to inhibit TAR RNA-Tat interaction. These include intercalators,12 (ethidium bromide13 and proflavine), DNA minor groove binders14 (Hoechst 33258, and DAPI), phenothiazine,15 argininamide,16 peptides,17 peptidomimetics,18 and aminoglycosides19.
Scheme 1.
Structures of small molecule ligands (neomycin and neomycin dimer) and TAR RNA used in the study.
Aminoglycosides are naturally occurring aminosugars linked through glycosidic linkages, known to bind to a wide variety of RNA structures.20 In the past few years, a number of aminoglycoside conjugates have been synthesized to achieve higher binding affinity and specificity towards RNA targets.20,21 In an attempt to achieve higher binding affinity and explore multiple binding sites on RNA targets, the homo and hetero dimeric units of aminoglycosides22,23 (tobramycin, neamine, neomycin B, and kanamycin A) have been synthesized with various linker length and functionalities through disulfide bond formation. These aminoglycosides exhibit higher binding affinity towards dimerized A-site 16 S construct and RRE RNA than their corresponding monomeric aminoglycoside units. Also, aminoglycoside dimers exhibit 20- to 12000- fold higher inhibitory effects towards the catalytic function of Tetrahymena ribozyme than the monomeric units.23 A neomycin neomycin dimer synthesized with an intercalator, naphthalenediimide, as linker, showed 35-fold higher affinity than neomycin towards A-site 16S RNA.24 We have recently reported that a neomycin dimer with a flexible thiourea linker showed nanomolar binding affinity towards AT rich DNA duplexes.25
Neomycin (Scheme 1), an aminoglycoside, is known to bind various RNA structures, such as the 5′-untranslated region of thymidylate synthase mRNA,26 rev response element (RRE) and the trans activation responsive RNA (TAR RNA) region of HIV-1,27,28 kissing loop formed by DIS (dimerization initiation site) of genomic HIV RNA,29 group I introns,30 ribonuclease P RNA,31 hairpin ribozyme32 and hammerhead ribozyme.33
Among all the aminoglycosides, neomycin has shown the highest inhibitory effect towards TAR RNA (Kd < 1 μM).19 ESI-MS34 and ribonuclease protection experiments35 have suggested that the probable binding site of neomycin lies in the stem region just below the tri-nucleotide bulge in TAR RNA. Further ESI-MS experiments and gel shift assays have revealed the existence of three neomycin binding sites in HIV TAR RNA.34 To achieve higher binding affinity and explore the multiple binding sites on HIV TAR RNA, we have designed and synthesized a series of neomycin dimers using click chemistry. Herein, we report synthesis and TAR RNA binding studies of a series of triazole linked neomycin dimers (for structure see Scheme 1) with various linker lengths and flexibility to optimize the binding affinity.
Synthesis of neomycin dimers using click chemistry
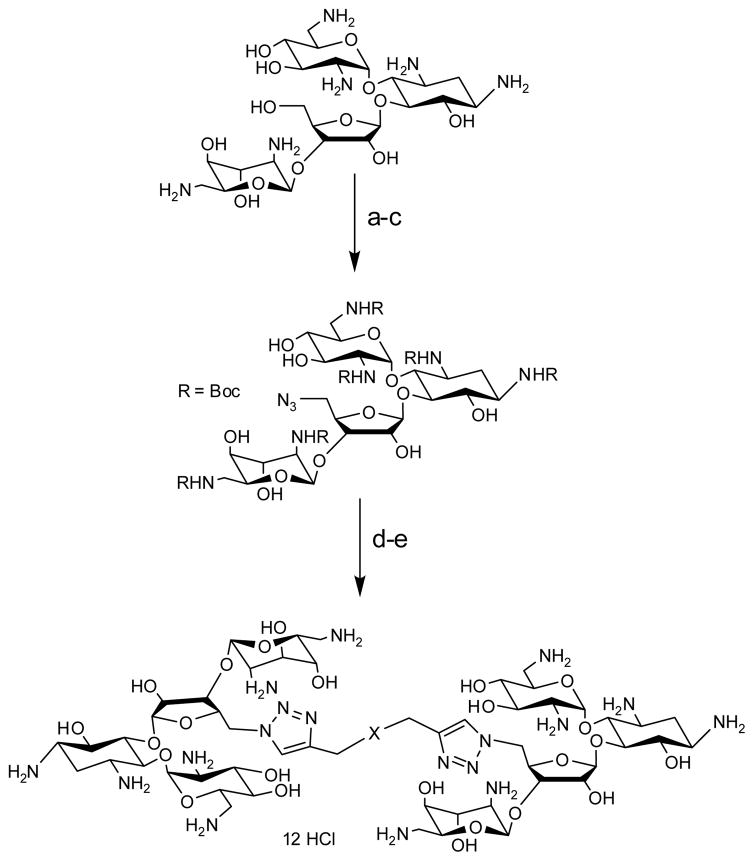
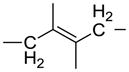
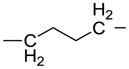
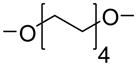
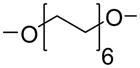
We have used click chemistry36 to synthesize a number of neomycin dimers. A number of commercially available dialkynes were reacted with 2 mole equivalent of boc-protected neomycin-5″-azide, in the presence of CuI, toluene, and DIPEA, to form the neomycin dimers (see Scheme 2). Two additional dialkynes were synthesized to achieve the longer linker length between two neomycin units (DPA58 and DPA60, Table 1).
Scheme 2.
Reagents and conditions: (a) (Boc)2O, DMF, H2O, Et3N, 60 C, 5 h, 60%. (b) TPS-Cl, pyridine, r.t., 40 h, 50%. (c) NaN3, DMF/H2O (10/1), 12h, 90 °C, 90%. (d) Toluene, CuI, DIPEA, 90 °C, 18 h, 85–92%. (e) 4 M HCl/dioxane, r.t., 5 min, 76–95%.
Table 1.
Table of UV thermal denaturation of ligand/HIV TAR RNA complexes at rdd = 1 and ΔTm. Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. HIV TAR RNA = 1 μM/strand. Neomycin dimer = 1 μM. Neomycin = 1 μM.
Neomycin dimers significantly enhance thermal stability of HIV TAR RNA
UV thermal denaturation studies were carried out to monitor the effect of neomycin dimers on HIV-1 TAR RNA. Figure 1 shows the representative plot of UV denaturation profile of HIV TAR RNA in the absence and presence of neomycin dimer (left) and neomycin (right) at a HIV-1 TAR RNA/ligand ratio of 1. The UV thermal denaturation temperature of HIV-1 TAR RNA and HIV-1 TAR RNA ligand complexes were determined using first derivative plots and the results are summarized in Table 1. The UV melting temperature (Tm) of HIV 1 TAR RNA in the absence of ligand is 68.9 °C (100 mM KCl, pH 6.8).
Figure 1.
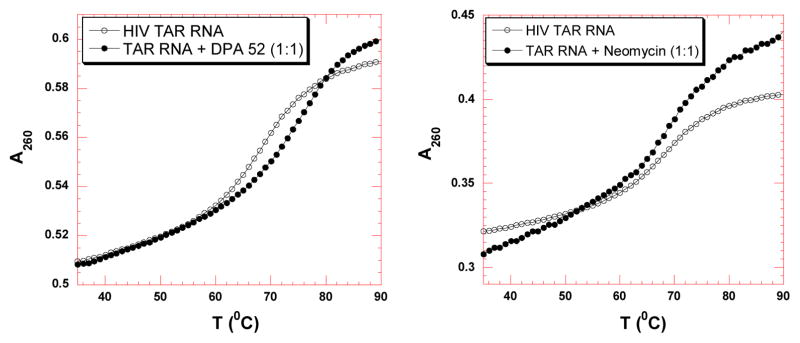
UV thermal denaturation data of HIV-1 TAR RNA with neomycin dimer (left) and neomycin (right). UV thermal denaturation data of HIV-1 TAR RNA in the absence (open circle) and presence (solid circle) of ligands (at rdd = 1). Buffer conditions: 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. HIV TAR RNA = 1 μM/strand. Neomycin dimer = 1 μM, neomycin = 1 μM
UV thermal denaturation of HIV-1 TAR RNA in the absence and presence of neomycin monomer and dimers was performed. A few trends can be identified: (1) There is very little change observed in the Tm of HIV-1 TAR RNA in the presence of neomycin (~0.2 °C), which lies within experimental error. (2) Neomycin dimers enhance the thermal stabilization of HIV TAR RNA from ~3–10 °C; depending on the linker length and structure (Table 1), (3) The UV thermal denaturation profiles of HIV-1 TAR RNA in the presence of neomycin dimers show only one transition, suggestive of specific binding at a single site. (4) A clear trend is observed between the linker length of neomycin dimers and the thermal stability of HIV 1 TAR RNA, such that the dimers with shorter linker length show higher thermal stabilization of HIV-1 TAR RNA, as opposed to neomycin dimers with longer linkers (Table 1). (5) There is very little further increase (1–2°C) in the thermal stabilization of HIV-1 TAR RNA at higher stoichiometric ratio of all neomycin dimers (Dimer: RNA ratio=2:1), indicative of a non-specific electrostatic interaction at higher concentrations of neomycin dimers (data not shown).
CD spectroscopy studies to characterize the binding between neomycin dimer and HIV-1 TAR RNA
Previous studies have revealed that there is a substantial change in the structure of HIV TAR RNA induced by Tat protein and other ligands which can be monitored by CD spectroscopy.37,38 Therefore, CD studies were carried out to monitor the binding interaction between neomycin dimer and HIV-1 TAR RNA. As depicted in Figure 2, the CD spectrum of HIV 1 TAR RNA alone is representative of an A-form RNA. The CD spectrum of HIV TAR RNA contains a strong positive band near 265 nm, a fairly intense negative band around 215 nm, and a weak negative band around 240 nm.39 A CD titration was performed between neomycin dimer and HIV TAR RNA by continuous addition of neomycin dimer in HIV TAR RNA solution (Figure 2A). The shape of both the CD spectra, in the absence and presence of neomycin dimer are similar suggesting that the overall conformation of HIV TAR RNA is conserved.
Figure 2.
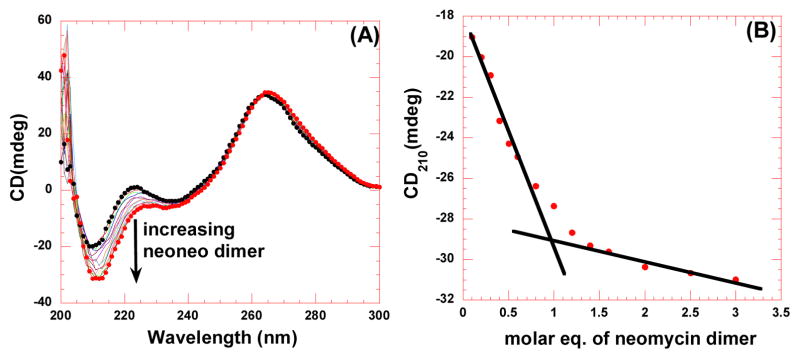
CD titration between neomycin dimer and HIV TAR RNA. (A) CD titration of HIV TAR RNA with increasing concentration of neomycin dimer DPA 51. The figure represents molar ellipticity for CD titration of HIV TAR RNA with neomycin dimer. The continuous changes in the CD spectra correspond to the incremental amount of neomycin dimer ranging from an rdd of 0 to 3. (B) A plot of normalized molar ellipticity versus rdd for CD titration of HIV TAR RNA with neomycin dimer. The continuous lines in the plot reflect the linear least squares fit of each apparent linear domain of the experimental data (filled circles) before and after the apparent inflection point. Molar ellipticity is per molecule of HIV TAR RNA, and rdd = ratio of the HIV TAR RNA/drug.
There are, however, some changes in the spectrum of HIV TAR RNA upon addition of neomycin dimer. The alteration of the CD spectrum of HIV TAR RNA is indicative of the interaction between HIV TAR RNA and neomycin dimer. The binding of neomycin dimer causes a gradual decrease in the peak at 210 nm and 225 nm upon incremental addition of neomycin dimer in the HIV TAR RNA solution. Contrary to Tat protein,39 there is a slight increase in the intensity at 265 nm along with a shift to longer wavelengths in the presence of neomycin dimer. The change in the molar ellipticity of HIV TAR RNA at 265 nm is typical of aminoglycosides-TAR binding.35 The peak at 265 nm has been attributed to the sensitivity of base stacking into the bulge region.35 This observation gives us an insight into the probable binding site of neomycin dimer to HIV TAR RNA. Tat protein binding to the bulge region of the HIV TAR RNA results in the modification of RNA base stacking and hence causes a significant decrease in the peak intensity around 265 nm. Neomycin dimer does not induce a significant change in the peak at 265 nm, suggesting that it does not bind in the same region as Tat. The CD data is consistent with the model of approximately one neomycin binding to the lower stem region of HIV TAR RNA (for structure see Scheme 1), and the second neomycin unit binding to upper stem or the hairpin region, though further studies need to be performed to confirm this hypothesis.
Ethidium bromide displacement titrations
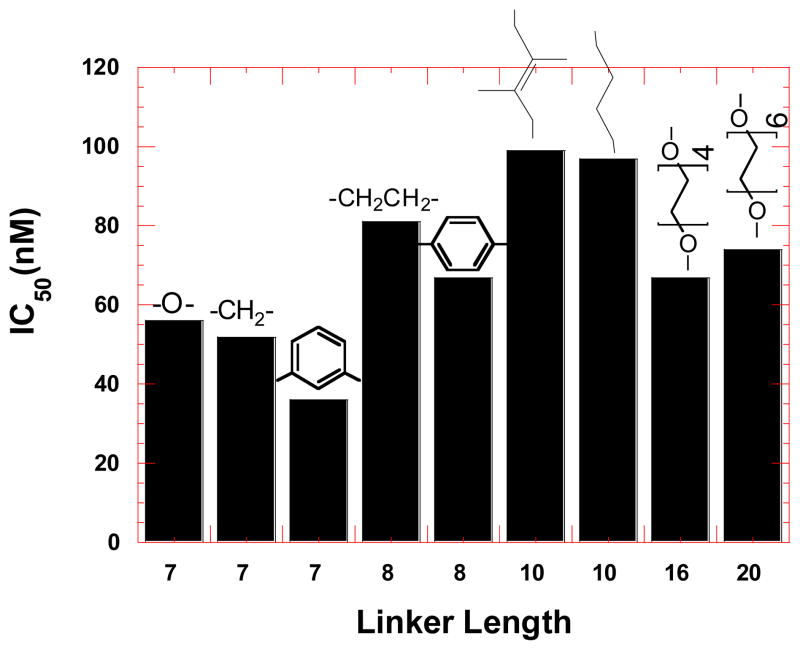
To further investigate the binding between HIV TAR RNA and neomycin dimers, ethidium bromide displacement experiments40,41 were used (Figure 3) and the results are summarized in Figure 4. The binding affinity of neomycin dimers is proportional to the amount of ethidium bromide displaced from HIV TAR RNA and reflected in the IC50 (the concentration of ligand required to displace 50 % ethidium bromide from HIV TAR RNA) values (Figure 3).
Figure 3.
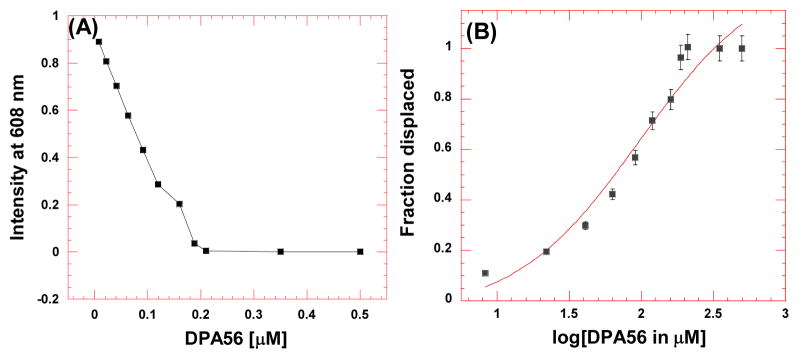
Representative example of ethidium bromide displacement assays. (A) Decrease in the fluorescence intensity of ethidium bromide-RNA complex with increase in the concentration of neomycin dimer. (B) The plot for fraction of ethidium bromide displaced from HIV TAR RNA versus the log of concentration of neomycin dimer. The data, shown with a sigmoidal fit, was used to determine the IC50 value.
Figure 4.
Bar graph representing the effect of linker length and structure of neomycin dimers on the binding affinity towards HIV-1 TAR RNA, determined using ethidium bromide displacement experiments.
Ethidium bromide displacement experiments indicate that in general, neomycin dimers with shorter linker lengths have higher affinity than neomycin dimers with longer linker towards HIV TAR RNA. The IC50 values for neomycin dimers fall within a range of 36–99 nM (see Figure 4), which is in the range of Tat-TAR binding (IC50=56 nM). In comparison to neomycin dimers, neomycin shows very low affinity towards HIV TAR RNA. The IC50 value of neomycin towards HIV TAR RNA is 417±115 nM. This trend is similar to what is observed with UV thermal denaturation experiments.
In conclusion, an efficient synthesis of neomycin dimers42 using click chemistry has been performed. Biophysical results from ethidium bromide displacement and UV thermal denaturation experiments show that the two neomycin binding sites on HIV TAR RNA are very close to each other as seen by the higher affinity of neomycin dimers with shorter linker length towards HIV TAR RNA. Further studies on the effect of these dimers on protein binding and HIV inhibition are ongoing and will be reported in due course.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health Grant (R15CA125724 to D.P.A.)
Footnotes
General synthesis procedures and characterization of one of the ligand is presented.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Draper DE. Annu Rev Biochem. 1995;64:593. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 2.Jones KA, Peterlin BM. Annu Rev Biochem. 1994;63:717. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout B, Jeang KT. J Virol. 1989;63:5501. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rana TM, Jeang K. Arch Biochem Biophys. 1999;365:175. doi: 10.1006/abbi.1999.1206. [DOI] [PubMed] [Google Scholar]
- 5.Muesing MA, Smith DH, Capon DJ. Cell. 1987;48:691. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 6.Rosen CA, Sodroski JG, Haseltine WA. Cell. 1985;41:813. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 7.Aboul-ela F, Karn J, Varani G. J Mol Biol. 1995;253:313. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 8.Feng S, Holland EC. Nature. 1988;334:165. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 9.Hauber J, Cullen BR. J Virol. 1988;62:673. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puglisi J, Tan R, Calnan B, Frankel A, Williamson Science. 1992;257:76. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 11.Thomas JR, Hergenrother PJ. Chem Rev. 2008;108:1171. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 12.Bailly C, Colson P, Houssier C, Hamy F. Nucleic Acids Res. 1996;24:1460. doi: 10.1093/nar/24.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratmeyer LS, Vinayak R, Zon G, Wilson WD. J Med Chem. 1992;35:966. doi: 10.1021/jm00083a024. [DOI] [PubMed] [Google Scholar]
- 14.Dassonneville L, Hamy F, Colson P, Houssier C, Bailly C. Nucleic Acids Res. 1997;25:4487. doi: 10.1093/nar/25.22.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind KE, Du Z, Fujinaga K, Peterlin BM, James TL. Chem Biol. 2002;9:185. doi: 10.1016/s1074-5521(02)00106-0. [DOI] [PubMed] [Google Scholar]
- 16.Tao J, Frankel AD. Proc Natl Acad Sci USA. 1992;89:2723. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S, Tamilarasu N, Ryan K, Huq I, Richter S, Still WC, et al. Proc Natl Acad Sci USA. 1999;96:12997. doi: 10.1073/pnas.96.23.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Biochemistry. 2007;46:741. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 19.Mei H, Mack DP, Galan AA, Halim NS, Heldsinger A, Loo JA, et al. Bioorg Med Chem. 1997;5:1173. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 20.Tor Y. Biochimie. 2006;88:1045. doi: 10.1016/j.biochi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Michael K, Tor Y. Chem Eur J. 1998;4:2091. [Google Scholar]
- 22.Wang H, Tor Y. Bioorg Med Chem Lett. 1997;7:1951. doi: 10.1016/s0960-894x(98)00663-5. [DOI] [PubMed] [Google Scholar]
- 23.Michael K, Wang H, Tor Y. Bioorg Med Chem. 1999;7:1361. doi: 10.1016/s0968-0896(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 24.Tok JB, Fenker J. Bioorg Med Chem Lett. 2001;11:2987. doi: 10.1016/s0960-894x(01)00602-3. [DOI] [PubMed] [Google Scholar]
- 25.Arya DP, Coffee RL, Jr, Xue L. Bioorg Med Chem Lett. 2004;14:4643. doi: 10.1016/j.bmcl.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Tok JB, Cho J, Rando RR. Biochemistry. 1999;38:199. doi: 10.1021/bi9819428. [DOI] [PubMed] [Google Scholar]
- 27.Mei H, Galan AA, Halim NS, Mack DP, Moreland DW, Sanders KB, et al. Bioorg Med Chem Lett. 1995;5:2755. [Google Scholar]
- 28.Zapp ML, Stern S, Green MR. Cell. 1993;74:969. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
- 29.Bernacchi S, Freisz S, Maechling C, Spiess B, Marquet R, Dumas P, et al. Nucleic Acids Res. 2007;35:7128. doi: 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Ahsen U, Davies J, Schroeder R. J Mol Biol. 1992;226:935. doi: 10.1016/0022-2836(92)91043-o. [DOI] [PubMed] [Google Scholar]
- 31.Mikkelsen NE, Brannvall M, Virtanen A, Kirsebom LA. Proc Natl Acad Sci USA. 1999;96:6155. doi: 10.1073/pnas.96.11.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Earnshaw DJ, Gait MJ. Nucleic Acids Res. 1998;26:5551. doi: 10.1093/nar/26.24.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermann T, Westhof E. Biopolymers. 1998;48:155. doi: 10.1002/(SICI)1097-0282(1998)48:2<155::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 34.Sannes-Lowery KA, Mei H, Loo JA. Int J Mass spectrom. 1999;193:115. [Google Scholar]
- 35.Wang S, Huber PW, Cui M, Czarnik AW, Mei H. Biochemistry. 1998;37:5549. doi: 10.1021/bi972808a. [DOI] [PubMed] [Google Scholar]
- 36.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Long KS, Crothers DM. Biochemistry. 1995;34:8885. doi: 10.1021/bi00027a041. [DOI] [PubMed] [Google Scholar]
- 38.Loret EP, Georgel P, Johnson WC, Ho PS. Proc Natl Acad Sci USA. 1992;89:9734. doi: 10.1073/pnas.89.20.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frankel AD. Curr Opin Genet Dev. 1992;2:293. doi: 10.1016/s0959-437x(05)80287-4. [DOI] [PubMed] [Google Scholar]
- 40.Luedtke NW, Hwang JS, Glazer EC, Gut D, Kol M, Tor Y. ChemBioChem. 2002;3:766. doi: 10.1002/1439-7633(20020802)3:8<766::AID-CBIC766>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 41.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J Am Chem Soc. 2001;123:5878. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 42.General procedure for synthesis of N-Boc protected neomycin dimers. To a solution of 5″-azide-neomycin B (62 mg, 0.05 mmol) in dry toluene (5 mL), diyne (0.025 mmol, 0.50 eq) was added followed by the addition of CuI (4.76 mg, 0.025 mmol) and DIPEA (6.46 mg, 0.05 mmol). The reaction mixture stirred at r.t. for 18 h in the atmosphere of argon. The volatiles were removed under vacuum. Purification by flash column chromatography (0 to 10 % ethanol in CH2Cl2) afforded the desired product as a white solid (80–89%).Deprotection of N-Boc protected neomycin dimers. To a solution of neomycin dimer (30 mg) in dioxane (3 mL), 4 M HCl/dioxane (1 mL) was added and the reaction started at room temperature. A white precipitate formed after 15 min. The reaction mixture was centrifuged and the solid was collected. The solid was washed with a solution of diethyl ether/hexane (3 × 5 mL each, v/v). The solid was then dissolved in water and lyophilized to afford a white solid (90–95 %). For characterization of one of the dimers, please see supporting information. Complete characterization of all the dimers will be reported elsewhere.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.