Abstract
Optineurin is a gene linked to glaucoma, amyotrophic lateral sclerosis, other neurodegenerative diseases, and Paget’s disease of bone. This review describes the characteristics of optineurin and summarizes the cellular and molecular biology investigations conducted so far on optineurin. Data from a number of laboratories indicate that optineurin is a cytosolic protein containing 577 amino acid residues. Interacting with proteins such as myosin VI, Rab8, huntingtin, transferrin receptor, and TANK-binding kinase 1, optineurin is involved in basic cellular functions including protein trafficking, maintenance of the Golgi apparatus, as well as NF-κB pathway, antiviral, and antibacteria signaling. Mutation or alteration of homeostasis of optineurin (such as overexpression or knockdown) results in adverse consequences in the cells, leading to the development of neurodegenerative diseases including glaucoma.
Keywords: Optineurin, Glaucoma, Amyotrophic lateral sclerosis, Mutations, Molecular structure, Localization, Functional consequence
1. Introduction
Optineurin, a 67-kDa protein, has attracted much attention in the scientific world in recent years. It was first isolated in 1998 by Li et al. in yeast two-hybrid screening using an adenovirus protein E3-14.7K (group C early transcription region 3 14.7-kDa protein) as a bait and was initially named as FIP-2 (14.7K-interacting protein 2). Its expression can be induced by tumor necrosis factor-α (TNF-α) in human 293 cells and adenocarcinoma MCF-7 cells. E3-14.7K protein has been shown to inhibit TNF-α-induced apoptosis and FIP-2 can reverse the E3-14.7K protective effect in human 293 cells (Li et al., 1998). Since optineurin has a strong homology to NF-κB essential molecule (NEMO), it was also named as NRP (NEMO-related protein) by Schwamborn et al. (2000).
The interest toward optineurin was appreciably elevated following the findings by Rezaie et al. (2002) that optineurin gene is associated with normal tension glaucoma (NTG), a subtype of primary open-angle glaucoma (POAG), one of the leading causes of irreversible bilateral blindness worldwide. It was named then as optineurin which stands for “optic neuropathy inducing” protein. More recently, mutations in optineurin are also found to be associated with amyotrophic lateral sclerosis (ALS; Maruyama et al., 2010). Optineurin is further noted to be localized in pathological structures in ALS, neurofibrillary tangles and dystrophic neuritis in Alzheimer’s disease, Lewy bodies and Lewy neuritis in Parkinson’s disease, ballooned neurons in Creutzfeldt–Jakob disease, glial cytoplasmic inclusions in multiple system atropy, and Pick bodies in Pick disease (Osawa et al., 2011). In addition, optineurin is identified as one of the genetic risk factor for Paget’s disease of bone (PDB) (Albagha et al., 2010; Chung et al., 2010).
This review describes the characteristics of optineurin protein and summarizes the cellular and molecular biology work performed on optineurin in recent years. Data from a number of laboratories indicate that optineurin is involved in basic cellular functions such as protein trafficking, maintenance of the Golgi apparatus, as well as NF-κB pathway and antiviral signaling. Mutation or alteration of homeostasis of optineurin (such as overexpression or knockdown) results in adverse consequences in the cells, leading to the development of neurodegenerative diseases including glaucoma.
2. Molecular Structure and Protein Characteristics
2.1. Genomic DNA
The human optineurin gene is located at chromosome 10:13142082-13180276 (gene ID: 10133) and spans about 37kb genomic region. Four different transcripts (NM_001008211.1, NM_001008212.1, NM_001008213.1, and NM_001008214.1) have been reported, each containing a different 5′-untranslated region (UTR), but all have the same open reading frame.
The detailed optineurin promoter structure is unknown. Sudhakar et al. (2009) cloned about 1kb of DNA fragment upstream of the human optineurin cDNA sequence. This putative promoter includes 221bp of exon 1. Analysis of this promoter showed several putative Sp1 sites and one NF-κB site located immediately upstream of the transcription start site. No TATA box or initiator element is present in this promoter but putative binding sites for heat shock factors, HSF1 and HSF2, MyoD, neuron-restrictive silencing factor (NRSF, also known as REST), and cyclic AMP response element-binding protein (CREB) were identified (Sudhakar et al., 2009). This putative promoter was activated upon treatment of HeLa and A549 cells with TNF-α. A smaller promoter from −136 to +221 was in addition made which contained the putative Sp1 and NF-κB sites. Similar to the 1kb full-length promoter, the smaller 0.36kb fragment can also be activated by TNF-α in A549 and HeLa cells. Interestingly, the smaller fragment showed more basal activity than the full-length 1kb promoter, suggesting that a negative regulatory element is present in sequences upstream of the minimal promoter.
2.2. mRNA and gene expression during embryogenesis
The human optineurin gene contains 3 noncoding exons in the 5′-UTR region and 13 exons that code for a 577-amino acid protein. Alternative splicing at the 5′-UTR generates at least four different isoforms, but all have the same open reading frame (Rezaie and Sarfarazi, 2005). The synonyms that have been used to describe this gene include: FIP-2, GLC1E, NRP, HIP7, HYPL, and TFIIIA-INTP.
Optineurin transcripts are ubiquitously present in various tissues in the adult mouse. It is expressed during early stages of eye development in mouse. At both 10.5 and 13.5 days postconception, a strong specific expression was detected by in situ hybridization in the optic vesicle (De Marco et al., 2006; Rezaie et al., 2007). During embryogenesis, optineurin may play a pivotal role in the overall development of the eye.
2.3. Protein
2.3.1. Domains
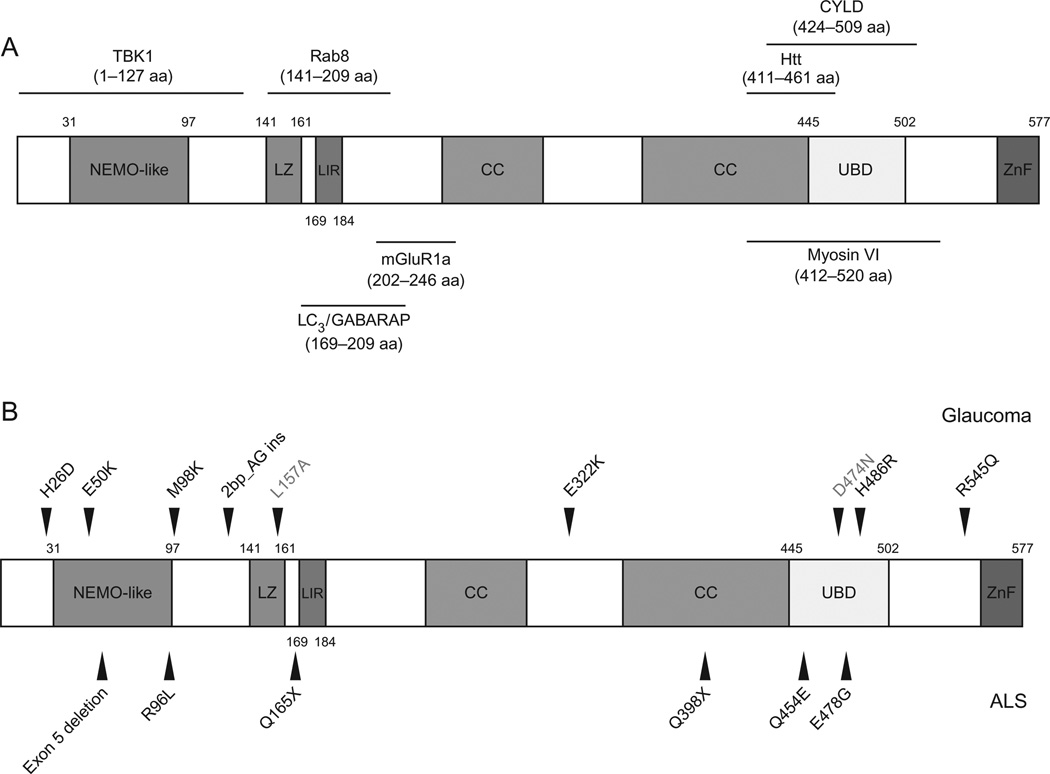
Figure 5.1 shows sequence alignment of optineurin protein from human, chimpanzee, cow, dog, rat, mouse, chicken, and zebrafish. Figure 5.2 presents phylogenetic tree to depict the distance between different species. A high amino acid homology exists in optineurin from different species (Rezaie and Sarfarazi, 2005; Rezaie et al., 2005). This protein contains several putative domains including a NEMO-like domain, at least one leucine zipper, multiple coiled-coil motifs, an ubiquitin-binding domain (UBD), a microtubule associated protein 1 light chain 3 (LC3)-interacting motif (LIR) (Wild et al., 2011), and a carboxyl (C)-terminal C2H2 type of zinc finger (Hattula and Peranen, 2000; Li et al., 1998; Schwamborn et al., 2000; Stroissnigg et al., 2002; Fig. 5.3A).
Figure 5.1.
Sequence alignment of optineurin protein from human, chimpanzee, cow, dog, rat, mouse, chicken, and zebrafish. Multiple sequence alignment is generated by ClustalW2 (http://www.ebi.ac.uk/msa/clustaw2/).
Figure 5.2.
Phylogenetic tree to show the distance of optineurin protein between different species. Phylogenetic tree is generated by ClustalW2 (http://www.ebi.ac.uk/msa/clustaw2/).
Figure 5.3.
(A) Schematic representation of human optineurin protein domains and the binding sites of optineurin-interacting proteins. CC, coiled-coil; LZ, leucine zipper domain; LIR, LC3 interacting motif; UBD, ubiquitin-binding domain; ZnF, zinc finger; aa, amino acid. (B) Optineurin mutations associated with glaucoma and ALS. Note that L157A is not disease associated but rather is a mutation used in the study to affect the leucine zipper domain. D474N also is not a disease associated mutation. It was used in studies to abolish ubiquitin binding. (For color version of this figure, the reader is referred to the Web version of this chapter.)
Optineurin was reported in earlier studies (Rezaie et al., 2002; Sarfarazi and Rezaie, 2003) to be present in the aqueous humor and culture medium and was suspected to be a secreted protein. However, Kroeber et al. (2006) did not detect any optineurin in the aqueous humor in their transgenic mice overexpressing wild-type optineurin under the control of lens specific βB1-crystalline promoter (βB1-crystallin-OPTN mice). Subsequent secretion assays also showed that optineurin is not secreted in human trabecular meshwork (TM) and retinal pigment epithelial (RPE) cultures, even under conditions in which secretion of proteins such as myocilin is promoted or when optineurin is overexpressed (Park et al., 2006). Optineurin is thus concluded not to be a secreted protein. The difference in results could be related to specificity of the antibody used.
2.3.2. Posttranslational modification
Optineurin is neither N- nor O-glycosylated (Ying et al., 2010). When subjected to membrane protein extraction, optineurin in total lysates of RGC5 cells (a transformed cell line used as a model for retinal ganglion cells or RGCs) distributed exclusively in the hydrophilic fraction. Optineurin is therefore not a membrane protein but rather a cytosolic protein, consistent with the prediction that the protein sequence contains no obvious transmembrane domains (Ying et al., 2010). Optineurin possesses an UBD, is ubiquitinated, and is processed through the ubiquitin–proteasome pathway (Shen et al., 2011; Ying et al., 2010).
The endogenous optineurin is phosphorylated (Ying et al., 2010). Using stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry, the endogenous optineurin was found to be phosphorylated at Ser177 which is adjacent to the optineurin LC3 interacting LIR site (Wild et al., 2011). Multiple phosphorylated LIR peptides with up to three phosphorylated groups were also identified in the presence of overexpressed TANK-binding kinase 1 (TBK1) in mouse embryo fibroblasts (Wild et al., 2011).
2.3.3. Oligomerization
By native blue gel electrophoresis, optineurin is shown to be capable of forming 420kDa homo-oligomers, which, based on the 67kDa monomer size, is estimated to be hexamers (Ying et al., 2010). In addition, optineurin interacts and associates with Rab8, myosin VI, and transferrin receptor (Chibalina et al., 2008; Ying et al., 2010), either in singly or in combination, to form supermolecular complexes with sizes larger than 400kDa.
2.3.4. Cellular localization
Optineurin is ubiquitously expressed in many tissues including the heart, brain, placenta, skeletal muscle, kidney, pancreas, adrenal cortex, liver, and the eye (Li et al., 1998; Rezaie and Sarfarazi, 2005; Rezaie et al., 2005). In the eye, optineurin is expressed in the TM, nonpigmented ciliary epithelium, and remarkably in retina (Rezaie and Sarfarazi, 2005). A study (Kroeber et al., 2006) using transgenic mice indicated that the retina staining is confined to neurons and that RGC is labeled with a high intensity. The strong RGC labeling was confirmed in another investigation (De Marco et al., 2006).
The endogenous optineurin generally is localized in the cytoplasm with a diffuse distribution pattern. Of note is that the immunostaining patterns may depend on the antibodies and washing procedures used. There are two commercially available antibodies, anti-C-terminal- and anti-INT-optineurin. The former is reactive to human, mouse, and rat optineurin, while the latter is reactive only to the human protein. In both RGC5 and human APRE-19 RPE cells, a diffuse, cytoplasmic distribution pattern of optineurin resulted when they were washed in glycine-containing solution and stained with anti-C-terminal-optineurin antibody. When glycine was omitted in the rinse, the cytoplasmic staining was low and optineurin appeared to locate largely in the perinuclear area overlapping with the Golgi marker GM130. Anti-INT-human optineurin antibody, presumably reactive against epitope(s) different from those of anti-C-terminal antibody, yielded the diffuse, cytosolic pattern regardless whether glycine was used for the rinse in human RPE cells. It was concluded that while optineurin has a diffuse, cytoplasmic distribution pattern, a population of the protein is associated with the Golgi apparatus. As optineurin is shown not to be a membrane protein, the optineurin–Golgi association is probably indirect via interactions of other Golgi-associated proteins such as huntingtin (Htt) and Rab8 (Ying et al., 2010).
When overexpressed after transfection, forced expressed exogenous optineurin accumulates, appearing as dots or granular structures, termed foci, around the Golgi complex. The foci, seemingly residing in close proximity, essentially do not colocalize with the Golgi complex (Park et al., 2006; Ying et al., 2010).
2.3.5. Cellular processing
By pulse chase experiments, the endogenous optineurin was found to be a relatively short-lived protein with a half life of about 8h (Ying et al., 2010) in RGC5 cells. The endogenous optineurin is ubiquitinated and its level in neuronal RGC5 and PC12 cells is increased by treatment of proteasomal inhibitors but not by autophagic and lysosomal inhibitors, indicating that the ubiquitin–proteasome system is the major pathway for endogenous optineurin processing. Autophagy and lysosomes have a rather minor role (Shen et al., 2011).
3. Optineurin-Binding Partners
Optineurin has been shown to interact with a number of proteins (Fig. 5.3A) including Rab8, huntingtin, myosin VI (Hattula and Peranen, 2000; Sahlender et al., 2005), transferrin receptor (Park et al., 2010), metabotropic glutamate receptor (Anborgh et al., 2005), transcription factor IIIA (TFIIIA) (Moreland et al., 2000), serine/threonine kinase receptor-interacting protein 1 (RIP1) (Zhu et al., 2007), and TBK1 (Morton et al., 2008) implicating roles of optineurin in multiple cellular functions.
3.1. Rab8
Rab proteins belong to a large family of small guanosine triphosphatases (GTPases) that participate and regulate various membrane transport pathways. Each Rab protein has a distinct location corresponding to the pathway it regulates (Stenmark and Olkkonen, 2001; Zerial and McBride, 2001). Rab8, a binding partner of optineurin, has been shown to regulate polarized membrane trafficking pathways from the Golgi complex to the cell surface (Huber et al., 1993a,b; Moritz et al., 2001) and promote changes in cell shape by reorganizing actin and microtubules (Huber et al., 1993b; Peranen et al., 1996). The constitutively active GTP-bound mutant form of Rab8-Q67L, but not the dominant-negative GDP-bound mutant form of Rab8-T22N, interacts with the amino (N)-terminal region (amino acids 141–209) of optineurin (Hattula and Peranen, 2000). This region of optineurin contains the leucine zipper domain. Optineurin and Rab8 are suggested to form a complex that participates in regulation of the post-Golgi transport of proteins, the sorting of which is mediated by the clathrin adaptor complex 1 (Au et al., 2007; Sahlender et al., 2005).
3.2. Huntingtin
Htt is a huge cytosolic protein (3144 amino acids) associated with Huntington’s disease. The abnormal polyglutamine expansion in the N-terminal region of Htt produces significant dysfunction and neural death, especially in the medium spiny neurons of the striatum (Kremer et al., 1994). Subcellular fractionation and microscopic studies have shown that Htt is associated with vesicles and microtubules (DiFiglia et al., 1995; Hoffner et al., 2002) by interacting with HAP1, Htt-associated protein 1, which is reported to form a complex with the dynactin subunit p150glued and modulate or regulate the dynein–dynactin complex. There is evidence that Htt participates in post-Golgi trafficking of proteins that follow the regulated secretory pathway (del Toro et al., 2006). Deletion experiments by Hattula and Peranen (2000) showed that Htt directly binds to a C-terminal (amino acids 411–461) region of optineurin. Optineurin colocalizes with Htt in the Golgi apparatus, linking Htt to Rab8 (Sahlender et al., 2005). Coex-pression of optineurin and Htt enhanced the recruitment of Htt to Rab8-positive vesicular structures. Post-Golgi trafficking to lysosomes is impaired in cells expressing mutant Htt by delocalizing optineurin/Rab8 complex from the Golgi apparatus (del Toro et al., 2009).
3.3. Myosin VI
Myosin VI is a multifunctional actin-based motor protein found in a number of intracellular compartments including endocytic vesicles (Aschenbrenner et al., 2003; Buss et al., 2001), membrane ruffles (Buss et al., 1998), the Golgi complex, and secretory vesicles (Buss et al., 1998). It plays a role in the basolateral delivery of membrane proteins (Au et al., 2007; Buss et al., 2001, 2004; Spudich et al., 2007; Warner et al., 2003). Using the tail of myosin VI as a bait in a yeast two-hybrid screening of human umbilical vein epithelial cell cDNA library, a 375-amino acid C-terminal fragment of optineurin was identified as a myosin VI-binding partner (Sahlender et al., 2005). Further deletion studies indicated that the myosin VI-binding site is between amino acids 412 and 520 which interacts with the Arg-Arg-Leu (RRL) sequence in the tail domain of myosin VI. Knockdown optineurin by siRNA caused a marked reduction in the amount of myosin VI associated with the Golgi complex, fragmentation of the Golgi, and a reduction in exocytosis of vesicular stomatitis virus G-protein to the plasma membrane. Optineurin was thus suggested to mediate the targeting of myosin VI to the Golgi complex and play a role in organization of the Golgi apparatus and exocytosis (Sahlender et al., 2005). Subsequent studies have established that both myosin VI and optineurin are required in the final stages of the secretory pathway for the fusion of secretory vesicles with the plasma membrane (Bond et al., 2011).
3.4. Transferrin receptor
Immunoprecipitation experiment established that transferrin receptor interacts with optineurin (Park et al., 2010). The binding domain is yet to be specified. It has been shown that sequestration of transferrin receptor upon overexpression of optineurin may be a factor that leads to impairment of the transferrin uptake in cells (Park et al., 2010).
3.5. Metabotropic glutamate receptors 1 and 5 (mGluR1 and mGluR5)
mGluRs that are linked to the activation of phospholipase C (PLC), increase in intracellular inositol 1,4,5-triphosphate (IP3) formation, and the release of Ca2+ stores inside the cells play an important role in regulating neuronal function (Conn and Pin, 1997). They belong to a subfamily of G-protein-coupled receptors and are divided into three groups based on the homology of their amino acid sequences. mGluR1 and mGluR5 are group I members and each has several splice variants [mGluR1 has four: a (or α), b (or β), c, and d. mGluR5 has two: a and b]. Optineurin has been shown to be a group I mGluR-interacting protein (both mGluR1 and mGluR5) (Anborgh et al., 2005) that functions to inhibit mGluR-G-protein coupling to PLC and IP3 signaling (Anborgh et al., 2005). Htt coprecipitates with mGluR1 (probably through optineurin) and when wild type (Q15) and mutant (Q135) Htt were coexpressed with optineurin, only the latter resulted in augmented optineurin binding to mGluR1a and increased optineurin-mediated attenuation of mGluR1a signaling (Anborgh et al., 2005).
3.6. Transcription factor IIIA (TFIIIA)
TFIIIA activates 5S ribosomal RNA gene transcription in eukaryotes. It is a single protein that contains zinc and possesses repetitive C2H2 zinc finger domains (Clemens et al., 1993; Theunissen et al., 1992). Optineurin protein was identified in the yeast two-hybrid system using the C-terminal portion of Xenopus TFIIIA as a bait (Moreland et al., 2000). The interaction was verified by co-immunoprecipitation in vitro and by cochromatography (Moreland et al., 2000). The TFIIIA-interacting region is likely within the central leucine-rich domain on optineurin. The complex of the two proteins (as evidenced by cochromatography) is stable at high salt inferring a hydrophobic nature for the TFIIIA–optineurin interaction (Moreland et al., 2000). The possible role optineurin in the 5S ribosomal RNA gene transcription remains to be determined.
3.7. Serine/threonine kinase receptor-interacting protein 1 (RIP1)
NF-κB is sequestered in the cytoplasm in a complex with inhibitory IκB protein. Upon simulation by cytokines such as TNF-α, signaling intermediates TRADD (TNF receptor-associated death domain), TRAF2 (TNF receptor-associated factor 2), and RIP1 are recruited to the TNF receptor (TNFR), resulting in the assembly of a signaling complex. In this complex, RIP1 is rapidly ubiquitinated with Lys(K)63-linked polyubiquitin (polyUb) chains and binds with the UBD domain of the regulatory subunit of IκB kinase (IKK) complex, IKKγ (also known as NEMO). This leads to activation of two catalytic subunits, IKKα and IKKβ, of IKK to trigger the phosphorylation, ubiquitination, and degradation of IκB NF-κB is then translocated into nucleus to induce transcription of target genes (Ea et al., 2006; Wu et al., 2006). NEMO is thus an essential component for NF-κB signaling. Optineurin has a high homology with NEMO (53%). It has a K63-linked polyUb-binding domain similar to that of NEMO. A point mutation Asp474 →Asn (D474N) (D311N in NEMO) in the UBD domain of optineurin abolishes its binding to K63-linked polyUb. Optineurin has been shown to compete with NEMO to bind with ubiquitinated RIP which is necessary for the efficient activation of NF-κB induced by TNF-α (Zhu et al., 2007). Overexpression of optineurin competitively inhibited TNF-α-induced activation of NF-κB. On the other hand, microRNA silencing of optineurin resulted in markedly enhanced TNF-α-induced NF-κB activation. This indicates that optineurin is a negative regulator of TNF-α-induced NF-κB activation (Zhu et al., 2007) via binding with polyUb RIP.
3.8. TAX1, TAX2, and TAX1BP1
TAX1 is one of the nonstructural genes human T-cell leukemia virus type 1 (HTLV-1) encoded. TAX2 is the equivalent of TAX1 in HTLV-2 and TAX1 and TAX2 have many shared activities. Both coprecipitate with optineurin and TAX1 has the same subcellular distribution as optineurin (Journo et al., 2009). The UBD domain of optineurin is required for its binding to TAX1. A point mutation D474N in the UBD domain of optineurin severely reduces the interaction of optineurin with TAX1 (Journo et al., 2009).
Ubiquitination of TAX1 is required for its interaction with optineurin. Binding with optineurin can stabilize TAX1 polyubiquitination and ubiquitination-defective TAX1 mutants exhibit impaired binding to optineurin. TAX1 has been known to bind NEMO, triggering the activation of IKKα and IKKβ (Chu et al., 1999; Harhaj and Sun, 1999;Jin et al., 1999) and in turn activation of NF-κB. Since optineurin is NEMO-like, it is not surprising that optineurin is also capable of potentiating the activation of NF-κB by TAX1.
Tax1-binding protein 1 (TAX1BP1) is a binding partner of TAX1 reported to interact with A20, Itch and ring finger protein 11 (RNF11) to form a functional ubiquitin-editing complex that regulates the ubiquitination of RIP1 and TRAF6 (Shembade et al., 2007). Interestingly, TAX1BP1 itself also interacts with optineurin. When TAX1BP1 is present, the interaction between optineurin and TAX1 is strongly induced and the interaction between optineurin and TAX1BP1 is also increased by coexpression of TAX1, suggesting that these three proteins interact with each other to form a ternary complex (Journo et al., 2009). RNF11 interacts with optineurin (Azmi and Seth, 2005) likewise. It is unclear though whether RNF11 is also present in the TAX1BP1/TAX1/optineurin complex and whether it regulates the TAX1 ubiquitination.
3.9. CYLD
CYLD was originally identified as a tumor suppressor gene mutated in familial cylindromas (turban tumor syndrome). This gene encodes a cytoplasmic protein that functions as a deubiquitinating enzyme. It is a NEMO-interacting protein and thus a negative regulator of NF-κB activation (Kovalenko et al., 2003; Trompouki et al., 2003). It has been demonstrated that via its deubiquitinase activity, CYLD specifically catalyses cleavage of K63-linked polyUb chains from its target proteins such as RIP, NEMO, and TRAFs to prevent NF-κB activation (Brummelkamp et al., 2003; Nagabhushana et al., 2011). Using full-length optineurin as a bait, CYLD was identified as optineurin-interacting protein by yeast two-hybrid screening (Nagabhushana et al., 2011). Deletion analysis showed that the C-terminal region of optineurin that contains an UBD domain (424–509 amino acids) is involved in binding to CYLD (Fig. 5.3A). A point mutation His468 →Arg (H486R) in the UBD domain of optineurin abolishes its interaction with CYLD. Deletions of various domains of CYLD revealed that amino acids 460–592 of CYLD which encompass a cytoskeleton associated protein glycine-rich (CAP) domain are sufficient for interaction with optineurin (Nagabhushana et al., 2011). Mediating the interaction between CYLD and polyUb RIP, optineurin may act as an adaptor protein bringing CYLD and the CYLD substrate RIP together to facilitate deubiquitination of ubiquitinated RIP by CYLD (Nagabhushana et al., 2011).
3.10. TANK (TRAF-associated NF-κB activator) binding kinase 1 (TBK1)
TBK1, TANK-binding kinase 1 or TNF-α-activated protein kinase 1, is a member of the IKK subfamily of protein kinase. TBK1 becomes activated in response to lipopolysaccharide or viral double-stranded DNA. It phos-phorylates the transcription factor interferon regulatory factor 3 (IRF3), induces its translocation to the nucleus, and stimulates transcription of the genes coding type-1 interferons (Hemmi et al., 2004; Matsui et al., 2006; McWhirter et al., 2004). TBK1 can also be recruited to the TNF receptor complex in response to TNF-α (Kuai et al., 2004) and activate NF-κB-directed transcription (Bonnard et al., 2000).
TBK1 was identified as an optineurin binding partner by yeast two-hybrid screens using optineurin as a bait (Morton et al., 2008). The TBK1 binding site is located between residues 1 and 127 of optineurin and the optineurin-binding domain is localized to C-terminal domain residues 601–729 of TBK1 (Morton et al., 2008). Protein sequence databases search disclosed that the amino acid sequence between residues 78 and 121 of optineurin is homologous to the TBK1-interaction domain present in three other binding partners for TBK1, TANK, NAP1 (IKK-related kinase- or NAK-associated protein 1), and SINTBAD (similar to NAP1, TBK1 adaptor). It is likely that optineurin, TANK, NAP1, and SINTBAD all bind to the same region of TBK1 at the common TBK1-binding domain.
Optineurin has been demonstrated to have a role in the inhibition of virus-triggered interferon-β induction (Mankouri et al., 2010). TBK1– optineurin interaction may be involved in the antiviral signaling pathways.
3.11. LC3/GABARAP
Microtubule associated protein 1 light chain 3 (LC3) is an autophagy protein (mammalian homolog of yeast Atg8) required in the formation of autophagosomes (Kabeya et al., 2000). Autophagy is initiated by the formation of a cup-shaped membrane (termed phagophore or isolation membrane) that has been recently noted to originate from the endoplasmic reticulum (Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009). The phagophore then enwraps parts of the cytoplasm to form a double-membrane vesicle (termed autophagosome), which eventually fuses with the lysosomes/vacuole to form autolysosomes (Weidberg et al., 2010). LC3 conjugates with phosphatidylethanolamine to form lipidated LC3 (LC3-II) before association with the membrane. It then localizes to the nascent and early autophagic vacuole membranes. After autophagosomes seal, the LC3 attached to the outer membrane is cleaved off and the LC3 on the inner membrane is trapped inside the sealed autophagosomes (Eskelinen, 2005). γ-Aminobutyric acid receptor-associated protein (GABARAP) is also an Atg8 homologue in the mammalian system. Similar to LC3, GABARAP subjects to a C-terminal modification process and is involved in autopha-gosomal membrane formation (Chakrama et al., 2010; Kabeya et al., 2004).
LC3/GABARAP was recently identified to be an optineurin binding partner (Wild et al., 2011). The specific interactions between optineurin and LC3/GABARAP proteins were verified by pull down assays in mammalian cells, directed yeast two-hybrid transformations, and in vitro pull down using purified proteins (Wild et al., 2011). Deletion experiment on optineurin pinpointed an LC3 interacting LIR motif which is located between the coiled-coil domains of optineurin encompassing amino acids 169–209. This LIR motif contains a linear tetrapeptide sequence which is also present in known autophagy receptors (such as NBR1, P62/SQSTM1, NIX/BNIP3L, and CALR) (Wild et al., 2011).
Optineurin is localized in LC3-positive vesicles upon induction of autophagy. A single point mutation of Phe178 →Ala (F178A) or Ile181 →Ala (I181A) abrogated the colocalization and the interaction between optineurin and LC3/GABARAP (Wild et al., 2011). A similar abrogation was also observed with ubiquitin binding-deficient optineurin variants. Optineurin is thus concluded to serve as an autophagy receptor that binds with LC3/GABARAP via a phenylalanine-containing LIR motif and ubiquitin via its UBD domain, allowing optineurin to be involved in selective autophagy of ubiquitin-coated cytosolic Salmonella enterica. TBK1 is shown to phosphorylate optineurin on Ser177, enhancing thereby the LC3 binding by optineurin and promoting autophagic clearance of Salmonella. Silencing optineurin or TBK1, or LC3- or ubiquitin-binding-deficient optineurin mutants impaired Salmonella autophagic clearance, resulting in increased bacterial proliferation (Wild et al., 2011).
3.12. Others
Other optineurin binding proteins may include: A20, ABIN1, TRAF3 (Mankouri et al., 2010), TBC1D17, UXT, ZBTB33, and BAT4. Further, investigations will be needed to elucidate the physiological and pathological significance of these interactions.
4. Optineurin Mutations Associated with Diseases (Fig. 5.3B)
4.1. Glaucoma
Glaucoma is a major cause of blindness characterized by degeneration of the optic nerve, RGC death (Kerrigan et al., 1997; Kuehn et al., 2005), and progressive axonal and visual field loss (Quigley, 2011). It is estimated that by the year of 2020, the number of affected population will rise to around 79.6 million (Quigley and Broman, 2006). It is a slow disease, usually bilateral but unequal in degree. The most common form of glaucoma, adult-onset POAG, is typically associated with an elevation of the intraocular pressure (IOP) and characteristic optic nerve degeneration. A subset of POAG patients, however, may manifest optic nerve damage even without an IOP increase, and this form of open-angle glaucoma is called normal tension glaucoma or NTG (Anderson, 2003; Hoyng and Kitazawa, 2002).
POAG is genetically heterogeneous caused by several susceptibility genes and perhaps also environmental factors (Allingham et al., 2009; Kwon et al., 2009; Wang et al., 2001; Wiggs, 2007). Currently, a total of 15 chromosomal loci, designated as GLC1A to GLC1O, have been linked to POAG. Four candidate genes identified so far include myocilin (GLC1A) (Stone et al., 1997), WD40-repeat36 (GLC1G) (Rao et al., 2011), optineurin (GLC1E) (Rezaie et al., 2002), and neurotrophin-4 (NTF-4) (GLC1O) (Rao et al., 2011).
Rezaie and coworkers in 2002 studied 54 families with autosomal dominantly inherited adult-onset POAG (the majority of these families presented with normal IOP) and identified the causative gene on chromosome 10p14 and designated it optineurin (OPTN). This gene was previously identified as FIP-2 (Li et al., 1998) and NRP (Schwamborn et al., 2000). DNA sequence analyses detected four mutations in patients with POAG: Glu50 →Lys (E50K), Met98 →Lys (M98K), Arg545 →Gln (R545Q), and 691_692insAG (2-bp “AG” insertion). These mutations were reported to be responsible for 16.7% of hereditary forms of NTG with an additional attributable risk factor of 13.6% in both familial and sporadic cases (Rezaie et al., 2002).
Subsequent studies indicated that optineurin sequence alterations do not have a strong correlation with high tension POAG (Alward et al., 2003; Ayala-Lugo et al., 2007; Wiggs et al., 2003) and that the frequency of coding and intronic polymorphisms within the optineurin gene in NTG populations varies with ethnic background (Baird et al., 2004; Caixeta-Umbelino et al., 2009; Hauser et al., 2006; Leung et al., 2003; Liu et al., 2008; Mukhopadhyay et al., 2005; Sarfarazi and Rezaie, 2003; Tang et al., 2003; Toda et al., 2004; Weisschuh et al., 2005; Yen et al., 2008). The 691_692insAG optineurin variant is rare (Ayala-Lugo et al., 2007). The R545Q variation is likely to be a non-disease-causing polymorphism. The M98K change may be associated with a fraction of NTG in patients of Japanese ethnicity (Alward et al., 2003). E50K is associated with NTG patients in Caucasian and Hispanic populations but the prevalence is found much lower than originally reported by Rezaie et al. (2002). Overall, the E50K frequency is reported to be between 0.1% and 13.5% (Ayala-Lugo et al., 2007) in POAG cases, afflicting an estimated 67,000 to 9 million patients. E50K mutation does, however, seem to be associated with a more progressive and severe disease (Aung et al., 2005; Hauser et al., 2006). In addition to the genetic link, a causal role of E50K in glaucoma is also implicated by a recent study of Chi et al. (2010) that loss of RGCs, reduction of retinal thickness, and excavation of the optic nerve head are observed in E50K-expressing normal tension transgenic mice.
Other optineurin gene alterations observed in various patient population include: a missense mutation Lys322 →Glu (E322K) in exon 10 of optineurin in Chinese POAG family (Xiao et al., 2009); His26 →Asp (H26D) alteration (Funayama et al., 2004; Fuse et al., 2004) in Japanese POAG families; Glu103 →Asp (E103D), Val148 →Val (V148V), and IVS13+21C→G in the sporadic Chinese patients with POAG (Leung et al., 2003), and H486R in POAG (Leung et al., 2003) and juvenile open-angle glaucoma (Willoughby et al., 2004) families.
4.2. Amyotrophic lateral sclerosis
ALS is a progressive disorder characterized by degeneration of motor neurons of the primary motor cortex, brainstem, and spinal cord (Leigh, 2007). It is a genetically heterogeneous disease. Genes that encode TAR DNA-binding protein of 43kDa (TDP-43) protein (Sreedharan et al., 2008), Cu/Zn superoxide dismutase (SOD-1) (Rosen et al., 1993), angio-genin (Greenway et al., 2006), vesicle-associated membrane protein (Nishimura et al., 2004), valosin-containing protein ( Johnson et al., 2010), and fused in sarcoma/translated in liposarcoma (FUS, also known as TLS) (Kwiatkowski et al., 2009; Vance et al., 2009) are reported to be responsible for some of the classic familial ALS.
Maruyama et al. (2010) reported three mutations in the gene encoding optineurin in Japanese familial or sporadic ALS patients: a homozygous deletion of exon 5, a homozygous nonsense Gln398 stop (Q398X, in exon 12), and a heterozygous missense Glu478 →Gly (E478G, in exon 14) mutation. In the gene with the deletion of exon 5, if there was a transcript, the transcript splicing from exon 4 to exon 6 would cause a frame shift and make a stop codon, which would be expected to translate a peptide 58 amino acids in length. Q398X mutation generates a premature stop codon at amino acid 398, truncating the 577 amino acid optineurin protein to one of 397 amino acids in length. This truncation results in a deletion of the coiled-coil 2 domain. E478G mutation is located in the UBD domain. This glutamic acid is highly conserved among optineurin proteins of a wide range of species and is situated within the DFxxER motif of the UBD domain shared among optineurin, NEMO, and A20 binding and inhibitor of NF-κB protein. The Q398X nonsense mutation and probably the exon 5 deletion mutation as well would cause a decrease in optineurin expression resulting from nonsense-mediated mRNA decay of the transcript carrying the nonsense mutations. The mechanism of recessive mutation causing ALS is expected to be loss of function. On the other hand, the E478G missense mutation increased the immunoreactivity for optineurin in the cell body and neurites. The increased amount and different distribution of the mutated protein might disturb neuronal functions, and accelerate the inclusion body formation in sporadic ALS.
A heterozygous nonsense 382_383insAG (2-bp “AG” insertion, also called 691_692insAG) mutation and a novel missense mutation Arg96→Leu (R96L) were reported in French familial ALS patients (Millecamps et al., 2011). The 382_383insAG was also previously described as a dominant mutation responsible for familial POAG in Japan (Rezaie et al., 2002) and Eastern Europe (Ayala-Lugo et al., 2007). This mutation presumably induces a premature stop codon in exon 6. The level of optineurin was decreased in patients with this mutation, and a loss of optineurin function was predicted. The missense mutation R96L on the contrary might lead to a gain of function, although no accumulation of the optineurin protein was detected in the patients’ lymphoblasts.
Van Blitterswijk et al. (2011) screened a large Dutch cohort of sporadic ALS patients and identified a nonsense Gln165 stop (Q165X) and a missense Gln454 →Glu (Q454E) mutations. Because the Q165X mutation would probably result in a 72% truncated optineurin protein, binding to Rab8, mGluR1a, TFIIIA, Htt, and myosin VI might be eradicated. The Q454E mutation is located near the area that contains binding sites for Htt and myosin VI. The localization of these two mutations and their predicted effects suggest that they are pathogenic. Mutations, however, were not detected in the Dutch familial ALS patients, underscoring that the genetic background of ALS may differ between different populations.
Optineurin was also shown to associate with another ALS-related protein, FUS (Ito et al., 2011) and myosin VI in the basophilic inclusions of familial ALS with FUS mutation and in neurodegenerative basophilic inclusion body disease. FUS has a role in intracellular trafficking in collaboration with myosin VI and is also known to act as a coactivator of NF-κB (Uranishi et al., 2001). Dysregulation of these pathways by FUS and optineurin is conceivably also an underlying factor for pathology.
4.3. Other neurodegenerative diseases
Optineurin was shown to be located not only in the skein-like inclusions and round hyaline inclusions in ALS but also in the senile plaques and neurofibrillary tangles in Alzheimer’s disease, Lewy bodies and Lewy neuritis in Parkinson’s disease, ballooned neurons in Creutzfeldt–Jakob disease, glial cytoplasmic inclusions in multiple system atrophy, and Pick bodies in Pick disease. This indicates that optineurin is widely distributed in neurodegenerative conditions (Osawa et al., 2011). The significance of such a finding, however, is unknown. Optineurin aggregates are generally found to be ubiquitin positive, and the optineurin aggregation may be the common process involved in neurodegeneration and cell death. Optineurin may be itself an aggregation-prone protein present in the affected neurons and glia. On the other hand, optineurin could also be just secondarily entrapped in the inclusion bodies in various conditions or in ubiquitin.
4.4. Paget’s disease of bone
PDB is one of the most frequent metabolic bone disorders (with a prevalence between 1% and 5%) affecting individuals above age 55 in Caucasian population (Altman et al., 2000; Chung et al., 2010; van Staa et al., 2002). This disease is characterized by focal areas of increased and disorganized bone remodeling that can cause bone pain, bone deformity, pathological fracture, deafness, and secondary osteoarthritis (Siris, 1998). The cause of PDB is not completely understood, although both genetic and environmental factors are implicated (Lucas et al., 2008). Between 15% and 40% of individuals with PDB have an affected first-degree relative (Morales-Piga et al., 1995). Current evidence suggests that this disease is genetically heterogeneous, resulting from mutations in several disease genes. One such gene, the ubiquitin-associated domain of Sequestosome 1 (SQSTM1), was identified and its mutations were noted to affect about 10% of the patients with sporadic or familial PDB (Beyens et al., 2004; Falchetti et al., 2004; Hocking et al., 2002; Laurin et al., 2002). Albagha et al. (2010), using a genome-wide association approach to screen genetic variants that predispose to PDB in individuals without SQSTM1 mutations, identified CSF1 gene at the 1p13 locus, optineurin gene at 10p13, and TNFRSF11A gene at 18q21 as candidate genes for disease susceptibility (Albagha et al., 2010). Optineurin may have an unrecognized role in regulating the bone metabolism.
5. Possible Cellular Function of Optineurin and Dysfunctions Caused by Optineurin Mutations
5.1. Neuroprotection or neurotoxicity?
Optineurin, when transfected into NIH3T3 cells, has been noted to increase cell survival, protecting them from hydrogen peroxide-induced cell death and blocking cytochrome C release from the mitochondria (De Marco et al., 2006). It was also suggested that wild-type optineurin may have a role in neuroprotection in the eye and the optic nerve head (Rezaie et al., 2002). However, E3–14.7K has been shown to inhibit TNF-α-induced apoptosis and optineurin can reverse the E3–14.7K protective effect in human 293 cells (Li et al., 1998). Overexpression of optineurin also did not protect against transforming growth factor-β (TGFβ)-induced apoptosis in βB1-crystallin-OPTN and βB1-crystallin-TGFβ1 double transgenic mice (Kroeber et al., 2006). Cell loss rather than protection was in addition observed upon overexpression of wild type and E50K optineurin in neuronal cells (Koga et al., 2010; Nagabhushana et al., 2010; Park et al., 2006). The cell loss was at least in part due to increased apoptosis, as evidenced by active caspase 3/7 staining (Koga et al., 2010). Apoptosis was moreover seen in RGCs of E50K transgenic mice (Chi et al., 2010). These different results from various investigations could be related to cell types/systems used, the level of optineurin expression, and/or the time points studied. Careful systematic investigations are needed to resolve the issue whether, or under what conditions, optineurin is neuroprotective or neurotoxic.
5.2. Maintenance of Golgi organization
The Golgi apparatus is a highly dynamic cellular organelle composed of cisternal stacks. It transports, processes, and sorts macromolecules such as proteins synthesized in the rough endoplasmic reticulum for cell secretion (exocytosis) or use within the cell (Farquhar and Palade, 1998; Marsh and Howell, 2002). The Golgi stacks are usually arranged as an interconnected network in the region around the centrosome due to a microtubule-dependent mechanism. The organization of Golgi complex is maintained by at least four systems: microtubules and microtubule-associated proteins, the actin-associated cytoskeleton, the Golgi matrix proteins, and proteins that ensure the targeting and fusion of transport vesicles to the correct compartment, such as GTP-binding proteins and SNAREs (soluble NSF [N-ethylmaleimide-sensitive fusion protein] attachment protein receptor) (Gonatas et al., 2006).
The dissociation of the Golgi apparatus or Golgi fragmentation is a physiological and pathological reaction. During mitosis, Golgi complex is disassembled in early prophase and is readily reassembled in telophase (Robbins and Gonatas, 1964). The Golgi apparatus is also found fragmented or dispersed in a variety of neurodegenerative diseases, such as Alzheimer’s disease (Baloyannis et al., 2009), Parkinson’s disease (Fujita et al., 2006), ALS (Fujita et al., 2008; Mourelatos et al., 1994; Stieber et al., 2000), corticobasal degeneration (Sakurai et al., 2000), spinocerebellar ataxia type 2, and Creutzfeldt–Jakob disease (Gonatas et al., 2006; Sakurai et al., 2000). The cytoplasmic aggregation of mutant protein is the common feature in neurodegenerative diseases. These mutant protein aggregations may directly or indirectly interact with any of one or more proteins involved in the maintenance of the structure of the Golgi apparatus and disrupt its structure and function (Gonatas et al., 2006).
Optineurin is reported to play a role in the maintenance of Golgi organization (Park et al., 2006; Sahlender et al., 2005). When optineurin was depleted from cells via RNA interference, the Golgi became fragmented. The ribbon structure of interconnected stacks of membrane cisternae was broken up and the disconnected Golgi stacks were dispersed throughout the cytoplasm (Sahlender et al., 2005). On the other hand, when optineurin was overexpressed in human TM, RPE, and RGC5 cells, the Golgi complex also was disconnected or fragmented (Park et al., 2006; Ying et al., 2010). As stated earlier, when cells were transfected to overexpress optineurin–GFP fusion protein, bright granular or punctuate structures termed foci were observed in perinuclear regions of all cell types. They are dynamic, moving around in both short and long ranges (Ying et al., 2010). Subsequent experimentation using nocodazole proved that the formation of optineurin foci is microtubule dependent (Park et al., 2006; Ying et al., 2010).
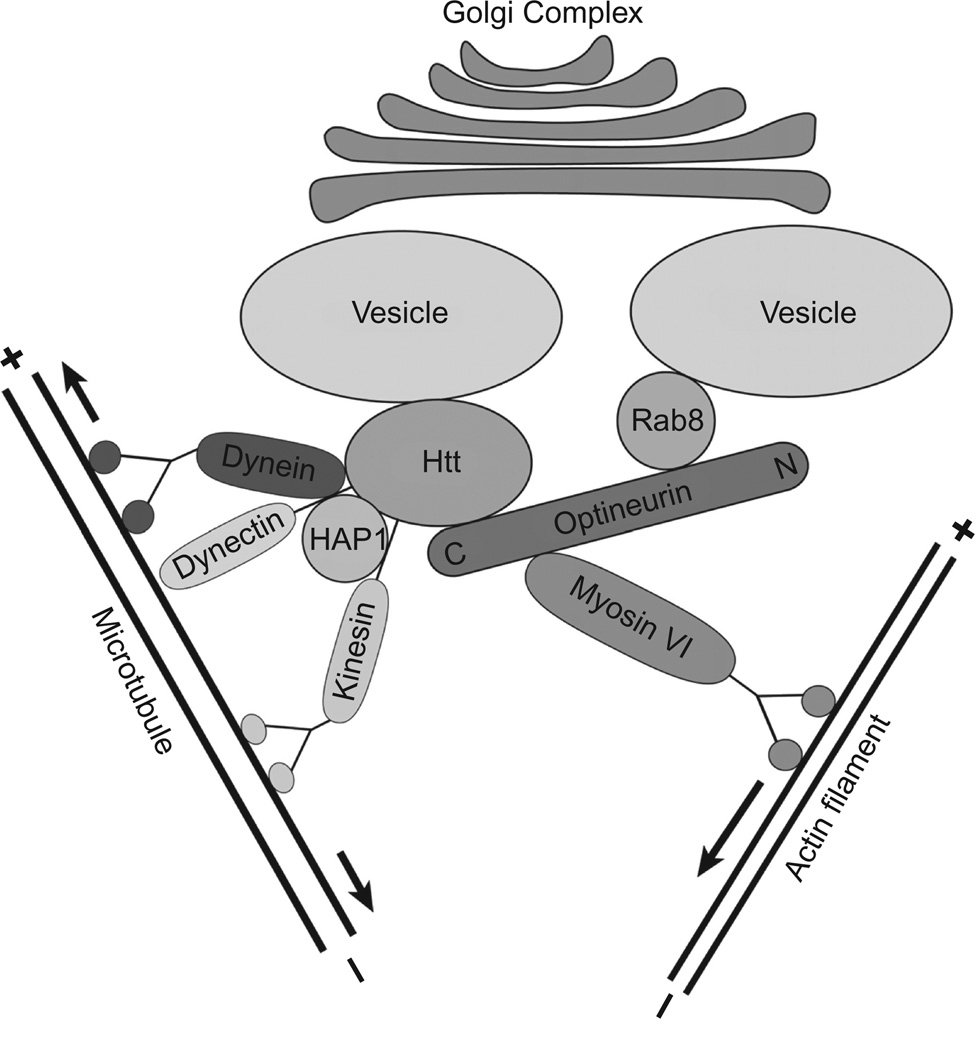
The mechanism as to how optineurin is involved in maintaining the Golgi integrity is unknown. One hypothesis forwarded by Sahlender et al. (2005) is that optineurin might coordinate the actin cytoskeleton and microtubule-based motor activities for Golgi maintenance through interactions with its binding partners. Myosin VI, an actin-based motor protein, might link via optineurin to Rab8 which is involved in sorting molecules in the exocytic pathway at the trans-Golgi network (TGN) and in membrane fusion at the plasma membrane. Optineurin also binds with Htt which has been shown to bind with dynein as well as HAP1, a protein that interacts with dynactin p150 subunit and kinesin. The motor protein complex formed by these proteins is engaged in both anterograde and retrograde transport process (Caviston and Holzbaur, 2009; Rong et al., 2007; Sahlender et al., 2005). In the case of optineurin deletion or overexpression, the interaction between optineurin and its binding partners (such as myosin VI, Rab8, and Htt) may be altered, and the balance between the motor proteins is disturbed to induce fragmentation of the Golgi complex (Fig. 5.4). The disturbance in balance may also be the basis why a reduction in the size of the Golgi complex was seen in Snell’s waltzer mice. It was reasoned that the absence of myosin VI in those mice might cause pulling of the Golgi membrane by the dynein motor complex toward the microtubule organization center, resulting in a smaller, more compact Golgi apparatus (Sahlender et al., 2005; Warner et al., 2003).
Figure 5.4.
Schematic model illustrating how optineurin may coordinate actin cytoskeleton and microtubule system for maintenance of the Golgi complex by interacting with various binding partners (modified from Sahlender et al., 2005 with updates). The C-terminal of optineurin binds with Htt which has been shown to bind with HAP1. HAP1 interacts with the plus (+)-end-directed microtubule motor protein dynein as well as dynein activator dynactin. The minus (−)-end-directed microtubule motor kinesin interacts with Htt and HAP1. Optineurin links Rab8 with myosin VI which is an actin-based motor protein. In scenarios of optineurin deletion or overexpression, the interaction between optineurin and its binding partners (such as myosin VI, Rab8, and Htt) may be altered and the balance between the motor proteins may be disturbed to induce fragmentation of the Golgi complex. (For color version of this figure, the reader is referred to the Web version of this chapter.)
Mutations in optineurin may also interfere with the optineurin interaction with its binding partners. When cells overexpressed a mutant that displays enhanced binding to Rab8 (Nagabhushana et al., 2010; Ying et al., 2010), such as E50K optineurin, the Golgi fragmentation was even more severe than the wild type (Park et al., 2006).
Again, much work remains to be done to unravel the mechanism(s). It does appear nevertheless that optineurin may need to be present in the cells in an optimal level such that all protein interactions are properly maintained. Disturbances of the various interactions, resulting from knockdown or mutation, or when optineurin is in excess, might have adverse consequences.
5.3. Vesicle trafficking
Vesicle trafficking plays a central role in the formation and maintenance of different intracellular compartments as well as in the communications between cells and their environment (Vassilieva and Nusrat, 2008). Major vesicle trafficking pathways consist of endocytosis and exocytosis. Protein trafficking by the endocytic pathway is an essential cellular mechanism critical for many functions including cell signaling, nutrient acquisition, and maintenance of the plasma membrane (Perret et al., 2005).
Optineurin-binding partners myosin VI, Htt, Rab8, and transferrin receptor are all involved in vesicular trafficking (del Toro et al., 2009; Hattula and Peranen, 2000; Park et al., 2010; Sahlender et al., 2005). The transferrin uptake, an indicator of the best characterized, clathrin-dependent, receptor-mediated endocytosis process, was examined in cells after transfection to overexpress wild-type optineurin. It was observed that the transferrin uptake was significantly decreased in optineurin-expressing cells (Park et al., 2010). The reduction was evident from the initial 2- and 5-min time points. Cotransfection with transferrin receptor, but not Rab8 or myosin VI, construct, rescued the optineurin inhibitory effect. The ectopic expression of optineurin caused transferrin receptor to colocalize with the optineurin foci (Nagabhushana et al., 2010; Park et al., 2010). Surface biotinylation experiments further showed that the surface level of transferrin receptor was lowered. It appears that the transferrin receptor molecules, via interactions with optineurin, are recruited and sequestered by the foci formed near the perinuclear area. This arrest may consequently lead to diminish availability of surface transferrin receptor and impeded transferrin uptake.
The E50K mutation, compared to the wild type, had an enhanced binding to transferrin receptor and yielded a more pronounced impairment in the transferrin uptake (Park et al., 2010). This mutation, generating much striking effects compared to the wild-type optineurin, is apparently a gain-of-toxicity mutation. The defective trafficking (Nagabhushana et al., 2010; Park et al., 2006), fragmentation of the Golgi complex (Park et al., 2010), and induction of apoptosis (Koga et al., 2010) are speculated to be the underlying bases why E50K mutation may render the patients predisposed to the glaucoma pathology.
A point mutation Leu157 →Ala (L157A) in the leucine zipper region with which foci formation is minimal and interaction with transferrin receptor is unaltered has no inhibitory effect on the transferrin uptake (Park et al., 2010). Another point mutation D474N in the UBD domain similarly displays few foci and reduced interaction with transferrin receptor. Transferrin molecules were internalized but were not trafficked to the perinuclear region (Nagabhushana et al., 2010). These findings imply that both the leucine zipper and UBD domains of optineurin may be required for the formation of foci as well as the interaction with transferrin receptor.
Using siRNA techniques, downregulation of optineurin was found to reduce significantly the transport of vesicular stomatitis virus G-protein to the cell surface and alter the morphology of Golgi complex in normal rat kidney and HeLa cells (Sahlender et al., 2005). Knocking down optineurin in RGC5 cells, as was seen with the overexpression, also compromised the transferrin uptake (Park et al., 2010).
There have been reports in the literature of various genes (Bache et al., 2003; Guilherme et al., 2004; Magadan et al., 2006; Morino et al., 2004; Park et al., 2009; Strick and Elferink, 2005) displaying identical or similar phenotypes under both overexpression and depletion conditions. For example, inhibition of transferrin internalization was seen to result from both overexpression and siRNA-mediated silencing of EHD2 (EH domain protein 2) (Guilherme et al., 2004). Genes Rab22a (Magadan et al., 2006), REP15 (Rab15 effector protein) (Strick and Elferink, 2005), and Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) (Bache et al., 2003; Morino et al., 2004) have also been noted to generate the same phenotype upon overexpression and knockdown: inhibited transferrin recycling, inhibited transferrin receptor recycling, and impaired degradation of epidermal growth factor, respectively. In the budding yeast, overexpression and deletion of Bud2 both resulted in a significant increase in the rate of actomyosin ring contraction (Park et al., 2009). In this last example, Bud2 overexpression was thought to act as a dominant negative. In the others, it was speculated that overexpression may lead to formation of unbalanced protein complexes, trapping proteins associated with endocytosis steps and inhibiting in turn their normal functions. This appears to be also the likely scenario involved in optineurin overexpression. Evidence is provided that the inhibition of transferrin uptake by overexpressed optineurin is mediated largely through the augmented optineurin–transferrin receptor interaction (Park et al., 2010). The precise mechanism involved in the depletion situation, however, still remains to be defined.
5.4. Regulation of NF-κB signaling
NF-κB is a transcription factor that controls a number of essential cellular functions including immune responses, cell proliferation, and antiapoptosis. NF-κB is retained in an inactive form in the cytoplasm. Upon activation, the inhibitory proteins of the IκB family are degraded following phosphorylation by the serine protein kinase IKK complex. NF-κB is then dissociated from the inhibitors and translocated to the nucleus to trigger transcription of target genes.
The TNF-α-stimulated NF-κB-dependent gene transcription is markedly enhanced when optineurin is downregulated (Zhu et al., 2009), indicating that optineurin functions normally as a negative regulator of NF-κB (Sudhakar et al., 2009; Zhu et al., 2009). The mechanism by which optineurin regulates NF-κB is, however, not completely defined. It may be related to NEMO, the catalytic unit known as IKKγ of the IKK complex, since optineurin shares a high amino acid homology with NEMO (Schwamborn et al., 2000). Optineurin has been shown to compete with NEMO to bind with ubiquitinated RIP which is necessary for efficient activation NF-κB induced by TNF-α (Zhu et al., 2007). Once bound to polyUb RIP, optineurin may bind with another binding partner, deubiquitinase CYLD, to deubiquitinate polyUb RIP and thereby block the downstream reaction in the NF-κB pathway, resulting in apoptosis (Nagabhushana et al., 2011). There appears to be a negative feedback loop; the optineurin gene expression is elevated by treatment of TNF-α via the NF-κB pathway, and the optineurin induced in turn inhibits the NF-κB activation, suggesting a physiological role for optineurin in dampening the TNF-α signaling (Sudhakar et al., 2009; Zhu et al., 2007).
TBK1, an optineurin binding protein, can be recruited to the TNF receptor complex in response to TNF-α (Kuai et al., 2004) and activate NF-κB -directed transcription (Bonnard et al., 2000). Whether or how optineurin is involved in the TBK1-related process is currently unknown. E50K optineurin has been reported to enhance the binding of optineurin to TBK1. Such an enhancement may lead to aberrant NF-κB regulation and contribute as another factor in the development of glaucoma.
5.5. Antiviral signaling
Viral infection has been documented to stimulate innate immune response to produce various proinflammatory cytokines (such as type I interferons: IFNα/β) and chemokines to elicit an effective antiviral defense. Loss of IFNβ signaling leads to severe immunodeficiency toward viral infection (Muller et al., 1994). Optineurin suppresses cytokine responses to viral infections. When optineurin was overexpressed in the cells followed by infection with RNA virus Sendai virus (SeV), the cells failed to express IFNβ. Depletion of optineurin with siRNA by contrast promoted virus-induced IFNβ production and decreased RNA virus replication (Mankouri et al., 2010). Immunoprecipitation and immunofluorescence studies identified optineurin in a protein complex containing the antiviral protein kinase TBK1 and ubiquitin ligase TRAF3. Mutagenesis studies (D474N mutation) showed that binding of ubiquitin was essential for both correct subcellular localization and negative regulation of IFNβ by optineurin (Mankouri et al., 2010).
In another study, optineurin was noted to have antiviral activity through the viral TAX oncoprotein which is known to activate the NF-κB pathway. When cells were infected with HTLV-1, optineurin was demonstrated to interact cooperatively with the TAX1 binding protein TAXBP1 to increase the ubiquitination of TAX1 along with TAX1-dependent NF-κB activation and antiviral activity (Journo et al., 2009).
5.6. Antibacteria signaling
Optineurin is recently recognized as an autophagy receptor, functioning in innate immunity against cytosolic bacteria by linking the TBK1 signaling pathway to autophagic elimination of cytosolic pathogens (Wild et al., 2011). Optineurin directly binds with autophagy modifiers LC3/ GABARAP proteins and ubiquitin chains (Wild et al., 2011). Both the binding of optineurin to LC3 and selective autophagy of ubiquitin-coated cytosolic S. enterica are promoted after phosphorylation of optineurin by TBK1 at Ser-177 (Wild et al., 2011).
It appears that most Salmonella upon infection reside in Salmonella-containing vacuoles (SCVs) in the cells. A fraction of Salmonella may escape from the SCVs to the host cell cytosol where they hyperproliferate and serve as a reservoir for dissemination (Knodler et al., 2010). As a cellular defense mechanism, cytosolic Salmonella is rapidly coated with ubiquitin (Perrin et al., 2004). Optineurin is then recruited to the ubiquitinated cytosolic Salmonella. Following subsequent phosphorylation by TBK1, the LC3-optineurin binding is increased, and the phosphorylated optineurin can in turn act as a molecular trigger to promote autophagic clearance of cytosolic bacteria (Wild et al., 2011).
5.7. Induction of autophagy
Autophagy is another major route besides the ubiquitin–proteasome system for protein and organelle clearance in eukaryotic cells (Glickman and Ciechanover, 2002; Kirkin et al., 2009; McCray and Taylor, 2008). It is an evolutionally conserved mechanism responsible for bulk degradation of long-lived proteins and cytoplasmic recycling of organelles during development, tissue homeostasis, and environmental stress such as starvation or amino acid depletion (Meijer and Codogno, 2009; Mizushima, 2007). There are three types of autophagy: macroautophagy, chaperone-mediated autophagy, and microautophagy (Todde et al., 2009). Among them, macro-autophagy (the most common form, often referred to as autophagy) is the one mediated by the organelle termed autophagosome. Upon an induction signal, macroautophagy (autophagy) starts when a flat membrane cistern wraps around a portion of cytosol and/or organelles, forming a closed double-membrane bound vacuole that contains cytoplasm. This vacuole, called autophagosome (Bao et al., 2010; Eskelinen, 2005; Mizushima et al., 2010), undergoes stepwise maturation processes that include fusing events with lysosomes to form autolysosomes (Dunn, 1990). The contents of autolysosomes are finally degraded by acidic lysosomal hydrolases and the degraded products are transported back to the cytoplasm.
Optineurin has been shown to bind with autophagic marker LC3 and is an autophagy receptor (Wild et al., 2011). In neuronal cells that overexpress wild type and E50K optineurin, the level of proteasome regulatory β5 subunit (indicative of proteasome activity) is reduced and the level of LC3 is increased compared with mock transfected or nontransfected controls. Autophagosome formation was also detected by electron microscopy. The number of foci formed in optineurin overexpressing cells was found increased upon treatment of an autophagic inhibitor, 3-methyladenine, but decreased by treatment of autophagic inducer, rapamycin. These results indicate that the ubiquitin–proteasome function is compromised when optineurin is upregulated or mutated, while autophagy is induced and comes into play to degrade the overexpressed or mutated optineurin (Shen et al., 2010). In consistence with the conjecture, the turnover of overexpressed wild type or E50K optineurin is dramatically slowed down compared to the endogenous optineurin (Ying et al., 2010).
6. Concluding Remarks and Open Questions
Optineurin has attracted ample attention from investigators in a wide variety of fields due, in part, to its association with diseases such as glaucoma and ALS. Impressive advances have been made in the past few years regarding optineurin, affording a better understanding of the protein characteristics, binding partners, as well as its possible involvement in the Golgi maintenance, protein trafficking, and signal transduction in the cells. There are, however, still questions to be addressed. For instance, it is far from fully understood how mutations identified might affect cellular functions of optineurin and lead to disease phenotypes. Golgi fragmentation has been shown to be one of the causes of neural death in neurodegeneration (Fan et al., 2008). The mechanism why optineurin silencing or overexpression would trigger Golgi defect is unknown. Is Ca2+ homeostasis a player? Is the microtubule system involved?
Optineurin has many binding partners and evidently has multiple functions. Are there other novel interacting partners and yet-discovered optineurin functions? How are the various functions regulated? Protein aggregation is a common phenomenon in the neurodegenerative diseases such as Lewy bodies (α-synuclein) in Parkinson’s disease, neuritic plaques (β-amyloid), and neurofibrillary tangles (hyperphosphorylated tau) in Alzheimer’s disease, as well as Htt aggregates in Huntington’s disease (Lee et al., 2011; Ross and Poirier, 2004; Selkoe, 2003). The endogenous optineurin can interact with itself to form hexamers and can also bind with partner proteins to form supermolecular complexes (Ying et al., 2010). Is optineurin an aggregation-prone protein? Will it form amyloid fibers? What are the conditions that drive optineurin to aggregation? Is the aggregate cytotoxic? What are the consequences? Finally, what is the role of autophagy in optineurin clearance and pathology?
Transgenic and optineurin knockout mice may be developed as powerful tools for further exploration of optineurin functions. Modern, emerging techniques such as proteomics and molecular biology manipulations will also greatly facilitate studies to resolve open questions and provide insights into the mechanisms directly involved in the disease development. Such information will be of critical importance in designs of target therapies and have high translational impact in treatment and prevention of optineurin-related diseases.
ACKNOWLEDGMENTS
The authors thank Ms. Lisa Birmingham for assistance in illustrations and preparations of Figs. 5.3 and 5.4. The work was supported by Grant EY018828 and Core Grant EY001792 from the National Eye Institute, National Institutes of Health, Bethesda, Maryland.
REFERENCES
- Albagha OM, Visconti MR, Alonso N, Langston AL, Cundy T, Dargie R, et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat. Genet. 2010;42:520–524. doi: 10.1038/ng.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allingham RR, Liu Y, Rhee DJ. The genetics of primary open-angle glaucoma: a review. Exp. Eye Res. 2009;88:837–844. doi: 10.1016/j.exer.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman RD, Bloch DA, Hochberg MC, Murphy WA. Prevalence of pelvic Paget’s disease of bone in the United States. J. Bone Miner. Res. 2000;15:461–465. doi: 10.1359/jbmr.2000.15.3.461. [DOI] [PubMed] [Google Scholar]
- Alward WL, Kwon YH, Kawase K, Craig JE, Hayreh SS, Johnson AT, et al. Evaluation of optineurin sequence variations in 1,048 patients with open-angle glaucoma. Am. J. Ophthalmol. 2003;136:904–910. doi: 10.1016/s0002-9394(03)00577-4. [DOI] [PubMed] [Google Scholar]
- Anborgh PH, Godin C, Pampillo M, Dhami GK, Dale LB, Cregan SP, et al. Inhibition of metabotropic glutamate receptor signaling by the huntingtin-binding protein optineurin. J. Biol. Chem. 2005;280:34840–34848. doi: 10.1074/jbc.M504508200. [DOI] [PubMed] [Google Scholar]
- Anderson DR. Collaborative normal tension glaucoma study. Curr. Opin. Ophthalmol. 2003;14:86–90. doi: 10.1097/00055735-200304000-00006. [DOI] [PubMed] [Google Scholar]
- Aschenbrenner L, Lee T, Hasson T. Myo6 facilitates the translocation of endocytic vesicles from cell peripheries. Mol. Biol. Cell. 2003;14:2728–2743. doi: 10.1091/mbc.E02-11-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au JS, Puri C, Ihrke G, Kendrick-Jones J, Buss F. Myosin VI is required for sorting of AP-1B-dependent cargo to the basolateral domain in polarized MDCK cells. J. Cell Biol. 2007;177:103–114. doi: 10.1083/jcb.200608126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung T, Rezaie T, Okada K, Viswanathan AC, Child AH, Brice G, et al. Clinical features and course of patients with glaucoma with the E50K mutation in the optineurin gene. Invest. Ophthalmol. Vis. Sci. 2005;46:2816–2822. doi: 10.1167/iovs.04-1133. [DOI] [PubMed] [Google Scholar]
- Ayala-Lugo RM, Pawar H, Reed DM, Lichter PR, Moroi SE, Page M, et al. Variation in optineurin (OPTN) allele frequencies between and within populations. Mol. Vis. 2007;13:151–163. [PMC free article] [PubMed] [Google Scholar]
- Azmi P, Seth A. RNF11 is a multifunctional modulator of growth factor receptor signaling and transcriptional regulation. Eur. J. Cancer. 2005;41:2549–2560. doi: 10.1016/j.ejca.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 2003;278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- Baird PN, Richardson AJ, Craig JE, Mackey DA, Rochtchina E, Mitchell P. Analysis of optineurin (OPTN) gene mutations in subjects with and without glaucoma: the Blue Mountains Eye Study. Clin. Experiment. Ophthalmol. 2004;32:518–522. doi: 10.1111/j.1442-9071.2004.00886.x. [DOI] [PubMed] [Google Scholar]
- Baloyannis SJ, Mauroudis I, Manolides SL, Manolides LS. Synaptic alterations in the medial geniculate bodies and the inferior colliculi in Alzheimer’s disease: a Golgi and electron microscope study. Acta Otolaryngol. 2009;129:416–418. doi: 10.1080/00016480802579074. [DOI] [PubMed] [Google Scholar]
- Bao XH, Naomoto Y, Hao HF, Watanabe N, Sakurama K, Noma K, et al. Autophagy: can it become a potential therapeutic target? Int. J. Mol. Med. 2010;25:493–503. doi: 10.3892/ijmm_00000369. [DOI] [PubMed] [Google Scholar]
- Beyens G, Van Hul E, Van Driessche K, Fransen E, Devogelaer JP, Vanhoenacker F, et al. Evaluation of the role of the SQSTM1 gene in sporadic Belgian patients with Paget’s disease. Calcif. Tissue Int. 2004;75:144–152. doi: 10.1007/s00223-004-0244-4. [DOI] [PubMed] [Google Scholar]
- Bond LM, Peden AA, Kendrick-Jones J, Sellers JR, Buss F. Myosin VI and its binding partner optineurin are involved in secretory vesicle fusion at the plasma membrane. Mol. Biol. Cell. 2011;22:54–65. doi: 10.1091/mbc.E10-06-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, et al. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kB-dependent gene transcription. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TP, Nijman SM, Dirac AM, Bernards R. Loss of cylindromatosis tumor suppressor inhibits apoptosis by activating NF-kB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Buss F, Kendrick-Jones J, Lionne C, Knight AE, Cote GP, PaulLuzio J. The localization of myosin VI at the Golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J. Cell Biol. 1998;143:1535–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Luzio JP, Kendrick-Jones J. Myosin VI, a new force in clathrin mediated endocytosis. FEBS Lett. 2001;508:295–299. doi: 10.1016/s0014-5793(01)03065-4. [DOI] [PubMed] [Google Scholar]
- Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu. Rev. Cell Dev. Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- Caixeta-Umbelino C, de Vasconcellos JP, Costa VP, Kasahara N, Della Paolera M, de Almeida GV, et al. Lack of association between optineurin gene variants T34T, E50K, M98K, 691_692insAG and R545Q and primary open angle glaucoma in Brazilian patients. Ophthalmic Genet. 2009;30:13–18. doi: 10.1080/13816810802502970. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Holzbaur EL. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrama FZ, Seguin-Py S, Le Grand JN, Fraichard A, Delage-Mourroux R, Despouy G, et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010;6:495–505. doi: 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- Chi ZL, Akahori M, Obazawa M, Minami M, Noda T, Nakaya N, et al. Overexpression of optineurin E50K disrupts Rab8 interaction and leads to a progressive retinal degeneration in mice. Hum. Mol. Genet. 2010;19:2606–2615. doi: 10.1093/hmg/ddq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibalina MV, Roberts RC, Arden SD, Kendrick-Jones J, Buss F. Rab8-optineurin-myosin VI: analysis of interactions and functions in the secretory pathway. Methods Enzymol. 2008;438:11–24. doi: 10.1016/S0076-6879(07)38002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ZL, Shin YA, Yang JM, DiDonato JA, Ballard DW. IKKγ mediates the interaction of cellular IκB kinases with the tax transforming protein of human T cell leukemia virus type 1. J. Biol. Chem. 1999;274:15297–15300. doi: 10.1074/jbc.274.22.15297. [DOI] [PubMed] [Google Scholar]
- Chung PY, Beyens G, Boonen S, Papapoulos S, Geusens P, Karperien M, et al. The majority of the genetic risk for Paget’s disease of bone is explained by genetic variants close to the CSF1, OPTN, TM7SF4, and TNFRSF11A genes. Hum. Genet. 2010;128:615–626. doi: 10.1007/s00439-010-0888-2. [DOI] [PubMed] [Google Scholar]
- Clemens KR, Wolf V, McBryant SJ, Zhang P, Liao X, Wright PE, et al. Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu. Rev. Pharmacol. Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J. Biol. Chem. 2006;281:16147–16156. doi: 10.1074/jbc.M601467200. [DOI] [PubMed] [Google Scholar]
- del Toro D, Canals JM, Gines S, Kojima M, Egea G, Alberch J. Mutant huntingtin impairs the post-Golgi trafficking of brain-derived neurotrophic factor but not its Val66Met polymorphism. J. Neurosci. 2006;26:12748–12757. doi: 10.1523/JNEUROSCI.3873-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro D, Alberch J, Lazaro-Dieguez F, Martin-Ibanez R, Xifro X, Egea G, et al. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNF-α requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- Falchetti A, Di Stefano M, Marini F, Del Monte F, Mavilia C, Strigoli D, et al. Two novel mutations at exon 8 of the Sequestosome 1 (SQSTM1) gene in an Italian series of patients affected by Paget’s disease of bone (PDB) J. Bone Miner. Res. 2004;19:1013–1017. doi: 10.1359/JBMR.040203. [DOI] [PubMed] [Google Scholar]
- Fan J, Hu Z, Zeng L, Lu W, Tang X, Zhang J, et al. Golgi apparatus and neurodegenerative diseases. Int. J. Dev. Neurosci. 2008;26:523–534. doi: 10.1016/j.ijdevneu.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 1998;8:2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Ohama E, Takatama M, Al-Sarraj S, Okamoto K. Fragmentation of Golgi apparatus of nigral neurons with α-synuclein-positive inclusions in patients with Parkinson’s disease. Acta Neuropathol. 2006;112:261–265. doi: 10.1007/s00401-006-0114-4. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Mizuno Y, Takatama M, Okamoto K. Anterior horn cells with abnormal TDP-43 immunoreactivities show fragmentation of the Golgi apparatus in ALS. J. Neurol. Sci. 2008;269:30–34. doi: 10.1016/j.jns.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Funayama T, Ishikawa K, Ohtake Y, Tanino T, Kurosaka D, Kimura I, et al. Variants in optineurin gene and their association with tumor necrosis factor-α polymorphisms in Japanese patients with glaucoma. Invest. Ophthalmol. Vis. Sci. 2004;45:4359–4367. doi: 10.1167/iovs.03-1403. [DOI] [PubMed] [Google Scholar]
- Fuse N, Takahashi K, Akiyama H, Nakazawa T, Seimiya M, Kuwahara S, et al. Molecular genetic analysis of optineurin gene for primary open-angle and normal tension glaucoma in the Japanese population. J. Glaucoma. 2004;13:299–303. doi: 10.1097/00061198-200408000-00007. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gonatas NK, Stieber A, Gonatas JO. Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J. Neurol. Sci. 2006;246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- Guilherme A, Soriano NA, Bose S, Holik J, Bose A, Pomerleau DP, et al. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J. Biol. Chem. 2004;279:10593–10605. doi: 10.1074/jbc.M307702200. [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Sun SC. IKKγ serves as a docking subunit of the IκB kinase (IKK) and mediates interaction of IKK with the human T-cell leukemia virus TAX protein. J. Biol. Chem. 1999;274:22911–22914. doi: 10.1074/jbc.274.33.22911. [DOI] [PubMed] [Google Scholar]
- Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- Hauser MA, Sena DF, Flor J, Walter J, Auguste J, Larocque-Abramson K, et al. Distribution of optineurin sequence variations in an ethnically diverse population of low-tension glaucoma patients from the United States. J. Glaucoma. 2006;15:358–363. doi: 10.1097/01.ijg.0000212255.17950.42. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, et al. The roles of two IκB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum. Mol. Genet. 2002;11:2735–2739. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- Hoffner G, Kahlem P, Djian P. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with b-tubulin: relevance to Huntington’s disease. J. Cell Sci. 2002;115:941–948. doi: 10.1242/jcs.115.5.941. [DOI] [PubMed] [Google Scholar]
- Hoyng PF, Kitazawa Y. Medical treatment of normal tension glaucoma. Surv. Ophthalmol. 2002;47(Suppl. 1):S116–S124. doi: 10.1016/s0039-6257(02)00322-3. [DOI] [PubMed] [Google Scholar]
- Huber LA, de Hoop MJ, Dupree P, Zerial M, Simons K, Dotti C. Protein transport to the dendritic plasma membrane of cultured neurons is regulated by Rab8p. J. Cell Biol. 1993a;123:47–55. doi: 10.1083/jcb.123.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J. Cell Biol. 1993b;123:35–45. doi: 10.1083/jcb.123.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fujita K, Nakamura M, Wate R, Kaneko S, Sasaki S, et al. Optineurin is co-localized with FUS in basophilic inclusions of ALS with FUS mutation and in basophilic inclusion body disease. Acta Neuropathol. 2011;122:223–229. doi: 10.1007/s00401-011-0809-z. [DOI] [PubMed] [Google Scholar]
- Jin DY, Giordano V, Kibler KV, Nakano H, Jeang KT. Role of adapter function in oncoprotein-mediated activation of NF-κB: human T-cell leukemia virus type I Tax interacts directly with IκB kinase γ. J. Biol. Chem. 1999;274:17402–17405. doi: 10.1074/jbc.274.25.17402. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journo C, Filipe J, About F, Chevalier SA, Afonso PV, Brady JN, et al. NRP/optineurin cooperates with TAX1BP1 to potentiate the activation of NF-κB by human T-lymphotropic virus type 1 tax protein. PLoS Pathog. 2009;5:e1000521. doi: 10.1371/journal.ppat.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kerrigan LA, Zack DJ, Quigley HA, Smith SD, Pease ME. TUNEL-positive ganglion cells in human primary open-angle glaucoma. Arch. Ophthalmol. 1997;115:1031–1035. doi: 10.1001/archopht.1997.01100160201010. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, et al. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. USA. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Shen X, Park J, Qiu Y, Park BC, Shyam R, et al. Effects of myocilin and optineurin, two glaucoma genes on neurite outgrowth. Am. J. Pathol. 2010;176:343–352. doi: 10.2353/ajpath.2010.090194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Kremer B, Goldberg P, Andrew SE, Theilmann J, Telenius H, Zeisler J, et al. A worldwide study of the Huntington’s disease mutation. The sensitivity and specificity of measuring CAG repeats. N. Engl. J. Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- Kroeber M, Ohlmann A, Russell P, Tamm ER. Transgenic studies on the role of optineurin in the mouse eye. Exp. Eye Res. 2006;82:1075–1085. doi: 10.1016/j.exer.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Kuai J, Wooters J, Hall JP, Rao VR, Nickbarg E, Li B, et al. NAK is recruited to the TNFR1 complex in a TNFa-dependent manner and mediates the production of RANTES: identification of endogenous TNFR-interacting proteins by a proteomic approach. J. Biol. Chem. 2004;279:53266–53271. doi: 10.1074/jbc.M411037200. [DOI] [PubMed] [Google Scholar]
- Kuehn MH, Fingert JH, Kwon YH. Retinal ganglion cell death in glaucoma: mechanisms and neuroprotective strategies. Ophthalmol. Clin. North Am. 2005;18:383–395. doi: 10.1016/j.ohc.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwon YH, Fingert JH, Kuehn MH, Alward WL. Primary open-angle glaucoma. N. Engl. J. Med. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am. J. Hum. Genet. 2002;70:1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lim HS, Masliah E, Lee HJ. Protein aggregate spreading in neurodegenerative diseases: problems and perspectives. Neurosci. Res. 2011;70:339–348. doi: 10.1016/j.neures.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh PN. Chapter 13 amyotrophic lateral sclerosis. Handb. Clin. Neurol. 2007;82:249–278. doi: 10.1016/S0072-9752(07)80016-9. [DOI] [PubMed] [Google Scholar]
- Leung YF, Fan BJ, Lam DS, Lee WS, Tam PO, Chua JK, et al. Different optineurin mutation pattern in primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 2003;44:3880–3884. doi: 10.1167/iovs.02-0693. [DOI] [PubMed] [Google Scholar]
- Li Y, Kang J, Horwitz MS. Interaction of an adenovirus E3 14.7-kilodalton protein with a novel tumor necrosis factor α-inducible cellular protein containing leucine zipper domains. Mol. Cell. Biol. 1998;18:1601–1610. doi: 10.1128/mcb.18.3.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Akafo S, Santiago-Turla C, Cohen CS, Larocque-Abramson KR, Qin X, et al. Optineurin coding variants in Ghanaian patients with primary open-angle glaucoma. Mol. Vis. 2008;14:2367–2372. [PMC free article] [PubMed] [Google Scholar]
- Lucas GJ, Riches PL, Hocking LJ, Cundy T, Nicholson GC, Walsh JP, et al. Identification of a major locus for Paget’s disease on chromosome 10p13 in families of British descent. J. Bone Miner. Res. 2008;23:58–63. doi: 10.1359/jbmr.071004. [DOI] [PubMed] [Google Scholar]
- Magadan JG, Barbieri MA, Mesa R, Stahl PD, Mayorga LS. Rab22a regulates the sorting of transferrin to recycling endosomes. Mol. Cell. Biol. 2006;26:2595–2614. doi: 10.1128/MCB.26.7.2595-2614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri J, Fragkoudis R, Richards KH, Wetherill LF, Harris M, Kohl A, et al. Optineurin negatively regulates the induction of IFNβ in response to RNA virus infection. PLoS Pathog. 2010;6:e1000778. doi: 10.1371/journal.ppat.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh BJ, Howell KE. The mammalian Golgi-complex debates. Nat. Rev. Mol. Cell Biol. 2002;3:789–795. doi: 10.1038/nrm933. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Matsui K, Kumagai Y, Kato H, Sato S, Kawagoe T, Uematsu S, et al. Cutting edge: role of TANK-binding kinase 1 and inducible IκB kinase in IFN responses against viruses in innate immune cells. J. Immunol. 2006;177:5785–5789. doi: 10.4049/jimmunol.177.9.5785. [DOI] [PubMed] [Google Scholar]
- McCray BA, Taylor JP. The role of autophagy in age-related neurodegeneration. Neurosignals. 2008;16:75–84. doi: 10.1159/000109761. [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in TBK1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Autophagy: regulation and role in disease. Crit. Rev. Clin. Lab. Sci. 2009;46:210–240. doi: 10.1080/10408360903044068. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Boillee S, Chabrol E, Camu W, Cazeneuve C, Salachas F, et al. Screening of OPTN in French familial amyotrophic lateral sclerosis. Neurobiol. Aging. 2011;32(557):e511–e553. doi: 10.1016/j.neurobiolaging.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Piga AA, Rey-Rey JS, Corres-Gonzalez J, Garcia-Sagredo JM, Lopez-Abente G. Frequency and characteristics of familial aggregation of Paget’s disease of bone. J. Bone Miner. Res. 1995;10:663–670. doi: 10.1002/jbmr.5650100421. [DOI] [PubMed] [Google Scholar]
- Moreland RJ, Dresser ME, Rodgers JS, Roe BA, Conaway JW, Conaway RC, et al. Identification of a transcription factor IIIA-interacting protein. Nucleic Acids Res. 2000;28:1986–1993. doi: 10.1093/nar/28.9.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino C, Kato M, Yamamoto A, Mizuno Z, Hayakawa A, Komada M, et al. A role of Hrs in endosomal sorting of ligand-stimulated and unstimulated epidermal growth factor receptor. Exp. Cell Res. 2004;297:380–391. doi: 10.1016/j.yexcr.2004.03.038. [DOI] [PubMed] [Google Scholar]
- Moritz OL, Tam BM, Hurd LL, Peranen J, Deretic D, Papermaster DS. Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus. rods. Mol. Biol. Cell. 2001;12:2341–2351. doi: 10.1091/mbc.12.8.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Hirano A, Rosenquist AC, Gonatas NK. Fragmentation of the Golgi apparatus of motor neurons in amyotrophic lateral sclerosis (ALS). Clinical studies in ALS of Guam and experimental studies in deafferented neurons and in b, b’-iminodipropionitrile axonopathy. Am. J. Pathol. 1994;144:1288–1300. [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Komatireddy S, Acharya M, Bhattacharjee A, Mandal AK, Thakur SK, et al. Evaluation of optineurin as a candidate gene in Indian patients with primary open angle glaucoma. Mol. Vis. 2005;11:792–797. [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Nagabhushana A, Chalasani ML, Jain N, Radha V, Rangaraj N, Balasubramanian D, et al. Regulation of endocytic trafficking of transferrin receptor by optineurin and its impairment by a glaucoma-associated mutant. BMC Cell Biol. 2010;11:4. doi: 10.1186/1471-2121-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagabhushana A, Bansal M, Swarup G. Optineurin is required for CYLD-dependent inhibition of TNFα-induced NF-κB activation. PLoS One. 2011;6:e17477. doi: 10.1371/journal.pone.0017477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Mizuno Y, Fujita Y, Takatama M, Nakazato Y, Okamoto K. Optineurin in neurodegenerative diseases. Neuropathology. 2011;31:569–574. doi: 10.1111/j.1440-1789.2011.01199.x. [DOI] [PubMed] [Google Scholar]
- Park BC, Shen X, Samaraweera M, Yue BYJT. Studies of optineurin, a glaucoma gene: Golgi fragmentation and cell death from overexpression of wild-type and mutant optineurin in two ocular cell types. Am. J. Pathol. 2006;169:1976–1989. doi: 10.2353/ajpath.2006.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Cable AE, Blair J, Stockstill KE, Shannon KB. BMC Cell Biol. 2009;10:43–54. doi: 10.1186/1471-2121-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BC, Ying H, Shen X, Park JS, Qiu Y, Shyam R, et al. Impairment of protein trafficking upon overexpression and mutation of optineurin. PLoS One. 2010;5:e11547. doi: 10.1371/journal.pone.0011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen J, Auvinen P, Virta H, Wepf R, Simons K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J. Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret E, Lakkaraju A, Deborde S, Schreiner R, Rodriguez-Boulan E. Evolving endosomes: how many varieties and why? Curr. Opin. Cell Biol. 2005;17:423–434. doi: 10.1016/j.ceb.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr. Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Quigley HA. Glaucoma. Lancet. 2011;377:1367–1377. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KN, Nagireddy S, Chakrabarti S. Complex genetic mechanisms in glaucoma: an overview. Indian J. Ophthalmol. 2011;59(Suppl):S31–S42. doi: 10.4103/0301-4738.73685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie T, Sarfarazi M. Molecular cloning, genomic structure, and protein characterization of mouse optineurin. Genomics. 2005;85:131–138. doi: 10.1016/j.ygeno.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- Rezaie T, Waitzman DM, Seeman JL, Kaufman PL, Sarfarazi M. Molecular cloning and expression profiling of optineurin in the rhesus monkey. Invest. Ophthalmol. Vis. Sci. 2005;46:2404–2410. doi: 10.1167/iovs.04-1243. [DOI] [PubMed] [Google Scholar]
- Rezaie T, Stoilov I, Sarfarazi M. Embryonic expression of the optineurin (glaucoma) gene in different stages of mouse development. Mol. Vis. 2007;13:1446–1450. [PubMed] [Google Scholar]
- Robbins E, Gonatas NK. The ultrastructure of a mammalian cell during the mitotic cycle. J. Cell Biol. 1964;21:429–463. doi: 10.1083/jcb.21.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J, Li SH, Li XJ. Regulation of intracellular HAP1 trafficking. J. Neurosci. Res. 2007;85:3025–3029. doi: 10.1002/jnr.21326. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A, Okamoto K, Fujita Y, Nakazato Y, Wakabayashi K, Takahashi H, et al. Fragmentation of the Golgi apparatus of the ballooned neurons in patients with corticobasal degeneration and Creutzfeldt-Jakob disease. Acta Neuropathol. 2000;100:270–274. doi: 10.1007/s004010000182. [DOI] [PubMed] [Google Scholar]
- Sarfarazi M, Rezaie T. Optineurin in primary open angle glaucoma. Ophthalmol. Clin. North Am. 2003;16:529–541. doi: 10.1016/s0896-1549(03)00061-0. [DOI] [PubMed] [Google Scholar]
- Schwamborn K, Weil R, Courtois G, Whiteside ST, Israel A. Phorbol esters and cytokines regulate the expression of the NEMO-related protein, a molecule involved in a NF-κB-independent pathway. J. Biol. Chem. 2000;275:22780–22789. doi: 10.1074/jbc.M001500200. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Liebl DJ, Harhaj EW. Essential role for TAX1BP1 in the termination of TNF-α-, IL-1- and LPS-mediated NF-κB and JNK signaling. EMBO J. 2007;26:3910–3922. doi: 10.1038/sj.emboj.7601823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ying H, Qiu Y, Park JS, Shyam R, Chi ZL, et al. Processing of optineurin in neuronal cells. J. Biol. Chem. 2011;286:3618–3629. doi: 10.1074/jbc.M110.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siris ES. Paget’s disease of bone. J. Bone Miner. Res. 1998;13:1061–1065. doi: 10.1359/jbmr.1998.13.7.1061. [DOI] [PubMed] [Google Scholar]
- Spudich G, Chibalina MV, Au JS, Arden SD, Buss F, Kendrick-Jones J. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat. Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieber A, Gonatas JO, Gonatas NK. Aggregation of ubiquitin and a mutant ALS-linked SOD1 protein correlate with disease progression and fragmentation of the Golgi apparatus. J. Neurol. Sci. 2000;173:53–62. doi: 10.1016/s0022-510x(99)00300-7. [DOI] [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- Strick DJ, Elferink LA. Rab15 effector protein: a novel protein for receptor recycling from the endocytic recycling compartment. Mol. Biol. Cell. 2005;16:5699–5709. doi: 10.1091/mbc.E05-03-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroissnigg H, Repitz M, Miloloza A, Linhartova I, Beug H, Wiche G, et al. FIP-2, an IκB-kinase-γ-related protein, is associated with the Golgi apparatus and translocates to the marginal band during chicken erythroblast differentiation. Exp. Cell Res. 2002;278:133–145. doi: 10.1006/excr.2002.5567. [DOI] [PubMed] [Google Scholar]
- Sudhakar C, Nagabhushana A, Jain N, Swarup G. NF-κB mediates tumor necrosis factor α-induced expression of optineurin, a negative regulator of NF-κB. PLoS One. 2009;4:e5114. doi: 10.1371/journal.pone.0005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Toda Y, Kashiwagi K, Mabuchi F, Iijima H, Tsukahara S, et al. The association between Japanese primary open-angle glaucoma and normal tension glaucoma patients and the optineurin gene. Hum. Genet. 2003;113:276–279. doi: 10.1007/s00439-003-0964-y. [DOI] [PubMed] [Google Scholar]
- Theunissen O, Rudt F, Guddat U, Mentzel H, Pieler T. RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell. 1992;71:679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- Toda Y, Tang S, Kashiwagi K, Mabuchi F, Iijima H, Tsukahara S, et al. Mutations in the optineurin gene in Japanese patients with primary open-angle glaucoma and normal tension glaucoma. Am. J. Med. Genet. A. 2004;125A:1–4. doi: 10.1002/ajmg.a.20439. [DOI] [PubMed] [Google Scholar]
- Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim. Biophys. Acta. 2009;1792:3–13. doi: 10.1016/j.bbadis.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Uranishi H, Tetsuka T, Yamashita M, Asamitsu K, Shimizu M, Itoh M, et al. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-κB p65-mediated transcription as a coactivator. J. Biol. Chem. 2001;276:13395–13401. doi: 10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- van Blitterswijk M, van Vught PWJ, van Es MA, Schelhaas HJ, van der Kooi AJ, de Visser M, et al. Novel optineurin mutations in sporadic amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.05.019. in press. [DOI] [PubMed] [Google Scholar]
- van Staa TP, Selby P, Leufkens HG, Lyles K, Sprafka JM, Cooper C. Incidence and natural history of Paget’s disease of bone in England and Wales. J. Bone Miner. Res. 2002;17:465–471. doi: 10.1359/jbmr.2002.17.3.465. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva EV, Nusrat A. Vesicular trafficking: molecular tools and targets. Methods Mol. Biol. 2008;440:3–14. doi: 10.1007/978-1-59745-178-9_1. [DOI] [PubMed] [Google Scholar]
- Wang N, Chintala SK, Fini ME, Schuman JS. Activation of a tissue-specific stress response in the aqueous outflow pathway of the eye defines the glaucoma disease phenotype. Nat. Med. 2001;7:304–309. doi: 10.1038/85446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner CL, Stewart A, Luzio JP, Steel KP, Libby RT, Kendrick-Jones J, et al. Loss of myosin VI reduces secretion and the size of the Golgi in fibroblasts from Snell’s waltzer mice. EMBO J. 2003;22:569–579. doi: 10.1093/emboj/cdg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophago-some biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisschuh N, Neumann D, Wolf C, Wissinger B, Gramer E. Prevalence of myocilin and optineurin sequence variants in German normal tension glaucoma patients. Mol. Vis. 2005;11:284–287. [PubMed] [Google Scholar]
- Wiggs JL. Genetic etiologies of glaucoma. Arch. Ophthalmol. 2007;125:30–37. doi: 10.1001/archopht.125.1.30. [DOI] [PubMed] [Google Scholar]
- Wiggs JL, Auguste J, Allingham RR, Flor JD, Pericak-Vance MA, Rogers K, et al. Lack of association of mutations in optineurin with disease in patients with adult-onset primary open-angle glaucoma. Arch. Ophthalmol. 2003;121:1181–1183. doi: 10.1001/archopht.121.8.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby CE, Chan LL, Herd S, Billingsley G, Noordeh N, Levin AV, et al. Defining the pathogenicity of optineurin in juvenile open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 2004;45:3122–3130. doi: 10.1167/iovs.04-0107. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. [corrected]. Nat. Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Meng Q, Tsai JC, Yuan H, Xu N, Li Y. A novel optineurin genetic mutation associated with open-angle glaucoma in a Chinese family. Mol. Vis. 2009;15:1649–1654. [PMC free article] [PubMed] [Google Scholar]
- Yen YC, Yang JJ, Chou MC, Li SY. Absence of optineurin (OPTN) gene mutations in Taiwanese patients with juvenile-onset open-angle glaucoma. Mol. Vis. 2008;14:487–494. [PMC free article] [PubMed] [Google Scholar]
- Ying H, Shen X, Park BC, Yue BYJT. Posttranslational modifications, localization, and protein interactions of optineurin, the product of a glaucoma gene. PLoS One. 2010;5:e9168. doi: 10.1371/journal.pone.0009168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFalpha-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Curr. Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]