Abstract
To characterize fluorescent enteric neurons labeled for expression of cytoplasmic markers in zebrafish mutants, we developed a new MATLAB-based program that can be trained by user input. We used the program to count enteric neurons and to analyze co-expression of the neuronal marker, Elavl, and the neuronal subtype marker, serotonin, in 3D confocal image stacks of dissected whole-mount zebrafish intestines. We quantified the entire population of enteric neurons and the serotonergic subpopulation in specific regions of the intestines of gutwrencher mutant and wild-type sibling larvae. We show a marked decrease in enteric neurons in gutwrencher mutants that is more severe at the caudal end of the intestine. We also show that gutwrencher mutants have the same number of serotonin-positive enteroendocrine cells in the intestine as wild types.
Introduction
We and others have identified several mutant zebrafish lines that exhibit enteric nervous system (ENS) defects and thus may serve as models of genetic diseases that affect ENS function.1–3 Understanding the roles of the mutant genes requires quantitative expression analysis at several different developmental stages for a number of known cell identity markers, for example, neurotransmitters that distinguish distinct types of enteric neurons.4 The process of counting enteric neurons in these mutants is very time-consuming, especially if one relies on manual identification of cells in sectioned animals, as we have done in the past.1 We and many other researchers have resorted to using cumbersome techniques when attempting to quantify cells in sectioned tissue (for examples and discussion of some of these techniques, see5,6). Without these techniques, fragments of cells in multiple sections would quickly lead to erroneous results. Another approach is to count cells in three-dimensional (3D) confocal image stacks from the entire organism or from the specific region of interest, in our case the intestinal tract. Although such whole-mount techniques bypass many of the issues associated with counting cells in sectioned material, determining cell counts from stacks of confocal images poses other problems. Here we describe analysis of the enteric nervous system of gutwrencherb1088 (gwr) mutants using a new method we developed for computer-assisted quantification of cells in whole-mount 3D confocal image stacks of dissected intestines.
gutwrencherb1088 (gwr) is a gene that appears to be pivotal for proper ENS development.1 Previous counts of enteric neurons in sectioned gutwrencher mutant zebrafish larvae revealed a 3.5-fold decrease in enteric neurons overall and a 6-fold decrease in the number of serotonin (5HT) positive enteric neurons compared to wild types.1 This observed decrease in enteric neurons has also been shown to correlate with dysfunctional gut motility.1 To better characterize gwr and other mutations that affect the enteric nervous system, additional co-expression analyses must be done to show whether all enteric neurons are affected equally, or whether a mutation preferentially affects specific types of enteric neurons.
We were unable to find counting programs that are appropriate for quantifying enteric neurons in whole-mount zebrafish intestines. Many programs used to quantify eukaryotic cell numbers, for example those described by Oberlaender et al.7 and DeCoster et al.8 rely on images of nuclear markers, which allow for straightforward image segmentation and watershed analysis algorithms to separate and identify individual cells quickly. However, these programs fail to separate cells with cytoplasmic labeling, such as those we use here. There are also a number of programs (for examples, see 9–11) that are capable of identifying and separating clusters of cells, but only in two-dimensional images. Whole-mount 3D image stacks of dissected larval zebrafish intestines with neurons fluorescently labeled for cytoplasmic markers are therefore inappropriate for either of these classes of programs. There may be other programs available that would suit our purposes, however, we decided to generate a new program that would be tailored to our specific needs. Here we describe the new program we generated and show that it accurately counts neurons labeled for expression of one or two markers in 3D image stacks of dissected zebrafish intestines. A feature of this program is that it can be trained by the user, and thus could be adapted to count other types of fluorescently-labeled cells in the intestine or other regions of whole-mount zebrafish embryos or larvae. Our counts of enteric neurons using this program reveal that even in wild types there are significantly fewer enteric neurons at the caudal end of the intestine than in the region of the mid-intestine in young larvae, and that this difference is magnified in gwr mutants. In addition, we provide counts of serotonergic enteroendocrine cells in the larval zebrafish intestine, and show that their numbers are similar in gwr mutant and wild-type larvae.
Materials and Methods
Animals
Animals were reared at 28.5°C according to standard zebrafish husbandry12 and staged by days postfertilization at 28.5°C (dpf).
Immunohistochemistry
Antibody staining for Elavl (1:10,000, Molecular Probes Inc., Eugene, OR, catalog number A-21271) and 5HT (1:10,000, Immunostar, Hudson, WI, catalog number 20080) was performed at 5 dpf as previously described.4 Secondary antigens were visualized with standard fluorophore-labeled antibodies for rabbit IgG (1:1,000, Molecular Probes Inc., catalog number A-11008) and mouse IgG (1:1,000, Molecular Probes Inc., catalog number A-11030). gwr mutants were separated from wild-type siblings at 5 dpf according to morphological characteristics.1
Manual cell counting
After immunohistochemistry, intestines were dissected and mounted in PBS on a cover slip. Z-stacks were acquired on a Zeiss LSM 5 Pascal confocal microscope and subsequently projections were made with the y-axis as turning axis, 180° projections and difference angle 2° using LSM 5 Pascal imaging software (see Supplementary Movie S1; Supplementary Data are available online at www.liebertpub.com/zeb). Counts of labeled cells were made at the level of the mid-intestine and the level of the vent (Fig. 1). In vent images, only the most aboral 200 μm were analyzed. In mid-intestine images, we examined a 200 μm region from the top of each image. We counted Elavl positive, Elavl and 5HT double positive, and 5HT positive cells, rotating the projections to ensure that we counted all cells. Counts are taken from five wild types and five mutants.
FIG. 1.
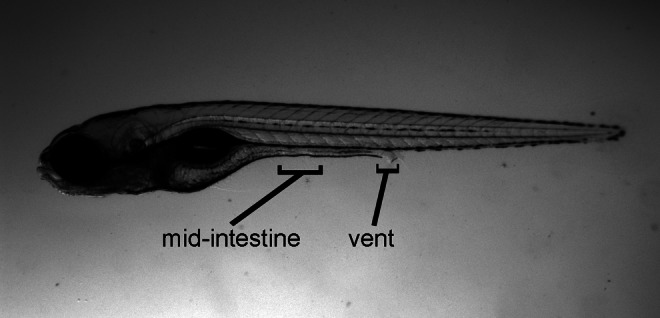
A representative 5 dpf wild-type larva showing the mid-intestine and vent regions where enteric neurons and enteroendocrine cells were counted.
Image segmentation and denoising algorithm
We identified fluorescence channels within each image as having either relatively high or low levels of background, corresponding in our case to 488 nm (5HT; Alexa Fluor 488) and 546 nm (Elavl; Alexa Fluor 546) channels, respectively. We processed each image channel separately, based on the wavelength being visualized (Fig. 2A). To reduce image noise and blurring, a pixelwise adaptive Wiener filter based on statistics estimated from a local 10-pixel neighborhood of each pixel was applied to each 2D matrix (Fig. 2B).13 These matrices corresponded to a single z-stack channel within a 3D confocal image.
FIG. 2.
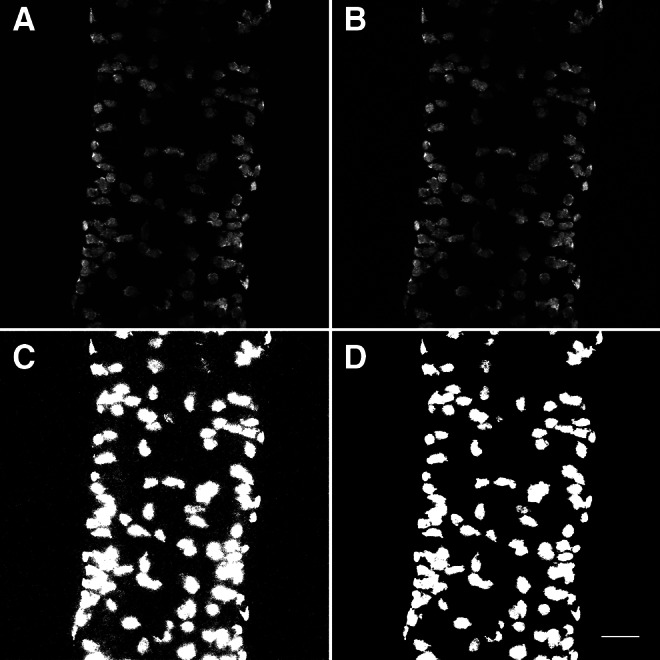
The algorithm presented here is capable of properly segmenting images of dissected whole-mount zebrafish intestines. (A) Maximum intensity projection of dissected, whole-mount, wild-type mid-intestine labeled with anti-Elavl antibody. (B) Maximum intensity projection of A after deblurring with Wiener filter. (C) Maximum intensity projection of output from Otsu's N-thresholding algorithm applied to B. (D) Maximum intensity projection of C after removal of all small pixel clusters. Scale bar=25 μm.
We thresholded each image channel using an automated determination of the threshold level14,15 (Fig. 2C). Clusters of less than 100 pixels, corresponding to noise or background signal, were deleted from both binary image channels (Fig. 2D). To merge punctate pixel clusters in the 488 nm channel, morphological opening and closing operations were performed. We found these morphological operations to be unnecessary in the 546 nm channel, because there was relatively low noise and clear labeling of complete cells in the 546 nm binary image compared to images from the 488 nm channel. Then, in both channels, morphological erosion was performed if any pixel clusters exceed six-times the volume of a single stereotyped ENS cell, or if more than 1,000 clusters remained, as these qualities indicate remaining noise or background signal in the binary images. The cells being analyzed have stereotyped sizes, thus, pixel clusters do not need to be separated to the point of containing only one cell, as cluster characteristics can be used to find the number of cells within a pixel cluster.
Cell type identification and co-expression analysis algorithm
We identified relevant cell types through analysis of binary images corresponding to individual channels from raw confocal images (Fig. 3A). All pixel clusters corresponding to enteric neurons were revealed by the 546 nm binary image (Fig. 3B). We constructed an image (C) consisting of pixels that co-localized in both the 488 nm and 546 nm binary images such that
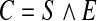 |
FIG. 3.
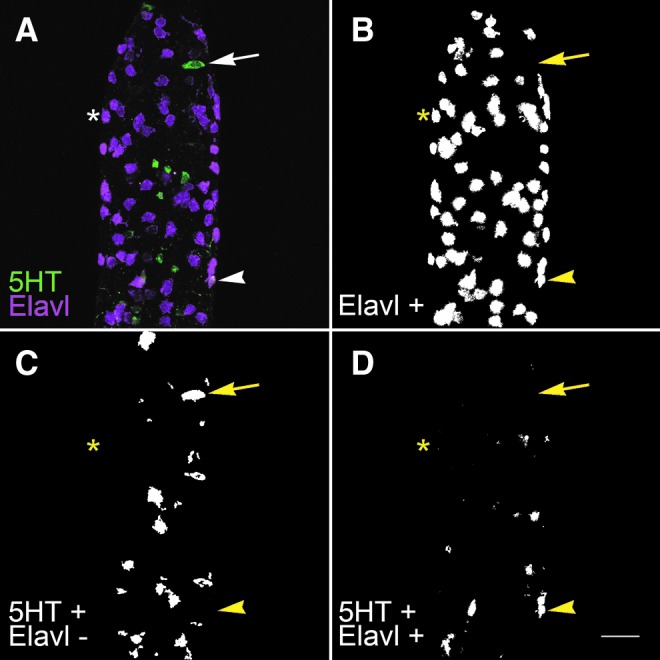
LsmNoDesktopSegment is capable of identifying relevant cell types without user input. (A) Maximum intensity projection of dissected, whole-mount, wild-type intestine at the level of the vent labeled with anti-Elavl (green) and anti-5HT (magenta) antibodies. (B) Binary image of all Elavl labeling revealing all enteric neurons shown in A. (C) Binary image of 5HT-positive clusters that do not co-localize with pixels in B, thus revealing only the serotonergic enteroendocrine cells seen in A. (D) Binary image of 5HT-positive clusters that contain pixels from B, which corresponds to serotonergic enteric neurons shown in A. (B–D) Images contain small artifacts that were later ignored during the counting process, allowing only true cells to be counted. Asterisk, nonserotonergic neuron (5HT-, Elavl+); arrowhead, serotonergic neuron (5HT+, Elavl+); arrow, serotonergic enteroendocrine cell (5HT+, Elavl-). Aboral end of the vent is visible at the top of the image. Scale bar=25 μm.
where S is the set of all pixels clusters in the 488 nm (5HT) binary image, E is the set of all pixel clusters in the 546 nm (Elavl) binary image, and ∧ represents the operation of identifying all pixels that located at the same coordinates in each image (Fig. 3D). To identify clusters that were unique to the 546 nm channel, we constructed a binary image (S’) such that
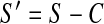 |
where all co-localized pixel clusters were removed from the 488 nm (5HT) binary image (Fig. 3C). Relevant cell types were thereby represented by binary images E, C, and S’, corresponding to enteric neurons, serotonergic neurons, and serotonergic enteroendocrine cells, respectively (Fig. 3B–D).
Cell cluster estimation and counting algorithm
We cropped raw and binary images to a region of interest (Fig. 4A and B). We then individually examined each pixel cluster using the binary image as a colored mask over the appropriate raw image. We viewed maximum intensity projections of only the image layers where the cluster of interest appeared. The cluster in question was given a red color, while all other visible clusters were colored blue (Fig. 4C).
FIG. 4.
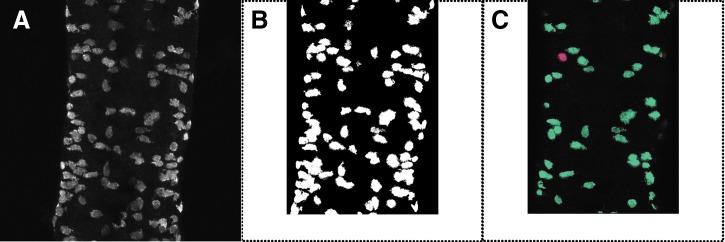
The algorithm presented here allows for identification of imaged cells within a specified region of interest. (A) Maximum intensity projection of dissected, whole-mount, wild-type mid-intestine labeled with anti-Elavl antibody. (B) Maximum intensity projection of cropped binary image corresponding to A. (C) Composite image using colorized B masking A projected through layers where the red cluster of interest occurs.
Data from previous analyses greatly informed the cluster size estimation and counting processes, because of the stereotyped size of cells being analyzed. If no previous cluster data was loaded, clusters were initially assumed to be single cells. If we loaded data from a previous analysis, we then approximated the probability of a given cluster being a single cell, or up to four closely joined cells. This estimation was made possible by comparing characteristics for each pixel cluster to characteristics of clusters containing different numbers of cells that had been previously processed. We automatically ignored any pixel clusters that were smaller than 85% of the smallest previously encountered cluster that we had identified as a cell. In addition, we ignored clusters if the raw image intensity in that region was lower than 75% of the least intense previously encountered cluster that we previously identified as a cell. We examined each cluster for its volume, maximum cross-sectional area, bounding box volume, and 2D bounding box area, as these four simple criteria accurately stratified clusters into one- through four-cell groups. For each cluster, we performed a z-test for each of these criteria, using data from previous analyses as reference distributions. The z-test probability (z) is given by
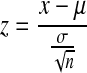 |
where x is the sample cluster value for a given characteristic, μ is the mean characteristic value for a given cluster size population, σ is the standard deviation of this population, and n is the population size. We then compared the products (p) of all z-test probabilities for each possible cluster size
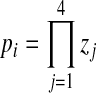 |
where zj is the z-test probability with the value of j referring to either cluster volume, maximum cross-sectional area, bounding box volume, or 2D bounding box area, and the value of i referring to the putative number of cells in a cluster.
The maximum value of p, corresponding with the most probable identity, became our initial guess (P) such that
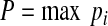 |
where pi is the z-test probability product for a given cluster size. We then either approved or denied the accuracy of P for each cluster. After all clusters were evaluated, we retrieved cell counts by calculating the sum of each cluster type for each binary image. Cluster data for each analysis is also saved and appended to previous cluster datasets, to assist with further analyses.
Computer-assisted cell counting
Computer-assisted cell counts were taken from the same z-stacks used for manual cell counting. All of the programs described in this article were written in MATLAB(v2012a). The computer-assisted cell counting programs described in this paper are available for download at: http://uoneuro.uoregon.edu/eisen/
Hardware and software
All programs were successfully tested on Windows 7 64-bit and Ubuntu 12.04 LTS laptop computers with Intel core i5-m430 processors and 4GB RAM in MATLAB(v2012a). Images were also processed on Linux supercomputer nodes featuring 12-core CPUs and 72GB RAM, running MATLAB(v2011b).
Different intestinal cell types can be accurately identified and counted by the new program
The LsmNoDesktopSegment program is able to rapidly and properly segment an entire directory of images with no user input necessary (Fig. 5). By implementing Otsu's image segmentation algorithm14,15 and simple binary image processing techniques, cells with fluorescent cytoplasmic labeling are separated from the background. LsmNoDesktopSegment is also capable of revealing specific cell types by comparing the segmented images for each fluorescent label (Fig. 5). All ENS neurons are Elavl positive,3 thus the Elavl and 5HT double positive cells are ENS neurons. The cells positive for only 5HT have previously been shown to be a subset of enteroendocrine cells in the intestinal epithelium.16 By simply identifying the marker shown in a given image channel, LsmNoDesktopSegment is capable of identifying these relevant cell types. All cell-like clusters of pixels in the Elavl binary image are identified as neurons, and then connected pixel clusters in the 5HT binary image are segregated by the presence or absence of co-localization with Elavl pixels. Ultimately, binary images for all neurons (Fig. 3B), enteroendocrine cells (Fig. 3C), and 5HT-positive neurons (Fig. 3D) are produced. These cell type-specific images are then passed to the LsmCounter program, where pixel clusters are finally identified as either cells, groups of cells, or background signal.
FIG. 5.
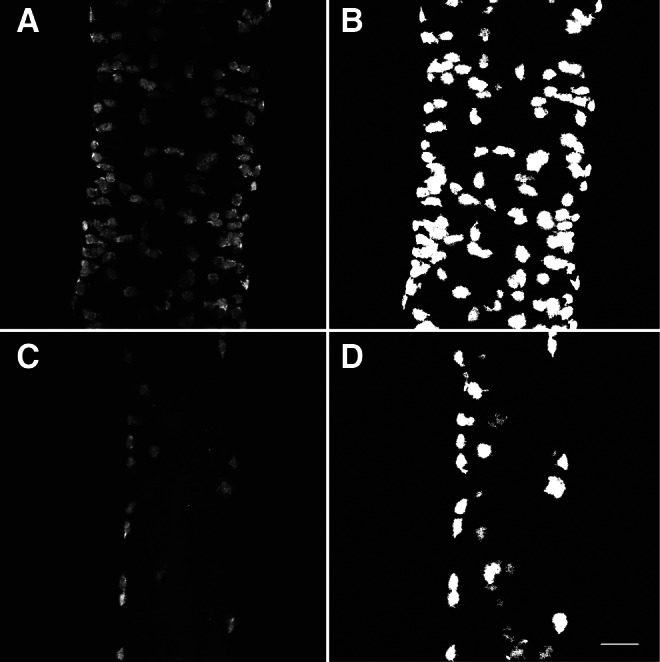
Segmentation of image stacks reveals isolated cells in dissected, whole-mount, zebrafish intestines. (A) Maximum intensity projection of dissected, whole-mount, wild-type mid-intestine labeled with anti-Elavl antibody. (B) Maximum intensity projection of the binary image stack produced by Otsu's N-thresholding algorithm applied to A. (C) Maximum intensity projection of whole-mount gutwrencher mid-intestine labeled with anti-Elavl antibody. (D) Maximum intensity projection of the binary image stack produced by Otsu's N-thresholding algorithm applied to C. Scale bar=25 μm.
LsmCounter saves descriptive data of each cell and cell cluster that it successfully counts, and these data can then be used to identify cells more efficiently. During the initial operation of LsmCounter, the false detection rate of cells is high, varying with the complexity of an image. On an initial run of the image presented in Figure 3A, approximately 33% of detections were correct, however analyzing the image with data from only one previous run raised the correct detection rate to 65%. In both cases, the final output counts were the same due to user guidance. During the initial run, the user effectively trains the program by indicating which pixel clusters are not cells. In subsequent rounds of analysis, LsmCounter ignores any pixel clusters of a size that is below a threshold determined by the smallest user-defined cell that was previously encountered. However, LsmCounter is designed to err on the side of false positives rather than false negatives, so that no real cells are missed, and because the user is always easily capable of denying a detection event.
Results
The new program accurately identified and counted cytoplasmically-labeled cells
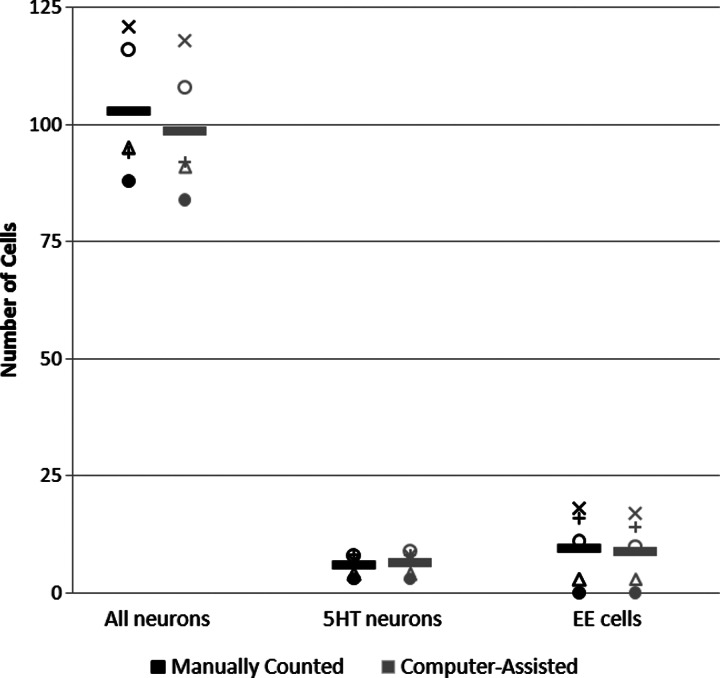
To be useful, our new program must be able to count cytoplasmically-labeled enteric neurons rapidly and accurately. To learn whether this was the case, we compared manual counts of enteric neurons from 3D confocal image stacks of dissected intestines with counts made by our new program. We found no statistical differences between the numbers of labeled cells detected by either manual or computerized means (Table 1 and Fig. 6). On average, manual and computer-assisted cell counts differed by less than one cell. Relative differences between cell count means from the two different counting methods did not differ significantly (p>> 0.05), as determined by two-tailed unpaired Student's t-test. We also found that LsmNoDesktopSegment counted cells much faster than they could be counted manually. Whereas manual counting takes approximately 5 minutes per dissected intestine for a researcher experienced in this method, LsmNoDesktopSegment can count the dissected intestine in 30 seconds or less when run on a standard laptop computer.
Table 1.
Manual and Computer-Assisted Cell Counts Are Not Statistically Different
| |
Manually counted cells |
LsmCounter output |
|
|
||
|---|---|---|---|---|---|---|
| Intestinal region | Mean | Std. dev. | Mean | Std. dev. | Relative difference | p value |
| Wild-type mid-intestine | ||||||
| All neurons | 102.8 | 14.7 | 98.6 | 14.0 | −4.2 | 0.655 |
| 5HT neurons | 6.0 | 2.3 | 6.4 | 2.7 | 0.4 | 0.809 |
| EE cells | 9.6 | 7.9 | 8.8 | 7.2 | −0.8 | 0.871 |
| Wild-type vent | ||||||
| All neurons | 65.0 | 10.0 | 66.8 | 10.1 | 1.8 | 0.784 |
| 5HT neurons | 1.6 | 0.5 | 1.8 | 0.8 | 0.2 | 0.667 |
| EE cells | 10.4 | 5.3 | 9.4 | 4.9 | −1.0 | 0.766 |
Cell counts were performed on 200 μm lengths of dissected intestine. 5HT-positive, Elavl-negative cells were counted as enteroendocrine cells (EE cells). Relative difference refers to the arithmetic difference between mean cell counts. P values were calculated via two-tailed unpaired Student's t-test. The overall mean relative difference is −0.6 cells. Abbreviations: Std.Dev., standard deviation.
FIG. 6.
The data for the wild-type mid-intestine in Table 1 are replotted here to show the high degree of correspondence between manual counts and computer-assisted counts. Each symbol represents an individual animal and the bars represent the mean. Variability between animals is discussed in the text.
gutwrencher mutants have fewer enteric neurons than wild types and this phenotype is more severe in the caudal intestine
The enteric neuron population of 5 dpf gutwrencher mutants is dramatically lower than that of wild-type siblings, with a greater difference in the vent than in the mid-intestine (Table 2). 5 dpf gutwrencher mutants exhibit nearly a 10-fold decrease of mid-intestine enteric neurons and a 6.4-fold decrease in 5HT-positive enteric neurons relative to their wild-type siblings. In the vent region, these differences are higher, with an over 50-fold decrease in enteric neurons in gutwrencher mutants and a complete absence of 5HT-positive enteric neurons. All of these observed trends are statistically significant, as determined by two-tailed unpaired Student's t-test (p<0.05).
Table 2.
gutwrencher Mutants Have Fewer Enteric Neurons Than Wild-Type Siblings
| |
|
Mean cell counts |
Fold change (p value) |
||||
|---|---|---|---|---|---|---|---|
| Intestinal region | Genotype | All neurons | 5HT neurons | EE cells | All neurons | 5HT neurons | EE cells |
| Mid-intestine | Wild-type | 98.6 | 6.4 | 8.8 | 9.5 (1.2e−6) | 6.4 (0.006) | 1.0 (0.959) |
| gutwrencher | 10.4 | 1.0 | 8.6 | ||||
| Vent | Wild-type | 66.8 | 1.8 | 9.4 | 56 (5.8e−7) | undef. (0.001) | 1.3 (0.665) |
| gutwrencher | 1.2 | 0.0 | 7.4 | ||||
Cell counts were performed on 200 μm lengths of dissected intestine. 5HT-positive, Elavl-negative cells were counted as enteroendocrine cells (EE cells). P values were calculated via two-tailed unpaired Student's t-test. Undef. represents the undefined value of any value divided by zero.
In both 5 dpf wild types and gutwrencher mutants, enteric neuron populations differ significantly along the length of the intestine (Table 3). Wild types exhibit a 1.5-fold reduction of enteric neurons in the vent region compared to the mid-intestine, and a roughly 3.6-fold decrease in 5HT-positive enteric neurons. In gutwrencher mutants, this reduction is exaggerated to an 8.7-fold reduction of enteric neurons and a complete lack of 5HT-positive neurons in the vent region.
Table 3.
gutwrencher Mutants Exhibit More Dramatic Reduction of Enteric Neurons in Vent Versus Mid-Intestine, Compared to Wild-Type Siblings
| |
Fold change (p value) |
||
|---|---|---|---|
| Genotype | All neurons | 5HT neurons | EE cells |
| Wild-type | 1.5 (0.003) | 3.6 (0.007) | 1.1 (0.882) |
| gutwrencher | 8.7 (0.014) | undef. (0.233) | 1.2 (0.789) |
Mean cell counts are presented in Table 2. P values were calculated via two-tailed unpaired Student's t-test. Undef. represents the undefined value of any value divided by zero.
Enteroendocrine populations appear constant along the intestine and do not differ between wild types and gutwrencher mutants
Enteroendocrine cell numbers do not differ significantly in any of our analyses. When gutwrencher mutants were compared to wild-type siblings, we saw no change in mid-intestine enteroendocrine cells and a 1.3 fold change in the vent region that is not statistically significant (Table 2). We also did not observe any significant differences between serotonergic enteroendocrine cell populations in the mid-intestine and vent regions of wild-type or gutwrencher mutant larvae (Table 3).
Discussion
We generated a new MATLAB-based program that enabled us to compare the number of cytoplasmically-labeled fluorescent enteric neurons in different intestinal regions and between wild types and mutants. The LsmNoDesktopSegment program is capable of rapidly processing 3D images of whole mount zebrafish intestines and LsmCounter is capable of assisting the user in quantifying the number of cells with a given label. Image processing with LsmNoDesktopSegment requires up to 30 seconds per image file when run on a standard laptop computer, and is easily capable of being run on a distributed computing network for even faster processing. The semi-guided nature of the LsmCounter program allows for oversight over the cell counting process, which means that the runtime is dictated by the researcher, image quality, and the number of pixel clusters in each binary image. The entire computer-assisted counting process typically requires less than 4 minutes per image stack, for a user familiar with the software. The cell counting algorithm presented here is also capable of assessing pixel clusters and estimating the number of cells in an image in approximately 30 seconds per image, though with reduced accuracy, due to the lack of user correction. Also, researchers may be reluctant to adopt fully-automated cell counting software due to a lack of transparency in the counting process, so we choose to maintain user oversight in the counting process, and thereby maintain maximum confidence in the cell counts produced.
In the process of characterizing intestinal 5HT and Elavl expression with LsmCounter, we demonstrated that the program is not limited to counting enteric neurons. Cells in the zebrafish intestine that express 5HT but do not express Elavl have previously been identified as enteroendocrine cells of the intestinal epithelium.16,17 Therefore, by simply subtracting the number of 5HT and Elavl co-expressing cells from the total 5HT-positive cells in a given image stack, we quantified serotonergic enteroendocrine cells in the mid-intestine and vent of wild types and gutwrencher mutant zebrafish (Fig. 3).
The techniques described here are likely to be easily adapted for DIC microscopy images. Because the image segmentation mechanics of LsmNoDesktopSegment simply require regions of high contrast, fluorescent images are unnecessary. LsmCounter tracks and counts objects that are brighter than the background, but this aspect of the program could be changed easily. Alternatively, inverted DIC images could be processed as fluorescent images.
Our results appear to suggest that the phenotype of gutwrencher mutants is more dramatic than was initially appreciated. gutwrencher mutants were previously described as exhibiting 3.5-fold fewer enteric neurons and 6-fold fewer 5HT-positive enteric neurons.1 Here we describe a similar 6.4-fold decrease in 5HT-positive enteric neurons, but total enteric neurons appear to be nearly 9.5-fold fewer in the mid-intestine and over 50-fold fewer in the vent of gutwrencher mutants (Table 2). Several circumstances may contribute to differences between the fold change of enteric neurons presented here and those previously described. In the current analysis, dissected whole-mount intestines of 5 dpf larvae were examined, whereas previous cell counts were performed on 4 dpf sectioned larvae, counted in alternating sections to prevent double counting.1 Enteric neurons are differentiating through this stage of development, thus our results suggest that the gutwrencher mutation affects differentiation of enteric neurons in the mid-intestine. Future studies will address whether this results from decreased proliferation of enteric progenitors. Our studies also raise the possibility that gutwrencher affects migration of enteric progenitors, because we see significantly fewer neurons at the caudal end of the intestine than in the mid-intestine. However, this could also result from depletion of the progenitor pool, something we can address in future studies.
Our results provide evidence that the serotonergic enteroendocrine population of gutwrencher mutants and wild-type siblings do not differ significantly (Table 2). Furthermore, serotonergic enteroendocrine cell numbers appear to remain constant between the mid-intestine and vent region (Table 3). The population of serotonergic enteroendocrine cells in 5 dpf wild types has previously been described as ranging from 10 to 18 cells in a 3 somite length region of the intestine immediately rostral to the vent,16 which translates to about 3–5 serotonergic enteroendocrine cells per 100 μm of intestinal length. Our numbers are very similar, at 3.7–4.7 serotonergic enteroendocrine cells per 100 μm of intestinal length. These results suggest that gutwrencher mutant phenotype does not affect the population of serotonergic enteroendocrine cells, further supporting the idea that gutwrencherb1088 is an ENS-specific gene. A caveat of this conclusion is that our results do not show whether other subpopulations of enteroendocrine cells are affected, nor do they rule out the possibility that enteroendocrine cell fate is altered. These questions can be addressed in future experiments designed to examine enteroendocrine cells in more detail in both wild types and gutwrencher mutants.
A possible limitation for furthering our understanding of enteric mutant phenotypes is that cells cannot easily be counted in the anterior intestine. This is not because our program cannot handle the counting, but rather because the tissue architecture prevents sufficient resolution on our confocal microscope. However, other microscopy methods, such as light sheet microscopy,18 should be able to solve this problem.
A striking observation was the variability in the number of specific types of enteric cells, even for animals of the same genotype. This is graphically illustrated in Figure 6. We believe that this variability is real, because we and others have found similar variability in the numbers of enteric neurons,1 the numbers of serotonin-positive enteric neuron,1 the numbers of serotonin-positive enteroendocrine cells,16 and the numbers of goblet cells,16 whether these cells were counted in whole mount or in sections. This variability calls into question the sensitivity of any counting method for detecting subtle phenotypic differences between wild types and mutants. For example, screening for enteric neuron mutants using a stereomicroscope, as in our initial screen,1 would likely overlook mutants with very slight decreases in enteric neurons. In any case, as in other situations in which there is variability, increasing the number of animals should provide more sensitivity in detection of subtle phenotypes.
Supplementary Material
Acknowledgments
We thank Raghuveer Parthasarathy for comments on the manuscript and algorithm, the University of Oregon MATLAB Club for comments on the program, and Dayna Lamb and the University of Oregon Zebrafish Facility for animal husbandry. Supported by NIH grants HD22486 (JSE) and HS007348 (LWS), a postdoctoral fellowship from the German Academic Exchange Service (DAAD) (JG), and a John Simon Guggenheim Memorial Foundation Fellowship (JSE).
Disclosure Statement
No competing financial interests exist.
References
- 1.Kuhlman J. Eisen JS. Genetic screen for mutations affecting development and function of the enteric nervous system. Devel Dyn. 2007;236:118–127. doi: 10.1002/dvdy.21033. [DOI] [PubMed] [Google Scholar]
- 2.Pietsch J. Delalande JM. Jakaitis B. Stensby JD. Dohle S. Talbot WS, et al. Lessen encodes a zebrafish trap100 required for enteric nervous system development. Development. 2006;133:395–406. doi: 10.1242/dev.02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepherd I. Eisen J. Development of the zebrafish enteric nervous system. Methods Cell Bio. 2011;101:143–160. doi: 10.1016/B978-0-12-387036-0.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uyttebroek L. Shepherd IT. Harrisson F. Hubens G. Blust R. Timmermans JP, et al. Neurochemical coding of enteric neurons in adult and embryonic zebrafish (Danio rerio) J Comp Neurol. 2010;518:4419–4438. doi: 10.1002/cne.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abercrombie M. Estimation of nuclear population from microtome sections. Anatom Record. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- 6.Williams RW. Rakic P. Three-dimensional counting: An accurate and direct method to estimate numbers of cells in sectioned material. J Comp Neurol. 1988;278:344–352. doi: 10.1002/cne.902780305. [DOI] [PubMed] [Google Scholar]
- 7.Oberlaender M. Dercksen VJ. Egger R. Gensel M. Sakmann B. Hege HC. Automated three-dimensional detection and counting of neuron somata. J Neurosci Methods. 2009;180:147–160. doi: 10.1016/j.jneumeth.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 8.DeCoster M. Maddi S. Dutta V. Microscopy and image analysis of individual and group cell shape changes during apoptosis. In: Méndez-Vilas A, editor; Díaz J, editor. Microscopy: Science, Technology, Applications and Education. Formatex; Spain: 2010. pp. 836–843. [Google Scholar]
- 9.Srinivasa G. Fickus MC. Guo YS. Linstedt AD. Kovacevic J. Active mask segmentation of fluorescence microscope images. Ieee T Image Process. 2009;18:1817–1829. doi: 10.1109/TIP.2009.2021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang QL. Niemi J. Tan CM. You LC. West M. Image degmentation and fynamic lineage analysis in single-cell fluorescence microscopy. Cytom Part A. 2010;77A:101–110. doi: 10.1002/cyto.a.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mihailescu M. Scarlat M. Gheorghiu A. Costescu J. Kusko M. Paun IA, et al. Automated imaging, identification, and counting of similar cells from digital hologram reconstructions. Appl Optics. 2011;50:3589–3597. doi: 10.1364/AO.50.003589. [DOI] [PubMed] [Google Scholar]
- 12.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th. University of Oregon Press; Eugene, OR: 2000. [Google Scholar]
- 13.Lim JS. Two-Dimensional Signal and Image Processing. Prentice Hall; Englewood Cliffs, NJ: 1990. [Google Scholar]
- 14.Otsu N. Threshold selection method from gray-level histograms. Ieee T Syst Man Cyb. 1979;9:62–66. [Google Scholar]
- 15.Garcia D. Fenech M. Qin Z. Soulez G. Cloutier G. Signal losses with real-time three-dimensional power Doppler imaging. Ultrasound Med Biol. 2007;33:1632–1639. doi: 10.1016/j.ultrasmedbio.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Bates JM. Mittge E. Kuhlman J. Baden KN. Cheesman SE. Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Devel Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 17.Olsson C. Holmberg A. Holmgren S. Development of enteric and vagal innervation of the zebrafish (Danio rerio) gut. J Comp Neurol. 2008;508:756–770. doi: 10.1002/cne.21705. [DOI] [PubMed] [Google Scholar]
- 18.Taormina MJ. Jemielita M. Stephens WZ. Burns AR. Troll JV. Parthasarathy R, et al. Investigation of bacterial-animal symbioses with light sheet microscopy. Biol Bull. 2012;223:7–20. doi: 10.1086/BBLv223n1p7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.