Abstract
Zebrafish (Danio rerio) have been proposed as a possible model organism for nutritional physiology. However, this potential has not yet been realized and studies on the field remain scarce. In this work, we investigated in this species the effect of a single meal as well as that of an increase in the ratio of dietary carbohydrates/proteins on the postprandial expression of several hepatic and muscle metabolism-related genes and proteins. Fish were fed once either a commercial diet (experiment 1) or one of two experimental diets (experiment 2) containing different protein and carbohydrate levels after 72 h of starvation. Refeeding induced the postprandial expression of genes of glycolysis (GK, HK1) and lipogenesis (FAS, G6PDH, ACCa) and inhibited those of gluconeogenesis (cPEPCK) and beta-oxidation (CPT1b) in the viscera. In the muscle, refeeding increased transcript levels of myogenesis (Myf5, Myogenin), inhibited those of Ub-proteasomal proteolytic system (Atrogin1, Murf1a, Murf1b), and induced the activation of key signaling factors of protein synthesis (Akt, 4EBP1, S6K1, S6). However, diet composition had a low impact on the studied factors. Together, these results highlight some specificity of the zebrafish metabolism and demonstrate the interest and the limits of this species as a model organism for nutritional physiology studies.
Introduction
The zebrafish (Danio rerio) is one of the most widely used animal models for developmental research, and it is now becoming an attractive model for drug discovery and toxicological screening.1 The completion of sequencing the zebrafish genome and the availability of full-length cDNAs and DNA microarrays for expression analysis, in addition to techniques for generating transgenic lines and targeted mutations, have made the zebrafish model even more attractive to researchers. Obviously, it has been found that zebrafish might be an interesting model organism in nutrition research.2 However, this potential has not yet been realized, and to our knowledge, studies on the field remain scarce and concern mainly the effect of long-term food deprivation and/or refeeding on hepatic, brain, and skeletal muscle transcriptomes.3,4 Recently, Robison et al. examined for the first time the effect of manipulating dietary macronutrient composition on hepatic gene expression in zebrafish.5 They demonstrated that long-term (12 weeks) manipulation of dietary carbohydrate levels had a significant effect on the pattern of expression of several hepatic genes during the postabsorptive period (24 h after the last meal). As such, no data is yet available for this species on the effect of macronutrient composition of the diet on the regulation of metabolism-related factors (genes and/or signaling pathways) during the postprandial period.
The regulation of intermediary metabolism often depends on the crosstalk between nutritional and hormonal signals. A well-known crosstalk is that committed by insulin and dietary amino acids. Amino acids are not only considered as precursors for the synthesis of proteins and other N-containing compounds. They are involved in the regulation of major metabolic pathways6 and are thus considered as signaling molecules. Amino acids regulate protein synthesis by activating the mammalian target of rapamycin/p70 S6 kinase transduction pathway, together with insulin.7,8 Recent studies indicate that amino acids play also an important role in controlling gene expression.9 In this regard, increasing evidence has emerged in recent years to show that amino acid availability affects the expression of genes involved in many cell functions, including lipid and glucose metabolisms,10–12 autophagy,13 myogenesis,14,15 and stress response.16–18 However, excessive levels of amino acids have also been observed to promote insulin resistance with underlying negative effects at the metabolic level.19
Dietary carbohydrates have also been observed to regulate the expression of several genes involved in both intermediary metabolism and growth-related factors.20 In this regard, glucose should not be uniquely considered as an energy fuel but also as a signaling molecule acting in synergy with hormones. Elevated glucose concentrations have thus been associated with an increase of the expression of several genes coding for enzymes needed for de novo lipogenesis such as acetyl CoA carboxylase (ACC), fatty acid synthase (FAS), and liver pyruvate kinase (L-PK),21–24 but also that of several myogenic regulatory factors coding genes.25 In recent years, great progress has been achieved in understanding the molecular mechanisms that couple glucose availability to gene transcription.20 However, these mechanisms have not been fully characterized to date and remain an important field of research for study of the regulation of physiological functions of individuals and/or animals living under conditions of restricted, imbalanced, or excessive food intake.
The goal of the study presented here was to make inroads in the understanding of the effect of manipulating dietary macronutrient composition on the postprandial expression of several key genes and proteins related to hepatic and muscle metabolism in zebrafish. With this goal, two experiments were carried out. In the former, we characterized the short-term postprandial response of metabolism-related genes and proteins to a single meal. Taking advantage of results obtained in the first experiment, we then monitored the effects of an increase in the ratio of dietary carbohydrates/proteins on the postprandial expression of the metabolism-related factors.
Materials and Methods
The experiments were carried out in accordance with French legislation governing the ethical treatment of animals, and the investigators were certified by the French government to carry out animal experiments.
Animals and experimental procedures
Two-month-old mixed-sex zebrafish (weights ranging from 200 to 250 mg) were used in this study. During the acclimation period, fish were maintained in three 20 L tanks (100 fish/tank) at 28°C in a 10 h:14 h light:dark photoperiod and fed ad libitum twice daily with a commercial diet (Gemma 300; crude protein, 61.41% dry matter; crude fat, 17.11% dry matter; gross energy, 20.98 kJ/g dry matter). In the single-meal experiment (experiment 1), fish were food deprived for 72 h, refed ad libitum, and sampled at 0.5, 2, 6, and 24 h after food administration. To limit handling stress in our successive samplings, the required number of fish (n=6, each sample corresponding to three fish) was withdrawn from one of the three tanks at each sampling time. As control, a group of fish (n=6, each sample corresponding to three fish) were sampled before refeeding. In the second experiment, fish were left unfed for 72 h and refed ad libitum with one of the two semipurified diets of high (H) or low (L) levels of protein (P) or carbohydrates (C) (high protein–low carbohydrates [HPLC] and low protein–high carbohydrates [LPHC], respectively) (Table 1). The amount of feed distributed per tank was measured to ensure that feed intake was similar between diets (around 3% of their body weight). The required number of fish (n=3 samples of three fish each) was sampled from each tank before feeding as well as 30 min, 2 h, 6 h, and 24 h after the meal. Fish were sacrificed by an overdose of isoeugénol (PHYTOSUNaroms; Omega Pharma). The viscera (liver, gastrointestinal tract, and diffuse pancreatic cells) and the muscle (posterior trunk without head) from each fish were dissected and immediately frozen in liquid nitrogen and kept at −80°C.
Table 1.
Composition of the Diets
| HPLC diet | LPHC diet | |
|---|---|---|
| Ingredients (g.kg−1) | ||
| Fish meala | 670 | 280 |
| Wheatb | 80 | 420 |
| Gelatinized starchc | 150 | 150 |
| Sunflower oild | 30 | 50 |
| Fish oile | 0 | 30 |
| Mineral premixf | 30 | 30 |
| Vitamin premixg | 30 | 30 |
| Binderh | 10 | 10 |
| Analytical composition | ||
| Dry matter (%) | 93.09 | 93.60 |
| Crude protein (% DM) | 52.88 | 27.81 |
| Crude fat (% DM) | 10.17 | 11.92 |
| NFEi | 29.95 | 53.27 |
| Gross energy (kJ/g DM) | 20.87 | 20.34 |
Sopropêche.
Sudouest aliment.
Roquette frères.
Lesieur.
Feedoil (North Sea Fish Oil, Sopropèche).
Mineral mixture (g or mg/kg diet): calcium carbonate (40% Ca), 2.15g; magnesium oxide (60% mg), 1.24 g; ferric citrate, 0.2 g; potassium iodide (75% I), 0.4 mg; zinc sulfate (36% Zn), 0.4 g; copper sulfate (25% Cu), 0.3 g; manganese sulfate (33% Mib), 0.3 g; dibasic calcium phosphate (20% Ca, 18%P), 5 g; cobalt sulfate, 2 mg; sodium selenite (30% Se), 3 mg; KCl, 0.9 g; NaCl, 0.4 g (Unité de Préparation des Aliments Expérimentaux).
Vitamin mixture (IU or mg/kg diet): DL-a tocopherol acetate, 60 IU; sodium menadione bi sulfate, 5 mg; retinyl acetate, 15,000 IU; DL-cholecalciferol, 3000 IU; thiamin, 15 mg; riboflavin, 30 mg; pyridoxine, 15 mg; B12, 0.05 mg; nicotinic acid, 175 mg; folic acid, 500 mg; inositol, 1000 mg; biotin, 2.5 mg; calcium panthotenate, 50 mg; choline chloride, 2000 mg (Unité de Préparation des Aliments Expérimentaux).
Sodium alginate GF 150 (Louis Francxois Exploitation)
Nitrogen-free extract (carbohydrate): 100 (crude protein+crude fat+ash).
DM, dry matter; HPLC, high protein–low carbohydrate; LPHC, low protein–high carbohydrate.
Chemical composition of the diets
The chemical composition of the diets was analyzed using the following procedures: dry matter was determined after drying at 105°C for 24 h, protein content (N×6.25) was determined by the Kjeldahl method after acid digestion,4 fat by petroleum ether extraction (Soxtherm), and gross energy in an adiabatic bomb calorimeter (IKA).
mRNA level analysis by real-time quantitative reverse transcriptase–polymerase chain reaction
mRNA level analysis was performed in fasted, 6- and 24-h refed fish, based on previous data identifying this time points as relevant for examining the postprandial response of genes to meal feeding in zebrafish.3 Total RNA was extracted from zebrafish viscera and muscle using TRIzol reagent (Invitrogen). Total RNA (1 μg) was reverse transcribed into cDNA with the Superscript III RNAse H Reverse Transcriptase kit (Invitrogen) using oligo(dT) primers. mRNA levels were determined by real-time quantitative reverse transcriptase–polymerase chain reaction (q-PCR) using the iCycler iQ (Bio-Rad). Analyses were performed on 10 μL of the diluted cDNA using the iQ SYBR Green Supermix (Bio-Rad), in a total PCR reaction volume of 25 μL, containing 200 nM of each primer. Primers were designed to overlap an intron if possible (Primer3 software) using sequences from the NCBI or Ensembl databases (Table 2). Relative quantification of the target gene transcript was done using 18S ribosomal RNA level, which was stably expressed in this experiment. Thermal cycling was initiated with incubation at 95°C for 90 s using hot-start iTaq DNA polymerase activation; 35 steps of PCR were performed, each one consisting of heating at 95°C for 20 s for denaturing, at 55°C for 10 s for primers annealing, and at 72°C for 10 s for the extension step. After the final PCR cycle, melting curves were systematically monitored (55°C temperature gradient at 0.5°C/s from 55 to 94°C) to ensure that only one fragment was amplified. Samples without RT and samples without RNA were run for each reaction as negative controls. For each sample, two RTs were performed, and for each RT two PCRs were done.
Table 2.
Primers for Real-Time Polymerase Chain Reaction
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Accession no.a |
|---|---|---|---|
| GKb | GCTGTGAAGTCGGCATGATA | CTTCAACCAGCTCCACCTTAC | BC122359.1 |
| HK1 | ACTTTGGGTGCAATCCTGAC | AGACGACGCACTGTTTTGTG | BC067330.1 |
| HK2b | CAACAACGCCACCGTCAAAATG | GCCCAGATCCAATGCCAAGAAA | NM_213066.1 |
| L-PK | TCCTGGAGCATCTGTGTCTG | GTCTGGCGATGTTCATTCCT | BC152219.1 |
| M-PK | TGGGCTTATTAAGGGCAGTG | TGCACCACCTTTGTGATGTT | BC165710.1 |
| cPEPCK | ATCACGCATCGCTAAAGAGG | CCGCTGCGAAATACTTCTTC | NM_214751.1 |
| mPEPCK | TGCCTGGATGAAATTTGACA | GGCATGAGGGTTGGTTTTTA | NM_213192.1 |
| G6Pase | TCACAGCGTTGCTTTCAATC | AACCCAGAAACATCCACAGC | BC164161.1 |
| FAS | GAGGGAAATCCGACAGTTGA | GACTCCAACAGAGCCTGAGC | XM_001923608.3 |
| G6PDH | CGTCTTTTGTGGCAGTCAGA | TGATGGGTGGTGTTTTCTCA | XM_694076.5 |
| SREBP1 | GAGCCACGACACAATCCTTC | CTGCAGCCAATTAATGACCA | NM_001105129.1 |
| ACCa | CACGATGCTCAGTTGTGTCC | CCATGACAGTGGACTTGACG | XM_001919780.3 |
| ACCb | CTTCAAAGGAAAGCAGACCG | TATGAGGGCAAATGAGAGGC | XM_678989.5 |
| CPT1ab | GCATTGATCGGCATCTCTTT | CAGTCTCCAAGGCTCTGACA | NM_001044854.1 |
| CPT1b | TCCATCTGGGATACACAGCA | CGATTCCCTTTGCAATCCTA | ENSDART00000083421 |
| MyoD | GGAGCGAATTTCCACAGAGACT | GTGCCCCTCCGGTACTGA | AF318503.2 |
| Myf5 | CCTCCCCAAGGTAGAGATCC | GTTCTCCACCTGTTCCCTGA | BC165074.1 |
| PCNA | GCCTTGGCACTGGTCTTTG | TGCCAAGCTGCTCCACATC | BC064299.1 |
| Myogenin | GGCCGCTACCTTGAGAGAGA | GAGCCTCAAAGGCCTCGTT | BC078421.1 |
| Atrogin1 | GACTTCTGCAGTGCCATCAA | GCCACTCCACTCAGAGAAGG | NM_200917.1 |
| Murf1a | TGTGAGACCCAAATGTTTGAA | TCCACTGAATTATCCAATGAAAA | NM_001002133.1 |
| Murf1b | CACCAACATGGACATTCAGC | TAGCACATCCTCGACACAGG | NM_201095.1 |
| LC3B | GTGGAGGATGTACGGCTGAT | GCAGTTGCTTCTCTCCCTTG | BC155206.1 |
| atg4b | GTCTGGATTTTGGGAAAGCA | CACCAATTGGCTGGAAGTTT | BC076463.1 |
| 18Sb | GAACGCCACTTGTCCCTCTA | GTTGGTGGAGCGATTTGTCT | FJ915075.1 |
Accession numbers are from www.ncbi.nlm.nih.gov/ or www.ensembl.org/
From Robison et al.5
Protein extraction and western blotting
Samples of frozen viscera and muscle (300 mg) were homogenized on ice with an Ultraturrax homogenizer in a buffer containing 150 mM NaCl, 10 mM 2-amino-2-hydroxymethyl-propane-1,3-diol, 1 mM ethylene-glycol-bis(a-aminoethyl)-N,N,N′,N′-tetra-acetic acid, 1 mM EDTA (pH 7.4), 100 mM sodium fluoride, 4 mM sodium pyrophosphate, 2 mM sodium orthovanadate, 1% Triton X-100, 0.5% NP-40-Igepal, and a protease inhibitor cocktail (Roche). Homogenates were centrifuged at 1000 g for 15 min at 4°C, and supernatant fractions were then centrifuged for 30 min at 20,000 g at 4°C. The resulting supernatant fractions were sampled and stored at −80°C. Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad). Lysates (40 μg protein) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting using the appropriate antibody: anti-phospho Akt (Ser473) (Cell Signaling Technologies, 9271), anti-carboxyl terminal Akt (Cell Signaling Technologies, 9272), anti-phospho S6K1 (Thr389) (Cell Signaling Technologies, 9205), anti-carboxyl terminal S6K1 (Cell Signaling Technologies, 9202), anti-phosho S6 (Ser235/Ser236) (Cell Signaling Technologies, 4856), anti-carboxyl terminal S6 (Cell Signaling Technologies, 2217), anti-phospho 4E-BP1 (Thr37/Thr46) (Cell Signaling Technologies, 9459), and anti-carboxyl terminal 4E-BP1 (Cell Signaling Technologies, 9452). After washing, membranes were incubated with an IRDye Infrared secondary antibody (LI-COR Inc. Biotechnology). Bands were visualized by Infrared Fluorescence using the Odyssey Imaging System (LI-COR Inc. Biotechnology) and quantified by Odyssey Infrared imaging system software (version 1.2; LI-COR Inc. Biotechnology).
Statistical analysis
Results are expressed as means±SEM and analyzed by one-way ANOVA followed by Tukey multiple-range test or Student's t-test as specified in figure legends. For all statistical analyses, the level of significance was set at p<0.05.
Results
Effect of a single meal on the expression of hepatic metabolism-related genes
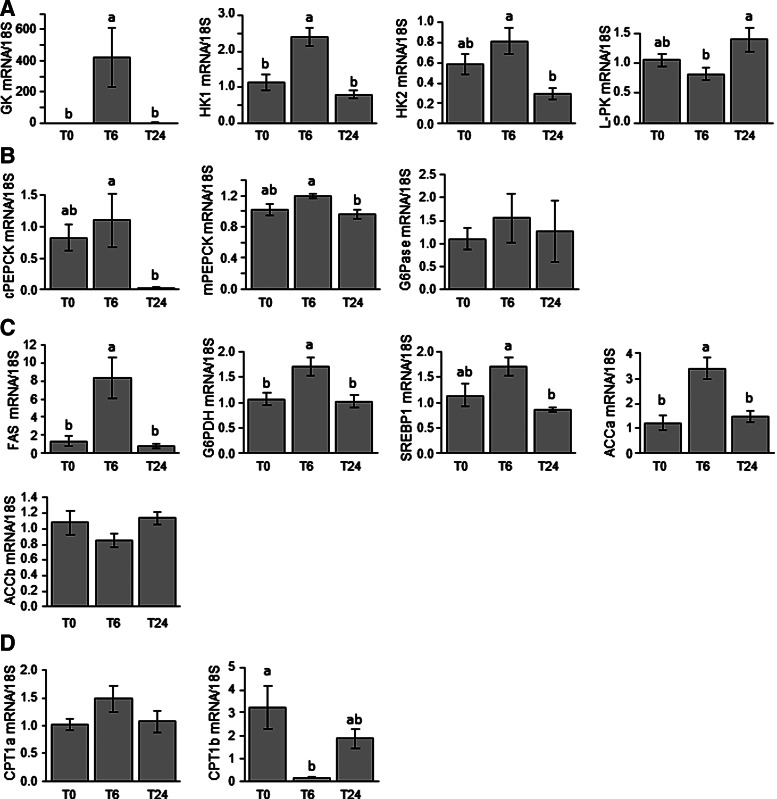
The effects of refeeding on the expression of hepatic metabolism-related genes were analyzed by comparing fasted and 6- and 24-h refed fish (Fig. 1). Refeeding led to an increase in the expression of glucokinase (GK) and HK1 mRNAs 6 h after the meal (Fig. 1A). Expression of HK2 and L-PK mRNAs remained unchanged between fasted and refed fish irrespective of the sampling time, but was significantly affected between 6- and 24-h refed fish. Similar observations were made for the gluconeogenic mRNAs cPEPCK and mPEPCK (Fig. 1B). In contrast, no effect of refeeding was monitored for G6Pase. Regarding lipid metabolism (Fig. 1C), expression of the lipogenic FAS, G6PDH, and ACCa (but not ACCb) genes significantly increased 6 h after refeeding and then declined to reach values found in fasted fish. The level of SREBP1 mRNA remained unchanged after refeeding whatever the sampling time considered, but was significantly affected between 6- and 24-h refed fish. Finally, we also investigated the expression of CPT1a and CPT1b (Fig. 1D) and recorded a significant decrease of the level of mRNA of the last isoform 6 h after refeeding.
FIG. 1.
Postprandial regulation of gene expression of hepatic enzymes involved in (A) glycolysis, (B) gluconeogenesis, (C) lipogenesis, and (D) lipolysis. Expression values are normalized with 18S-expressed transcripts. The mean and standard error (SE) of n=6 samples per group are shown. Data were analyzed using one-way analysis of variance (ANOVA), followed by the Tukey post hoc test. Different letters indicate a significant difference at p<0.05.
Effect of a single meal on the expression of muscle metabolism- and growth-related factors
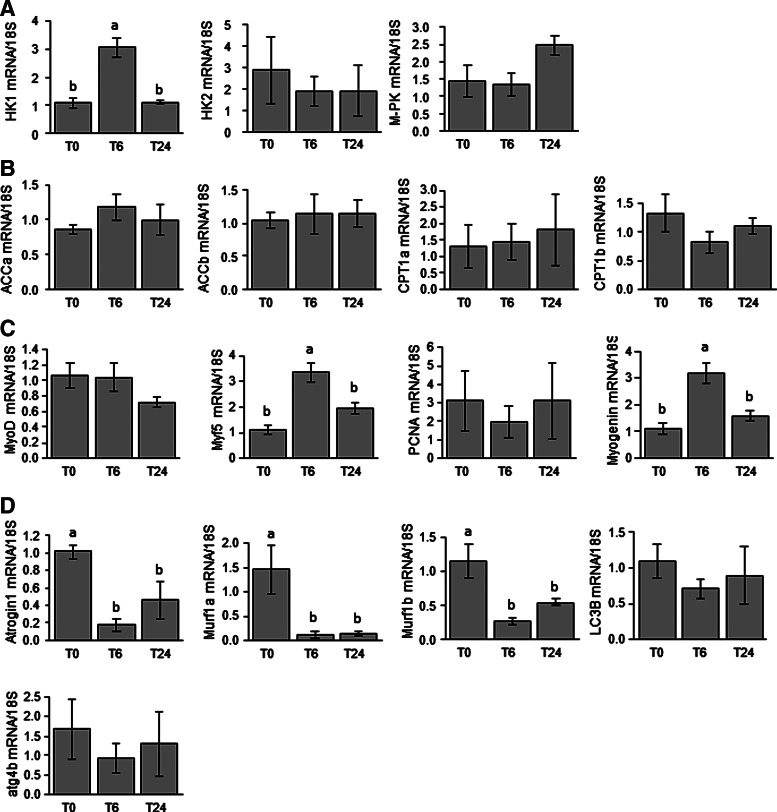
The effects of refeeding on the expression of several muscle metabolism- and growth-related genes were analyzed by comparing fasted and 6- and 24-h refed fish (Fig. 2). Refeeding led to an increase in the expression of HK1 mRNA 6 h after the meal (Fig. 2A). In contrast, expression of HK2 and M-PK mRNA remained unchanged between fasted and refed fish. Similarly, no effect of refeeding was observed for any of the monitored genes involved in energy metabolism (Fig. 2B). By investigating the regulation of expression of myogenic genes (Fig. 2C), we found that mRNA levels of the genes encoding Myf5 and Myogenin were upregulated 6 h after refeeding, whereas those of MyoD and PCNA remained unchanged. We also focused our attention on the mRNA levels of genes involved in the two main muscle proteolytic systems, namely, ub-proteasomal and autophagy-lysosomal systems (Fig. 2D). Transcript levels of genes involved in the former proteolytic system (atrogin1, murf1a, murf1b) were subjected to significant reduction by refeeding. In contrast, the autophagy-related genes (LC3B and atg4b) remained unaffected. Finally, we investigated the effect of refeeding on the phosphorylation of the main proteins (Akt, 4EBP1, S6K1, S6) of the muscle growth promoting Akt/TOR signaling pathways by comparing fasted and 0.5- and 2-h refed fish. As shown in Figure 3, we recorded an induction of the phosphorylation of all monitored proteins by refeeding.
FIG. 2.
Postprandial regulation of gene expression of muscle proteins involved in (A) glycolysis, (B) energy metabolism, (C) myogenesis, and (D) proteolysis. Expression values are normalized with 18S-expressed transcripts. The mean and SE of n=6 samples per group are shown. Data were analyzed using one-way ANOVA, followed by the Tukey post hoc test. Different letters indicate a significant difference at p<0.05.
FIG. 3.
Postprandial profile of components of the Akt/TOR signaling pathway in muscle of trout as determined by western blot densitometry. Twenty micrograms of total protein per lane was loaded on the gel. A representative blot is shown. Graphs represent the ratio between the phosphorylated protein and the total amount of the targeted protein. Results are mean±SE (n=6) and were analyzed using one-way ANOVA followed by the Tukey test for multiple comparisons. Mean values differ for a selected group not sharing a common letter (p<0.05).
Effect of dietary carbohydrate to protein ratio on the expression of hepatic and muscle metabolism- and growth-related factors
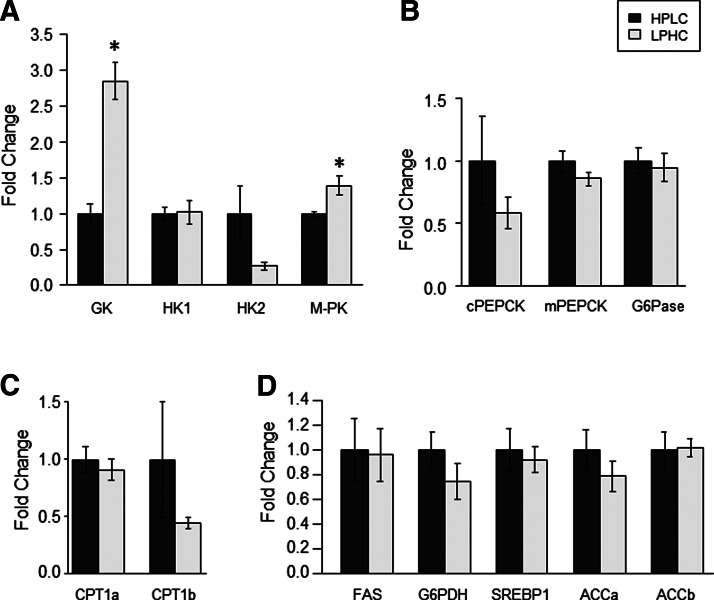
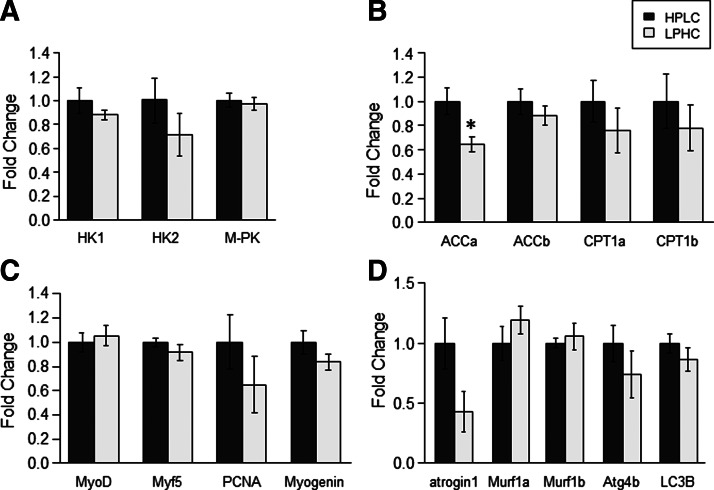
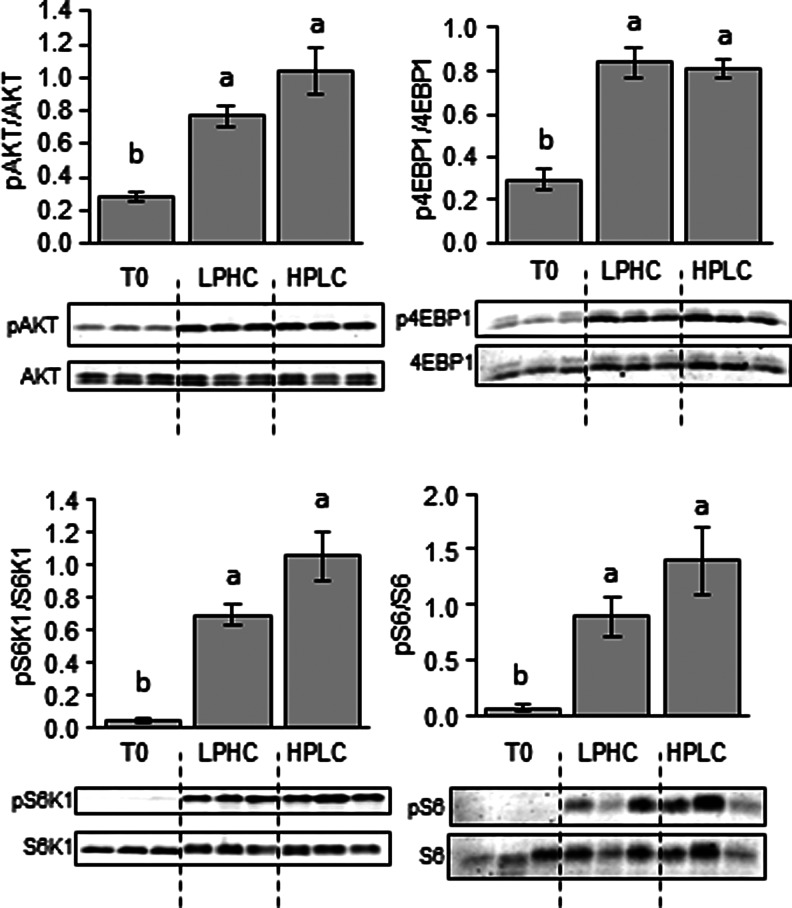
We investigated the effects of feeding the two experimental diets (HPLC and LPHC) on the expression of hepatic and muscle metabolism- and growth-related factors in fish sampled 2 h (for Akt/TOR signaling) and 6 h (for gene expression) after the meal. Of the 14 genes explored in viscera, GK and L-PK were the only genes affected by the carbohydrate to protein ratio in the diet, both of them displaying higher mRNA levels in fish refed the LPHC diet compared to the one refed the HPLC diet (Fig. 4). Similarly, in the muscle, except ACCa, whose expression decreased in LPHC-diet refed fish compared with their HPLC counterparts, the mRNA levels of all other genes analyzed in this study remained the same between the two groups of fish (Fig. 5). Likewise, no effect of carbohydrate-to-protein ratio in the diet was observed in the activation of the Akt/TOR signaling pathway (Fig. 6).
FIG. 4.
Gene expression of hepatic enzymes involved in (A) glycolysis, (B) gluconeogenesis, (C) lipolysis, and (D) lipogenesis in high protein–low carbohydrate- (HPLC) and low protein–high carbohydrate (LPHC)-diet 6-h refed rainbow trout. Expression values were normalized with 18S-expressed transcripts. Results are expressed as fold of the HPLC group and presented as means±SE (n=6). *Significant difference from HPLC group (p<0.05, Student's t-test).
FIG. 5.
Gene expression of muscle proteins involved in (A) glycolysis, (B) energy metabolism, (C) myogenesis, and (D) proteolysis in HPLC- and LPHC-diet 6-h refed rainbow trout. Expression values were normalized with 18S-expressed transcripts. Results are expressed as fold of the HPLC group and presented as means±SE (n=6). *Significant difference from HPLC group (p<0.05, Student's t-test).
FIG. 6.
Western blot analysis of Akt, S6K1, S6, and 4E-BP1 protein phosphorylation in muscle of fasted (T0) and HPLC- and LPHC-diet 2-h-refed rainbow trout. Twenty micrograms of total protein per lane was loaded on the gel. A representative blot is shown. Graphs represent the ratio between the phosphorylated protein and the total amount of the targeted protein. Results are means±SE (n=6) and were analyzed using one-way ANOVA followed by the Tukey test for multiple comparisons. Mean values for a selected group not sharing a common letter differ (p<0.05).
Discussion
The goals of our study were (a) to characterize the effect of a single meal on the postprandial expression of several hepatic and muscle metabolism-related genes and proteins and (b) to analyze the effects of an increase in the ratio of dietary carbohydrates/proteins on the expression of these metabolism-related factors in zebrafish.
Glucose metabolism
Glycolysis represents the only metabolic pathway leading to glucose catabolism in all organisms, including fish.26 All enzymes involved in the glycolysis have been reported in fish.27 Hexokinases catalyze the first reaction of glycolysis by phosphorylating glucose into glucose-6-phosphate. Four closely related hexokinases (HK I–IV) have been described in mammals.28,29 In the present study, we confirm that HK-I, HK-II, and HK-IV, known as GK, are also expressed in zebrafish. Refeeding increased expression of visceral GK and HK1 but has few effects on HK2 gene expression. When the proportion of carbohydrates is increased at the expense of proteins, the level of GK mRNA significantly increased. Unlike mammals in which insulin is a major regulator of GK gene expression,30 zebrafish GK gene expression is probably mainly controlled by carbohydrates, as suggested by Robison et al.5 and confirmed by González-Alvarez et al. after intraperitoneal administration of glucose to zebrafish.31 Several data obtained in other fish species tend toward the same conclusion that carbohydrates are the main regulator of GK gene expression in fish.32 In this regard, in vivo studies showed that GK mRNA expression increases in rainbow trout and gilthead sea bream according to the proportion of dietary carbohydrate content,32–36 and in vitro experiments demonstrated that glucose enhances accumulation of GK mRNA in rainbow trout hepatocytes.37 Zebrafish GK gene expression was also proposed to be subjected to a sexually dimorphic regulation in the postprandial state.5 Since fish of the present study were sexually immature, the influence of sex difference on transcriptional response was probably weak.
Despite a strong induction of GK gene expression, that of the downstream PK was poorly regulated by refeeding in zebrafish as previously observed in rainbow trout.38,39 In mammals, PK is submitted to dietary and hormonal regulations mainly committed by carbohydrates, insulin, and glucagon. Such regulations occur at the transcriptional level40 and also through post-transcriptional modifications of the protein, including phosphorylation and dephosphorylation process.41 It is therefore possible that zebrafish PK is mainly controlled by refeeding at the level of its activity through post-translational mechanisms as it has been previously proposed in rainbow trout.42 On the other hand, the positive effect of high carbohydrate diet on zebrafish L-PK gene expression is in agreement with what is observed in mammals.40
When looking at genes related to gluconeogenesis in viscera, we found that cytosolic and mitochondrial PEPCK mRNA levels were inhibited during refeeding. The activity of PEPCK is only controlled at the level of transcription as there are no known allosteric modifiers.43 In mammals, the gene encoding the cytosolic isoform is under nutritional and hormonal control, which is not the case of the mitochondrial isoform, known to be constitutively expressed. This feature may probably be extended to zebrafish inasmuch as cytosolic PEPCK gene expression exhibited to a 30-fold decrease between 6 and 24 h after the meal, whereas mPEPCK is poorly downregulated during the same period (1.2-fold increase). The increase of dietary carbohydrate does not affect the level of expression of the cytosolic PEPCK gene as previously observed in common carp, another Cyprinidae fish species.44
G6Pase was the second gluconeogenic enzyme investigated in the current study. We observed in zebrafish that G6Pase gene expression was affected by neither refeeding nor dietary carbohydrate proportion, whereas in mammals, G6Pase gene expression is downregulated by carbohydrate intake through transcriptional mechanisms.45 The absence of postprandial inhibition of G6Pase gene expression is also observed in rainbow trout.36,46 The authors suggest that this absence of inhibition might, at least partly, explain the prolonged postprandial hyperglycemic phenotype of carnivorous fish fed a high-carbohydrate diet.36,46 Our results are not in accordance with previous studies indicating that zebrafish, like other omnivores, metabolize glucose faster than carnivorous teleosts.47,48
Lipogenesis
In mammals, the two key enzymes required to divert glycolytic carbon flux into lipid biosynthesis are ACLY and FAS. ACLY converts cytosolic citrate into acetyl-CoA and oxaloacetate, thereby supplying the essential metabolite for lipid biosynthesis. FAS catalyzes all of the reaction steps involved in the conversion of acetyl-CoA and malonyl-CoA, supplied by ACCa, the hepatic isoform of ACC, to palmitate. For these reactions, FAS uses NADPH, generated by enzymes of the pentose phosphate shunt such as G6PDH. All these enzymes are also expressed in zebrafish and are positively regulated by the nutritional status as previously observed in mammals, where, for example, the rate of FAS gene expression is induced by re-feeding in rats.49
SREBP1 is essential for the transcriptional control of genes encoding enzymes of lipid biosynthesis such as FAS and ACLY. Except a decrease between 6 and 24 h after refeeding, SREBP1 mRNA expression was poorly regulated in zebrafish by the meal. Previous results obtained by Craig and Moon also demonstrated that 2 weeks of fasting is needed to significantly decrease SREBP1 gene expression in zebrafish.50 The expression of SREBP1 in zebrafish is potentially less sensitive to nutritional control than mammalian and avian species and even trout, where SREBP1 gene expression is repressed by short-term fasting.36,51–53
The data of the present work show that 50% reduction of dietary protein content in favor to carbohydrates does not change the postprandial level of expression of all investigated lipogenic genes. This absence of adaptation of the lipogenic pathway to dietary protein-to-carbohydrate ratio is surprising since modifications have been previously observed in both mammals and fish. In rats, high-protein diet lead to a reduction in the expression of lipogenic enzymes,54–56 whereas opposite effects were observed in rainbow trout.36 Further analysis of lipogenic enzyme activities would be of interest to investigate if regulations occur at this level. Whether a longer feeding trial than that presented here could affect the mRNA levels of lipogenic genes is also worth investigating.
Fatty acid β-oxidation
The β-oxidation of long-chain fatty acids plays a central role in the production of energy in situations of food deprivation or exercise where glucose is limited or need to be spared. CPT1 is responsible for the entry of long chain fatty acids into the mitochondria. It is considered as a limiting enzyme of β-oxidation.57,58 This enzyme is inhibited by malonyl-CoA, mainly produced by the reaction of ACCa and ACCb57 in the liver and the muscle, respectively. We observed in zebrafish that the gene encoding the b isoform of CPT1 is downregulated in visceral tissues 6 h after refeeding, in agreement with the increased expression of ACCa. On the contrary, muscle expressions of isoforms a and b of both CPT1 and ACC remain stable, suggesting that muscle β-oxidation is poorly controlled by fasting and refeeding compared with hepatic β-oxidation. This hypothesis needs to be carefully confirmed by a more precise time-course postprandial analysis of CPT1 and ACC muscle gene expression. We found that modification of the dietary protein to carbohydrate ratio has little effect on CPT1 and ACC gene expression in zebrafish, whereas inhibition of CPT1 gene expression is preferentially observed after a HPLC meal in rainbow trout.36,59 These results suggest a different control mechanism of energy metabolism between these two fish species.
Myogenesis
Myogenesis is regulated by a family of transcription factors called myogenic regulatory factors that are expressed in a temporally distinct pattern during determination, activation, and proliferation of muscle precursor cells (Myf5 and Myod) and in cells entering the terminal differentiation program (myogenin).60 Proliferative muscle precursor cells express also PCNA, which is a protein functioning as a cofactor of DNA polymerase-δ and is necessary for cell cycle progression and cell proliferation.61 We reported here that the expression of Myf5 and myogenin (but not that of MyoD and PCNA) is upregulated in zebrafish of 200–250 mg body weight (20–25 mm long) 6 h after a single commercial meal after a short starvation period (72 h). These results contradict previously published data in zebrafish of 460–530 mg body weight (30–33 mm long) showing no change in expression of these genes 3 and 6 h after a 3 h bloodworms feeding period after a starvation period of 7 days.3 This discrepancy between results may be caused by a different muscle growth mechanism of the zebrafish used in the two studies, that is, only by hypertrophy (increase in muscle fiber size) in 30-mm-long fish and by hypertrophy and hyperplasia (increase in the number of muscle fibers) in 20-mm-long ones.62 However, the differences of the experimental procedures between both studies (mainly the starvation and feeding times and the given diets) likely account for much of the observed differences. In this regard, changes in food availability are known to affect the expression of myogenin in the zebrafish63 as in rainbow trout,64,65 and Atlantic salmon,66 and that of Myf5 in rainbow trout65 and Atlantic salmon.67
The lack of effect of dietary carbohydrate-to-protein ratio in the expression of myogenic regulatory factors coding genes contradicts also with a recent study in rainbow trout, showing that carbohydrate levels in the diet affect the expression of Myf5 and myogenin.25 However, these results were obtained after 12 weeks of feeding, possibly reflecting that the effect of manipulating dietary macronutrient composition on the expression of these genes appears at long-term but not during the postprandial period. Furthermore, in contrast to zebrafish, rainbow trout displays an indeterminate muscle growth pattern allowed by a continuous production of muscle fibers (hyperplasia) in addition to an increase of fiber size (hypertrophy). This difference between these two piscine species could also explain the observed differences in gene expression.
Overall, these findings highlight the magnitude of the mechanisms involved in muscle growth in different fish species (indeterminate vs. determinate growth, long-term vs. short-term nutritional effects). These findings also demonstrate the importance of these species in comparative physiology and help gain better understanding of mechanisms involved in muscle growth.
Protein turnover
Protein accretion also reflects dynamic changes between synthesis and degradation. The former depends on the activity of the above-mentioned Akt/TOR signaling pathway.68 We, and others, reported previously in mammals and piscine species that the macronutrient composition of the diets affects the Akt/TOR signaling axis after a single meal, or, more precisely, reduction of the dietary protein content in favor of carbohydrates impairs the activation of the Akt/TOR signaling pathway.36,69 In the present study, several main proteins of this signaling route (Akt, 4EBP1, S6K1, S6) were activated by refeeding, but were not affected by macronutrient composition of the diet. A possible explanation for these results is that the protein levels of the LPHC diet, although marginal compared with the levels estimated for optimal growth,2 are sufficient to activate the Akt/TOR signaling pathway. However, the exact nutritional requirements are unknown for this species, and the only available data are approximations drawn from studies of cyprinid species such as the common carp (Cyprinus carpio) and goldfish (Carasius auratus).2 Further studies are warranted to precisely determine the nutritional requirements of zebrafish.
The degradation of proteins depends on the activity of the main proteolytic routes that are the ubiquitin-proteasomal and the autophagy-Lysosomal pathways. As already observed in zebrafish3 and other fish species,36,67,70–72 genes encoding E3-ubiquitin ligase, atrogin1/MAFbx and MuRF1, were inhibited by feeding. However, dietary carbohydrate-to-protein ratio had no impact on the expression of these genes. In mammals, the regulation of atrogin1/MAFbx and MuRF1 has been shown to be associated to the activity of the FoxO1/3 transcription factors, the downstream targets of the PI3K/Akt signaling pathway.73–75 In this regard, the correlation between the phosphorylation of Akt and the pattern of expression of the two E3-ubiquitine ligase coding genes in our study suggests a strong conservation of the mechanisms involved in the regulation of the Ub-proteasome-dependent proteolytic pathway between higher and lower vertebrates and confirms the previous observation in rainbow trout.71,76,77
In contrast, no relationship was observed between the activation of Akt and the expression of autophagy-related genes. In mammals, the expression of these genes is recognized to be also dependent on the PI3K/Akt signaling pathway via the activation of the transcription factor FoxO.78,79 However, recent in vitro and in vivo studies in rainbow trout indicate that IGF1 and insulin induce activation of Akt and FoxO but have low or no effect on autophagy-related transcripts levels, suggesting a minor role for this signaling axis on the autophagic/lysosomal pathway in this species.76,77 Whether the observed lack of feeding effect on the expression of autophagy-related genes is specific to the studied piscine species so far or due to the experimental conditions is worth investigating.
Conclusions and significance
The understanding of the role of nutrients as signaling molecules remains relatively limited, although significant progress has been achieved during the past few years, in particular with regard to amino acid control of physiological functions. Zebrafish has been proposed as a possible model organism to make inroads in nutritional physiology.2 However, to our knowledge, studies on the field remain scarce and deal mainly with the effect of fasting and/or refeeding.3,4 Here, we investigated in this species not only the effect of a single meal on the postprandial expression of several hepatic and muscle metabolism-related genes and proteins but also that of manipulating proteins and carbohydrates content of the diet. Our data show that following the genes or functions considered, the responses obtained with this species are similar to those observed in carnivorous species such as rainbow trout (e.g., lack of refeeding effect on the expression of PK), similar to those usually attributed to rodent species (e.g., effect of refeeding on cPEPCK gene expression) or specific to this model (e.g., no significant modification of SREBP1 mRNA expression between fasted and refed fish). Together, these specific features of its metabolism, along with its numerous well-recognized advantages (short generation time interval, capacity to produce numerous offspring, wide variety of molecular tools and information available for genomic analysis, a sequenced genome), make this species particularly relevant in comparative physiology. However, several gaps remain in the precise knowledge of the nutritional physiology of this species (e.g., nutritional requirement, regulation of feed intake, optimum feeding levels, maximum growth rate), limiting its use as a model organism in the field. In the future, one of the major challenges will be to fill these gaps.
Acknowledgments
We thank M.J. Borthaire and E. Plagnes-Juan for technical assistance.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lessman CA. The developing zebrafish (Danio rerio): a vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res C Embryo Today. 2011;93:268–280. doi: 10.1002/bdrc.20212. [DOI] [PubMed] [Google Scholar]
- 2.Ulloa PE. Iturra P. Neira R. Araneda C. Zebrafish as a model organism for nutrition and growth: towards comparative studies of nutritional genomics applied to aquacultured fishes. Rev Fish Biol Fish. 2011;21:649–666. [Google Scholar]
- 3.Amaral IP. Johnston IA. Insulin-like growth factor (IGF) signalling and genome-wide transcriptional regulation in fast muscle of zebrafish following a single-satiating meal. J Exp Biol. 2011;214:2125–2139. doi: 10.1242/jeb.053298. [DOI] [PubMed] [Google Scholar]
- 4.Drew RE. Rodnick KJ. Settles M. Wacyk J. Churchill E. Powell MS, et al. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio) Physiol Genomics. 2008;35:283–295. doi: 10.1152/physiolgenomics.90213.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robison BD. Drew RE. Murdoch GK. Powell M. Rodnick KJ. Settles M, et al. Sexual dimorphism in hepatic gene expression and the response to dietary carbohydrate manipulation in the zebrafish (Danio rerio) Comp Biochem Physiol Part D Genomics Proteomics. 2008;3:141–154. doi: 10.1016/j.cbd.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meijer AJ. Amino acids as regulators and components of nonproteinogenic pathways. J Nutr. 2003;133:2057S–2062S. doi: 10.1093/jn/133.6.2057S. [DOI] [PubMed] [Google Scholar]
- 7.Avruch J. Long X. Ortiz-Vega S. Rapley J. Papageorgiou A. Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–E602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimball SR. Jefferson LS. Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J Nutr. 2006;136:227S–231S. doi: 10.1093/jn/136.1.227S. [DOI] [PubMed] [Google Scholar]
- 9.Chaveroux C. Lambert-Langlais S. Cherasse Y. Averous J. Parry L. Carraro V, et al. Molecular mechanisms involved in the adaptation to amino acid limitation in mammals. Biochimie. 2010;92:736–745. doi: 10.1016/j.biochi.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Lansard M. Panserat S. Plagnes-Juan E. Dias K. Seiliez I. Skiba-Cassy S. L-leucine, L-methionine, and L-lysine are involved in the regulation of intermediary metabolism-related gene expression in rainbow trout hepatocytes. J Nutr. 2011;141:75–80. doi: 10.3945/jn.110.124511. [DOI] [PubMed] [Google Scholar]
- 11.Lansard M. Panserat S. Plagnes-Juan E. Seiliez I. Skiba-Cassy S. Integration of insulin and amino acid signals that regulate hepatic metabolism-related gene expression in rainbow trout: role of TOR. Amino Acids. 2010;39:801–810. doi: 10.1007/s00726-010-0533-3. [DOI] [PubMed] [Google Scholar]
- 12.Porstmann T. Santos CR. Griffiths B. Cully M. Wu M. Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seiliez I. Gabillard JC. Riflade M. Sadoul B. Dias K. Averous J, et al. Amino acids downregulate the expression of several autophagy-related genes in rainbow trout myoblasts. Autophagy. 2012;8:364–375. doi: 10.4161/auto.18863. [DOI] [PubMed] [Google Scholar]
- 14.Averous J. Gabillard JC. Seiliez I. Dardevet D. Leucine limitation regulates myf5 and myoD expression and inhibits myoblast differentiation. Exp Cell Res. 2012;318:217–227. doi: 10.1016/j.yexcr.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Erbay E. Park IH. Nuzzi PD. Schoenherr CJ. Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003;163:931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Averous J. Bruhat A. Jousse C. Carraro V. Thiel G. Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J Biol Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 17.Averous J. Lambert-Langlais S. Cherasse Y. Carraro V. Parry L. B'Chir W, et al. Amino acid deprivation regulates the stress-inducible gene p8 via the GCN2/ATF4 pathway. Biochem Biophys Res Commun. 2011;413:24–29. doi: 10.1016/j.bbrc.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Carraro V. Maurin A-C. Lambert-Langlais S. Averous J. Chaveroux C. Parry L, et al. Amino acid availability controls TRB3 transcription in liver through the GCN2/eIF2α/ATF4 pathway. PLoS One. 2010;5:e15716. doi: 10.1371/journal.pone.0015716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremblay F. Lavigne C. Jacques H. Marette A. Role of dietary proteins and amino acids in the pathogenesis of insulin resistance. Annu Rev Nutr. 2007;27:293–310. doi: 10.1146/annurev.nutr.25.050304.092545. [DOI] [PubMed] [Google Scholar]
- 20.Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab. 2005;16:489–494. doi: 10.1016/j.tem.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 21.da Silva Xavier G. Rutter GA. Diraison F. Andreolas C. Leclerc I. ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic beta-cells. J Lipid Res. 2006;47:2482–2491. doi: 10.1194/jlr.M600289-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Dentin R. Girard J. Postic C. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie. 2005;87:81–86. doi: 10.1016/j.biochi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Dentin R. Pegorier JP. Benhamed F. Foufelle F. Ferre P. Fauveau V, et al. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem. 2004;279:20314–20326. doi: 10.1074/jbc.M312475200. [DOI] [PubMed] [Google Scholar]
- 24.Vaulont S. Vasseur-Cognet M. Kahn A. Glucose regulation of gene transcription. J Biol Chem. 2000;275:31555–31558. doi: 10.1074/jbc.R000016200. [DOI] [PubMed] [Google Scholar]
- 25.Chapalamadugu KC. Robison BD. Drew RE. Powell MS. Hill RA. Amberg JJ, et al. Dietary carbohydrate level affects transcription factor expression that regulates skeletal muscle myogenesis in rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 2009;153:66–72. doi: 10.1016/j.cbpb.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Cowey C. Walton M. Walton M. Intermediary metabolism. In: Cowey C, editor; Intermediary Metabolism. Academic Press; New York: 1989. pp. 259–329. [Google Scholar]
- 27.Walton MJ. Cowey CB. Gluconeogenesis by isolated hepatocytes from rainbow-trout salmo-gairdneri. Comp Biochem Physiol B-Biochem Mol Biol. 1979;62:75–79. [Google Scholar]
- 28.Cardenas ML. Cornish-Bowden A. Ureta T. Evolution and regulatory role of the hexokinases. Biochim Biophys Acta. 1998;1401:242–264. doi: 10.1016/s0167-4889(97)00150-x. [DOI] [PubMed] [Google Scholar]
- 29.Wilson JE. Hexokinases. Rev Physiol Biochem Pharmacol. 1995;126:65–198. doi: 10.1007/BFb0049776. [DOI] [PubMed] [Google Scholar]
- 30.Iynedjian PB. Jotterand D. Nouspikel T. Asfari M. Pilot PR. Transcriptional induction of glucokinase gene by insulin in cultured liver cells and its repression by the glucagon-cAMP system. J Biol Chem. 1989;264:21824–21829. [PubMed] [Google Scholar]
- 31.González-Alvarez R. Ortega-Cuellar D. Hernandez-Mendoza A. Moreno-Arriola E. Villasenor-Mendoza K. Galvez-Mariscal A, et al. The hexokinase gene family in the zebrafish: structure, expression, functional and phylogenetic analysis. Comp Biochem Physiol B Biochem Mol Biol. 2009;152:189–195. doi: 10.1016/j.cbpb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Panserat S. Medale F. Blin C. Breque J. Vachot C. Plagnes-Juan E, et al. Hepatic glucokinase is induced by dietary carbohydrates in rainbow trout, gilthead seabream, and common carp. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1164–R1170. doi: 10.1152/ajpregu.2000.278.5.R1164. [DOI] [PubMed] [Google Scholar]
- 33.Capilla E. Medale F. Navarro I. Panserat S. Vachot C. Kaushik S, et al. Muscle insulin binding and plasma levels in relation to liver glucokinase activity, glucose metabolism and dietary carbohydrates in rainbow trout. Regul Pept. 2003;110:123–132. doi: 10.1016/s0167-0115(02)00212-4. [DOI] [PubMed] [Google Scholar]
- 34.Caseras A. Meton I. Vives C. Egea M. Fernandez F. Baanante IV. Nutritional regulation of glucose-6-phosphatase gene expression in liver of the gilthead sea bream (Sparus aurata) Br J Nutr. 2002;88:607–614. doi: 10.1079/BJN2002701. [DOI] [PubMed] [Google Scholar]
- 35.Meton I. Caseras A. Fernandez F. Baanante IV. Molecular cloning of hepatic glucose-6-phosphatase catalytic subunit from gilthead sea bream (Sparus aurata): response of its mRNA levels and glucokinase expression to refeeding and diet composition. Comp Biochem Physiol B Biochem Mol Biol. 2004;138:145–153. doi: 10.1016/j.cbpc.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Seiliez I. Panserat S. Lansard M. Polakof S. Plagnes-Juan E. Surget A, et al. Dietary carbohydrate-to-protein ratio affects TOR signaling and metabolism-related gene expression in the liver and muscle of rainbow trout after a single meal. Am J Physiol Regul Integr Comp Physiol. 2011;300:R733–R743. doi: 10.1152/ajpregu.00579.2010. [DOI] [PubMed] [Google Scholar]
- 37.Plagnes-Juan E. Lansard M. Seiliez I. Medale F. Corraze G. Kaushik S, et al. Insulin regulates the expression of several metabolism-related genes in the liver and primary hepatocytes of rainbow trout (Oncorhynchus mykiss) J Exp Biol. 2008;211:2510–2518. doi: 10.1242/jeb.018374. [DOI] [PubMed] [Google Scholar]
- 38.Panserat S. Plagnes-Juan E. Kaushik S. Nutritional regulation and tissue specificity of gene expression for proteins involved in hepatic glucose metabolism in rainbow trout (Oncorhynchus mykiss) J Exp Biol. 2001;204:2351–2360. doi: 10.1242/jeb.204.13.2351. [DOI] [PubMed] [Google Scholar]
- 39.Skiba-Cassy S. Lansard M. Panserat S. Medale F. Rainbow trout genetically selected for greater muscle fat content display increased activation of liver TOR signaling and lipogenic gene expression. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1421–R1429. doi: 10.1152/ajpregu.00312.2009. [DOI] [PubMed] [Google Scholar]
- 40.Yamada K. Noguchi T. Nutrient and hormonal regulation of pyruvate kinase gene expression. Biochem J. 1999;337(Pt 1):1–11. [PMC free article] [PubMed] [Google Scholar]
- 41.Assimacopoulos-Jeannet F. Jeanrenaud B. Insulin activates 6-phosphofructo-2-kinase and pyruvate kinase in the liver. Indirect evidence for an action via a phosphatase. J Biol Chem. 1990;265:7202–7206. doi: 10.1016/0261-5614(90)90109-6. [DOI] [PubMed] [Google Scholar]
- 42.Kirchner S. Kaushik S. Panserat S. Effect of partial substitution of dietary protein by a single gluconeogenic dispensable amino acid on hepatic glucose metabolism in rainbow trout (Oncorhynchus mykiss) Comp Biochem Physiol A Mol Integr Physiol. 2003;134:337–347. doi: 10.1016/s1095-6433(02)00267-2. [DOI] [PubMed] [Google Scholar]
- 43.Hanson RW. Patel YM. Phosphoenolpyruvate carboxykinase (GTP): the gene and the enzyme. Adv Enzymol Relat Areas Mol Biol. 1994;69:203–281. doi: 10.1002/9780470123157.ch6. [DOI] [PubMed] [Google Scholar]
- 44.Panserat S. Plagnes-Juan E. Kaushik S. Gluconeogenic enzyme gene expression is decreased by dietary carbohydrates in common carp (Cyprinus carpio) and gilthead seabream (Sparus aurata) Biochim Biophys Acta. 2002;1579:35–42. doi: 10.1016/s0167-4781(02)00501-8. [DOI] [PubMed] [Google Scholar]
- 45.Pilkis SJ. Granner DK. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol. 1992;54:885–909. doi: 10.1146/annurev.ph.54.030192.004321. [DOI] [PubMed] [Google Scholar]
- 46.Panserat S. Medale F. Breque J. Plagnes-Juan E. Kaushik S. Lack of significant long-term effect of dietary carbohydrates on hepatic glucose-6-phosphatase expression in rainbow trout (Oncorhynchus mykiss) J Nutr Biochem. 2000;11:22–29. doi: 10.1016/s0955-2863(99)00067-4. [DOI] [PubMed] [Google Scholar]
- 47.Eames SC. Philipson LH. Prince VE. Kinkel MD. Blood sugar measurement in zebrafish reveals dynamics of glucose homeostasis. Zebrafish. 2010;7:205–213. doi: 10.1089/zeb.2009.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stone DAJ. Dietary carbohydrate utilization by fish. Rev Fisheries Sci. 2003;11:337–369. [Google Scholar]
- 49.Paulauskis JD. Sul HS. Cloning and expression of mouse fatty acid synthase and other specific mRNAs. Developmental and hormonal regulation in 3T3-L1 cells. J Biol Chem. 1988;263:7049–7054. [PubMed] [Google Scholar]
- 50.Craig PM. Moon TW. Fasted zebrafish mimic genetic and physiological responses in mammals: a model for obesity and diabetes? Zebrafish. 2011;8:109–117. doi: 10.1089/zeb.2011.0702. [DOI] [PubMed] [Google Scholar]
- 51.Horton JD. Bashmakov Y. Shimomura I. Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci U S A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang PH. Ko YH. Chin HJ. Hsu C. Ding ST. Chen CY. The effect of feed restriction on expression of hepatic lipogenic genes in broiler chickens and the function of SREBP1. Comp Biochem Physiol B Biochem Mol Biol. 2009;153:327–331. doi: 10.1016/j.cbpb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y. Hillgartner FB. Starvation and feeding a high-carbohydrate, low-fat diet regulate the expression sterol regulatory element-binding protein-1 in chickens. J Nutr. 2004;134:2205–2210. doi: 10.1093/jn/134.9.2205. [DOI] [PubMed] [Google Scholar]
- 54.Blouet C. Mariotti F. Azzout-Marniche D. Bos C. Mathe V. Tome D, et al. The reduced energy intake of rats fed a high-protein low-carbohydrate diet explains the lower fat deposition, but macronutrient substitution accounts for the improved glycemic control. J Nutr. 2006;136:1849–1854. doi: 10.1093/jn/136.7.1849. [DOI] [PubMed] [Google Scholar]
- 55.Brito SM. Moura MA. Kawashita NH. Festuccia WT. Garofalo MA. Kettelhut IC, et al. Adaptation to a high protein, carbohydrate-free diet induces a marked reduction of fatty acid synthesis and lipogenic enzymes in rat adipose tissue that is rapidly reverted by a balanced diet. Can J Physiol Pharmacol. 2005;83:477–482. doi: 10.1139/y05-035. [DOI] [PubMed] [Google Scholar]
- 56.Stepien M. Gaudichon C. Fromentin G. Even P. Tome D. Azzout-Marniche D. Increasing protein at the expense of carbohydrate in the diet down-regulates glucose utilization as glucose sparing effect in rats. PLoS One. 2011;6:e14664. doi: 10.1371/journal.pone.0014664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartlett K. Eaton S. Mitochondrial beta-oxidation. Eur J Biochem. 2004;271:462–469. doi: 10.1046/j.1432-1033.2003.03947.x. [DOI] [PubMed] [Google Scholar]
- 58.Eaton S. Control of mitochondrial beta-oxidation flux. Prog Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- 59.Skiba-Cassy S. Panserat S. Larquier M. Dias K. Surget A. Plagnes-Juan E. Kaushik S. Seiliez I. Apparent low ability of liver and muscle to adapt to variation of dietary carbohydrate:protein ratio in rainbow trout (Oncorhynchus mykiss) Br J Nutr. 2013;109:1359–1372. doi: 10.1017/S0007114512003352. [DOI] [PubMed] [Google Scholar]
- 60.Sabourin LA. Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- 61.Bravo R. Frank R. Blundell PA. Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- 62.Johnston IA. Lee HT. Macqueen DJ. Paranthaman K. Kawashima C. Anwar A, et al. Embryonic temperature affects muscle fibre recruitment in adult zebrafish: genome-wide changes in gene and microRNA expression associated with the transition from hyperplastic to hypertrophic growth phenotypes. J Exp Biol. 2009;212:1781–1793. doi: 10.1242/jeb.029918. [DOI] [PubMed] [Google Scholar]
- 63.Masuda Y. Oku H. Okumura T. Nomura K. Kurokawa T. Feeding restriction alters expression of some ATP related genes more sensitively than the RNA/DNA ratio in zebrafish, Danio rerio. Comp Biochem Physiol B Biochem Mol Biol. 2009;152:287–291. doi: 10.1016/j.cbpb.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 64.Chauvigne F. Gabillard JC. Weil C. Rescan PY. Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen Comp Endocrinol. 2003;132:209–215. doi: 10.1016/s0016-6480(03)00081-9. [DOI] [PubMed] [Google Scholar]
- 65.Johansen KA. Overturf K. Alterations in expression of genes associated with muscle metabolism and growth during nutritional restriction and refeeding in rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 2006;144:119–127. doi: 10.1016/j.cbpb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 66.Bower NI. Taylor RG. Johnston IA. Phasing of muscle gene expression with fasting-induced recovery growth in Atlantic salmon. Front Zool. 2009;6:18. doi: 10.1186/1742-9994-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valente LM. Bower NI. Johnston IA. Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br J Nutr. 2012;108:2148–2157. doi: 10.1017/S0007114512000396. [DOI] [PubMed] [Google Scholar]
- 68.Kimball SR. The role of nutrition in stimulating muscle protein accretion at the molecular level. Biochem Soc Trans. 2007;35:1298–1301. doi: 10.1042/BST0351298. [DOI] [PubMed] [Google Scholar]
- 69.Chotechuang N. Azzout-Marniche D. Bos C. Chaumontet C. Gausseres N. Steiler T, et al. mTOR, AMPK, and GCN2 coordinate the adaptation of hepatic energy metabolic pathways in response to protein intake in the rat. Am J Physiol Endocrinol Metab. 2009;297:E1313–E1323. doi: 10.1152/ajpendo.91000.2008. [DOI] [PubMed] [Google Scholar]
- 70.Cleveland BM. Evenhuis JP. Molecular characterization of atrogin-1/F-box protein-32 (FBXO32) and F-box protein-25 (FBXO25) in rainbow trout (Oncorhynchus mykiss): expression across tissues in response to feed deprivation. Comp Biochem Physiol B Biochem Mol Biol. 2010;157:248–257. doi: 10.1016/j.cbpb.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Seiliez I. Panserat S. Skiba-Cassy S. Fricot A. Vachot C. Kaushik S, et al. Feeding status regulates the polyubiquitination step of the ubiquitin-proteasome-dependent proteolysis in rainbow trout (Oncorhynchus mykiss) muscle. J Nutr. 2008;138:487–491. doi: 10.1093/jn/138.3.487. [DOI] [PubMed] [Google Scholar]
- 72.Wang J. Salem M. Qi N. Kenney PB. Rexroad CE., 3rd Yao J. Molecular characterization of the MuRF genes in rainbow trout: potential role in muscle degradation. Comp Biochem Physiol B Biochem Mol Biol. 2011;158:208–215. doi: 10.1016/j.cbpb.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 73.Sacheck JM. Hyatt JP. Raffaello A. Jagoe RT. Roy RR. Edgerton VR, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 74.Sandri M. Sandri C. Gilbert A. Skurk C. Calabria E. Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stitt TN. Drujan D. Clarke BA. Panaro F. Timofeyva Y. Kline WO, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 76.Seiliez I. Gutierrez J. Salmeron C. Skiba-Cassy S. Chauvin C. Dias K, et al. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss) Comp Biochem Physiol B Biochem Mol Biol. 2010;157:258–266. doi: 10.1016/j.cbpb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 77.Seiliez I. Panserat S. Skiba-Cassy S. Polakof S. Effect of acute and chronic insulin administrations on major factors involved in the control of muscle protein turnover in rainbow trout (Oncorhynchus mykiss) Gen Comp Endocrinol. 2011;172:363–370. doi: 10.1016/j.ygcen.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 78.Mammucari C. Milan G. Romanello V. Masiero E. Rudolf R. Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 79.Zhao J. Brault JJ. Schild A. Cao P. Sandri M. Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]