Abstract
Plastid genomes are an invaluable resource for plant biological studies. However, the number of completely sequenced plant plastid genomes is still small compared with the vast number of species. To provide an alternative generalized approach, we designed a set of 138 pairs of universal primers for amplifying (termed “short-range PCR”) and sequencing the entire genomes of the angiosperm plastid genomes. The universality of the primers was tested by using species from the basal to asterid angiosperms. The polymerase chain reaction (PCR) success rate was higher than 96%. We sequenced the complete chloroplast genome of Liquidambar formosana as an example using this method and compared it to the genomes independently determined by long-range PCR (from 6.3 kb to 13.3 kb) and next-generation sequencing methods. The three genomes showed that they were completely identical. To test the phylogenetic efficiency of this method, we amplified and sequenced 18 chloroplast regions of 19 Saxifragales and Saxifragales-related taxa, as a case study, to reconstruct the phylogeny of all families of the order. Phylograms based on a combination of our data, together with those from GenBank, clearly indicate three family groups and three single families within the order. This set of universal primers is expected to accelerate the accumulation of angiosperm plastid genomes and to make faster mass data collection of plastid genomes for molecular systematics.
Keywords: plastid genome, universal primers, Saxifragales
Introduction
Complete plastid genomes have found extensive applications in sequence-based phylogeny. The systematically uncertain families, such as Vitaceae and Nelumbonaceae, have thus been pinpointed (Jansen et al. 2006). The plastid genome information has also proven very useful in studying evolution within families, for example, Pinaceae (Lin et al. 2010) and Pinus (Parks et al. 2009). Recently, plastid genomes have served as reservoirs of microsatellites (Scarcelli et al. 2011; Xue, Wang, et al. 2012) and high-resolution DNA barcodes (Kumar, Hahn, et al. 2009; Doorduin et al. 2011).
It has been shown that the reliability of a phylogeny is heavily dependent upon the number of informative characters. For instance, 10 kb of sequence is needed to resolve the familial relationship of Lamiales (Wortley et al. 2005). The best way to reach a convincing result is to add more genes to the data set. The rapid accumulation of the whole plastid genome sequences has made phylogenomics sensu O'Brien and Stanyon (1999). However, although the sequences of 269 plastid genomes of green plants have been deposited into the GenBank up to the end of 2012, this is still a very small number when compared with the 314,600 identified plant species (Mora et al. 2011). Plastid genome information is indispensable in biology, and easy and cheap methods for sequencing plastid genomes are urgently needed.
Three major strategies are usually used to sequence plastid genomes. The first approach is to isolate the plastids by the sucrose-gradient method. The plastid DNA is sometimes enriched by rolling circle amplification, long-range polymerase chain reaction (PCR), and bacterial artificial chromosome or fosmid library constructions. The long DNA is fragmented by endonucleases, ultrasonic devices, or other methods. The fragments are then cloned for sequencing (Goremykin et al. 2005; Wu et al. 2007; Cattolico et al. 2008; Lin et al. 2010; Wu, Lin, et al. 2011; Wu, Wang, et al. 2011). The second method is to first isolate total genomic DNA, construct a genomic DNA library, and sequence the library by the high-throughput next-generation sequencing (NGS) machines such as Illumina/Solexa, Roche/454, Danaher/Polonator, or ABI/SoLiD (Moore et al. 2006; Atherton et al. 2010; Lin et al. 2012). In the third method, genome-walking primers are designed for conserved regions, and the whole genome is sequenced by the Sanger process. The genomes of Amborella trichopoda (Goremykin et al. 2003b), Calycanthus fertilis (Goremykin et al. 2003a), Nymphaea alba (Goremykin et al. 2004), Acorus calamus (Goremykin et al. 2005), Cucumis sativus (Chung et al. 2007), Lemna minor (Mardanov et al. 2008), Dendrocalamus latiflorus and Bambusa oldhamii (Wu et al. 2009), Coix lacryma-jobi (Leseberg and Duvall 2009), and Oncidium (Wu et al. 2010) were determined using this protocol.
Isolation of chloroplast genomic DNA through the separation of chloroplasts appears to be reliable and was widely adopted in early studies. However, chloroplast isolation is time consuming and sometimes troublesome. Additionally, the resulting chloroplast DNA may not be pure, as it may be contaminated by material from other genomes. The determination of a plastid genome sequence through the NGS method is becoming increasingly popular because of the power of bioinformatics, the simplicity of the method, and the high throughput of data. The genome-walking method has attracted some attention. For example, partially universal primers have been proposed for the large single copy (LSC) region (Grivet et al. 2001) and for the inverted repeat (IR) sequence (Dhingra and Folta 2005). A chloroplast primer database has collected approximately 700 primers (Heinze 2007). Unfortunately, those primers are not intended for sequencing the whole plastid genomes, as they are not readily usable for such a purpose.
Many regions of the angiosperm plastid genomes are highly conserved. This conservativeness makes it possible to design universal primers for amplifying fragments covering almost the whole plastid genome, and the fragments can be sequenced using the Sanger method. We designed universal primers and utilized them on the species Sedum sarmentosum and Nelumbo spp. (Xue, Dong, et al. 2012; Xue, Wang, et al. 2012). These primers proved to be very useful. This article reports the development of those universal primers for short-range PCR, confirms their usefulness in sequencing the whole chloroplast genome using Liquidambar formosana as an example, and compares the three genomes determined using short-range PCR, long-range PCR, and the NGS methods. We also demonstrate the application of the universal primers in phylogenetic reconstruction of families in Saxifragales. We hope that these primers will accelerate the determination of the completely sequenced plastid genomes of flowering plants.
Materials and Methods
Genome Sequence Manipulation, Primer Design, and Universality Tests
Forty-eight complete plastid genomes of angiosperm species were downloaded from GenBank (supplementary table S1, Supplementary Material online). The coding genes (excepting tRNA) were extracted from the genome and were aligned with Clustal X ver. 2.0 (Larkin et al. 2007) and adjusted manually with Se-Al ver.2.0 (Rambaut 1996). Primer pairs were designed with Primer 5.0 using default settings to amplify fragments of approximately 1.5 kb. The overlap length was set to approximately 100 bp if possible. The primer pairs were synthesized by Sangon Biotech (Shanghai) Co. Ltd. (Beijing, China). Eight samples (supplementary table S2, Supplementary Material online) representing basal angiosperms, monocots, basal eudicots, Saxifragales, and rosid and asterid angiosperms were used to test the universality of the primers.
Confirmation of Single-Copy IR Boundary Regions
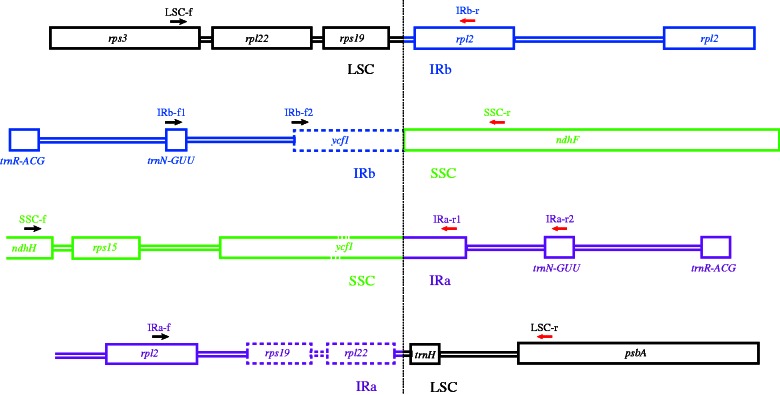
The IRa sequence is usually based on that of the IRb. The transition from LSC to IRb, IRb to small single copy (SSC), SSC to Ira, and IRa to LSC can be confirmed by the sequences of fragments amplified with region-specific primer pairs (fig. 1).
Fig. 1.—
Priming sites of the single-copy IR boundary region-specific primer pairs. The joining point is indicated by a vertical dotted line. The LSC, IRa, IRb, and SSC regions are indicated by different colors.
Determination of the Genome of L. formosana
Approximately 11 g of young, fresh leaves of L. formosana was homogenized in buffer A (Li et al. 2013), filtered into 50 µl centrifuge tubes, and centrifuged at 500 × g for 2 min. The supernatant was decanted to new tubes, and the precipitate was discarded to remove the nuclei. The new tubes were centrifuged at 1,000 × g to collect the chloroplasts, and the supernatant was discarded to remove the DNA and mitochondria. The precipitate (chloroplasts) was resuspended in buffer A and centrifuged to wash away the remaining DNA and mitochondria. DNA was extracted from the precipitate using the mCTAB method (Li et al. 2013) and purified using the Wizard DNA Clean-Up System (A7280, Promega Corporation, Madison, WI).
Two-thirds of the DNA was sent to the National Center for Gene Research, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China, to sequence the genome on the Illumina/Solexa Genome Analyzer II. Before assembling the paired-end Illumina/Solexa reads, the script fastq_quality_filter included in the FASTAX-Toolkit version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/download.html) was facilitated to remove reads of low quality (the minimum quality score to keep is 20). Reads that retained both ends were extracted using the Solexa Reads Preprocessor. The genome was assembled de novo using Velvet Assembler version 1.0.18 (Zerbino and Birney 2008) with the following parameters: a hash length of 57, minimum average coverage of 18, minimum contig length of 100 bp, expected coverage in short reads of unique sequences of 35, and insert length of 500 bp.
The concentration of the remaining DNA was diluted to approximately 5 ng/µl, for short- and long-range PCR. The long-range PCR followed the protocol of Wu et al. (2007), and the fragments were sequenced by the Shanghai Majorbio Pharm Technology Co., Ltd. The procedure for the short-range PCR was similar to Dong et al. (2012). A single annealing temperature of 55 °C was used for all primer pairs. Primer pair cp053f/cp053r and cp054f/cp054r failed, and a species-specific primer pair (Liqu053f 5′-CGG AAC GCG ATT GGT GTC TAA GAT-3′ and Liqu054r 5′-CCA TTC CCG ACG CAT CAT CCT CAT T-3′) was designed based on the flanking sequences to bridge the gaps. The PCR products were purified using PEG8000 and sequenced using BigDye Terminator v3.1 on an ABI prism 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). Sequence files from the DNA analyzer were checked and assembled with Sequencher v4.7 (Gene Codes, Ann Arbor, MI). The single-copy IR boundary regions were confirmed using region-specific primer pairs. The assembled genomes were annotated on the Dual Organellar Genome Annotator (DOGMA) (Wyman et al. 2004).
Variability Tests and Suitability Evaluations of Genes for Phylogeny
The variability of all protein-coding genes was parameterized by the number of polymorphic sites (S), nucleotide diversity per site (π), average number of nucleotide differences (k), and number of parsimony-informative characters (Npi), which were calculated using DnaSP 5.0 (Librado and Rozas 2009).
Phylogenetic Application of Plastid Genome Information: A Case Study on Saxifragales
We selected Saxifragales as an example because there are some systematic uncertainties about the families that could be solved using the plastid genome sequence. Incomplete sampling of families can cause phylogenetic uncertainty. We sampled 20 species (supplementary table S3, Supplementary Material online) representing all Saxifragales-related families, including some families previously belonging to Hamamelidales and major lineages within some of these families. Because of the unavailability of materials for analysis, the sequences of six Saxifragales-related taxa were downloaded from GenBank (supplementary table S4, Supplementary Material online). Nineteen plastid genomes of eudicots from GenBank (supplementary table S4, Supplementary Material online) were used as outgroups. Taking the length of fragments, S, π, k, Npi, and the availability of sequences in GenBank into consideration, 18 genes were used to construct the phylogeny of Saxifragales.
The best substitution model for each of the 18 genes was selected by ModelTest version 3.7 (Posada and Crandall 1998) under the Akaike information criterion. Maximum parsimony (MP) analysis, maximum likelihood (ML) analysis, and Bayesian inference (BI) were conducted on the concatenate data set of 45 species. Heuristic searches with 10,000 random-addition replicates were performed for MP using PAUP* 4.0b10 (Swofford 2002) with TBR branch swapping and the Multrees options. Nonparametric bootstrap support was evaluated for 1,000 replicates, each with 100 random-addition replicates, using the same settings. The nucleotide sequences were converted into amino acid sequences, and we then conducted MP analyses using PAUP. ML analysis was run with RAxML version 7.2.6 (Stamatakis 2006), following the procedure outlined in Moore et al. (2010). ML analyses were conducted by first running 100 bootstrap replicates. Every tenth bootstrap tree was used as a starting tree for a full ML search. The best tree resulting from those searches was considered to be the ML tree. A nonparametric bootstrap of 1,000 replicates was conducted to assess the uncertainty of the ML trees. MrBayes 3.2 (Ronquist et al. 2012) was used for BI. Two parallel analyses of four chains each were run for 50,000,000 generations. A burn-in of 25% was used for both analyses. The ML and Bayesian analyses were performed on the freely available Bioportal server (www.bioportal.uio.no) (Kumar et al. 2009).
Results
Primers and Their Universality
A total of 138 pairs of primers (supplementary table S5, Supplementary Material online) were designed to cover the whole plastid genome (supplementary fig. S1, Supplementary Material online). Among the primer pairs used were 75 pairs of amplifying protein-coding genes, 43 pairs of amplifying intergenic spacers and RNA genes, and 20 pairs of amplifying introns. The PCR products were designed to be 0.8–1.5 kb, a length suitable for PCR amplification and sequencing. Long fragment amplifications were performed in case no desirable priming sites exist, such as around trnT-psbD, rpoB-trnC, ycf1, and accD. Approximately 100 bp of overlap was considered sufficient to generate a reliable assembly. The sequences of some tRNA genes are too short to allow consideration of overlaps, and small gaps therefore needed to be bridged (supplementary fig. S1, Supplementary Material online).
Most of the primer sets produced single strong bands on the eight test species representing major lineages of angiosperm (supplementary fig. S2, Supplementary Material online). The PCR success rate was greater than 96% (supplementary table S2, Supplementary Material online). Sixteen pairs of primers (supplementary table S6, Supplementary Material online) were used for long-range PCR. The lengths of the fragments showed that they were from 6,704 bp to 13,298 bp.
Determination of Single-Copy IR Boundary Sites
The transitions of LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC (fig. 1) were confirmed by sequences amplified with the region-specific primers (supplementary table S7, Supplementary Material online). Some of the primer sequences were the same but annealed to the antisense strand. To avoid confusion, the primers were given new names (supplementary table S7, Supplementary Material online). Primer LSC-f (=cp096F) on rps3 of LSC with primer IRb-r (=cp096R) on rpl2 of IRb amplifies the single-copy IR boundary region fragment of LSC and IRb. Primer IRb-f2 (=cp124F) on the pseudogene ycf1 of IRb and primer SSC-r (=cp124R) on ndhF of SSC determines the single-copy IR boundary region of IRb and SSC. If the pseudogene ycf1 is lost in the taxon being tested, the combination of primer IRb-f1 (=cp123F) on trnNguu and primer SSC-r will work. The transitional area of SSC and IRa is more difficult to assess due to the long and variable ycf1 sequence. A reaction including primer SSC-f (=cp138F) on ndhH of SSC with primer IRa-r2 (=cp123F) on trnNguu or primer IRa-r1on ycf1 of IRa does not work efficiently due to the length of the fragment. Long-range PCR is usually necessary. Primer IRa-f (=cp096R) on rpl2 of IRa and primer LSC-r on psbA of LSC amplify the transitional region of IRa and LSC.
Complete Chloroplast Genome of L. formosana
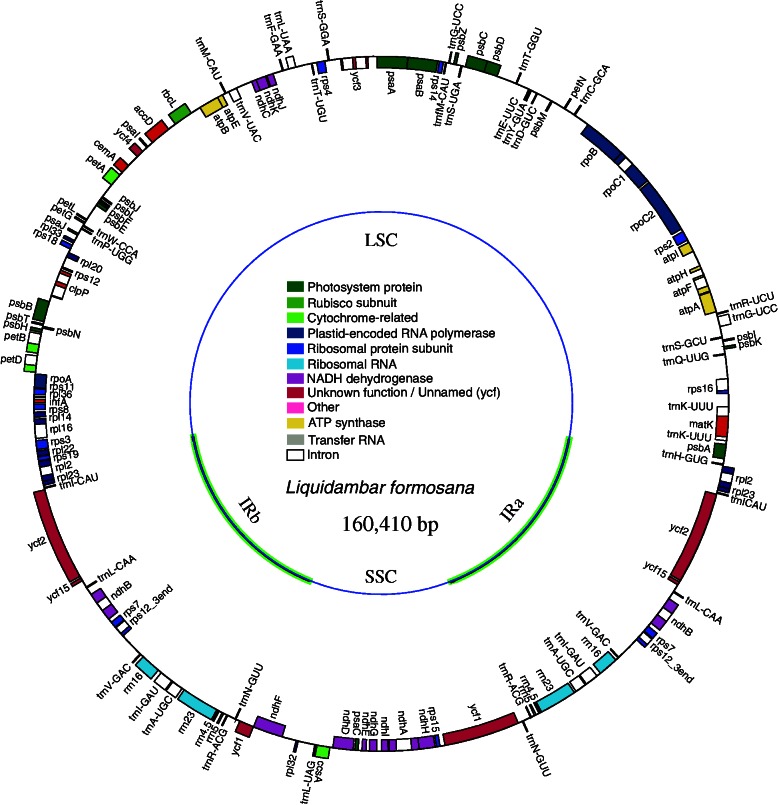
The chloroplast genomes of L. formosana sequenced independently, using the three methods, were completely identical (KC588388). A total of 311 fragments were used, covering the whole genome 1.966×. The chloroplast genome of L. formosana (fig. 2) had a length of 160,410 bp. The length of the LSC was 88,945 bp, the SSC was 18,917 bp, and the IRs were 26,274 bp. The IRa region terminates with the rpl2 gene. There were 131 genes in total (supplementary table S8, Supplementary Material online), including 79 protein-coding genes, 30 tRNA genes, 4 rRNA genes, and 1 pseudogene (ycf15). Sixteen genes have one intron each, and two genes (clpP and ycf3) have two introns. The G + C content was 37.95% for the whole genome and 42.08% in IR regions (table 1).
Fig. 2.—
Gene map of the Liquidambar formosana chloroplast genome. The thick lines indicate the extent of the IRs (IRa and IRb), which separate the genome into SSC and LSC regions. Genes on the outside of the map are transcribed clockwise, whereas genes on the inside of the map are transcribed counterclockwise. More detailed information is given in table 1 and supplementary table S8, Supplementary Material online.
Table 1.
Characteristics of the Chloroplast Genome of Liquidambar formosana
| Length (bp) | Whole genome | 160,410 |
| LSC | 88,945 | |
| IR | 26,274 | |
| SSC | 18,917 | |
| GC content (%) | Whole genome | 37.95 |
| IR region | 43.08 | |
| LSC region | 36.10 | |
| SSC region | 32.42 | |
| Coding regions | 40.39 | |
| Number of genes | Total | 131 |
| IR region (including rps12, ycf1) | 18 | |
| tRNA genes | 30 | |
| rRNA genes | 4 | |
| Pseudogene | 1 | |
| With one intron | 16 | |
| With two introns | 2 |
Relative Variability of Genes in Saxifragales
The variability of genes differs within the Saxifragales group (supplementary table S9, Supplementary Material online). The top 20 genes in terms of S, k, and Npi are accD, atpA, atpB, ccsA, matK, ndhA, ndhD, ndhF, ndhH, petA, psaA, psaB, psbB, psbC, rbcL, rpoA, rpoB, rpoC1, rpoC2, and ycf1. The top 10 candidates are matK, ndhD, ndhF, rpoB, rpoC1, rpoC2, and ycf1. These genes are usually longer than 1 kb. In terms of π, accD, ccsA, matK, ndhA, ndhD, ndhF, rpoA, rpoC2, and ycf1 rank in the top 20, and ccsA, matK, ndhD, ndhF, and ycf1 rank in the top 10. Therefore, matK, ndhD, ndhF, and ycf1 are the most variable chloroplast genes in Saxifragales.
Phylogeny of Saxifragales
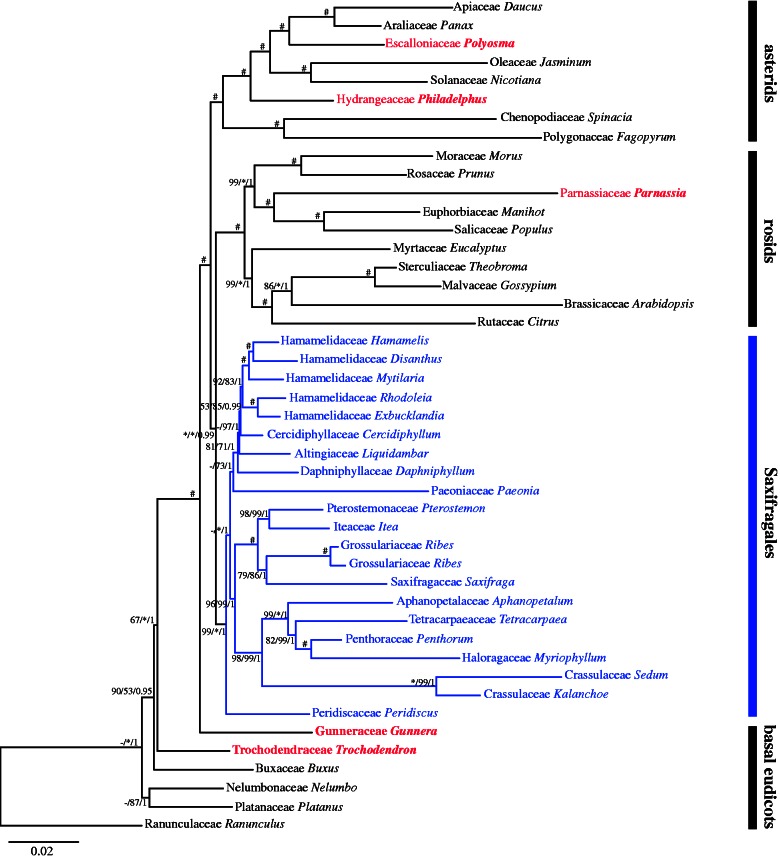
The combined data set of 18 plastid genes (supplementary table S10, Supplementary Material online) of all 45 accessions reached a length of 28,686 bp after the exclusion of rare insertions and some unreliably aligned sites. There are 6,578 parsimony-informative sites in the data set. MP, ML, and BI trees suggest that Gunneraceae belong to the basal eudicots, Hydrangeaceae belong to asterids, and Parnassiaceae belong to rosids. The monophyly of Saxifragales is highly supported (fig. 3, supplementary fig. S3, Supplementary Material online), and its close relationship with rosids is indicated. Within Saxifragales, there are six well-supported lineages: 1) Hamamelidaceae (including Altingiaceae, Cercidiphyllaceae, and Daphniphyllaceae); 2) Saxifragaceae (including Grossulariaceae, Iteaceae, and Pterostemonaceae); 3) Crassulaceae; 4) Haloragaceae (including Aphanopetalaceae and Penthoraceae); (5) Paeoniaceae; and 6) Peridiscaceae.
Fig. 3.—
Phylogram of the best ML tree based on 18 chloroplast regions with 6,578 parsimony-informative sites from a total length of 26,686 bp. Numbers associated with branches are bootstrap supports (>50%) and Bayesian posterior probabilities (>0.5) for the branches in the order MP/ML/BI. *Indicates 100, and #indicates 100/100/1.
Discussion
The PCR-based method (genome walking) had been used in the early attempts to sequence many plastid genomes such as A. trichopoda (Goremykin et al. 2003b), C. fertilis (Goremykin et al. 2003a), N. alba (Goremykin et al. 2004), Oryza nivara (Masood et al. 2004), and Aco. calamus (Goremykin et al. 2005). The field was silent for a few years due to concerns about incorporating extra fragments into the plastid genomes and producing incorrect genome structure because of plastid genome substructuring or high divergence (Jansen et al. 2005). Realizing that such mistakes are unlikely, the same strategy was soon used extensively on plants for which the reference genomes were available (Ibrahim et al. 2006; Mardanov et al. 2008; Wu et al. 2009). However, the same concern remains for the taxa without a reliable reference. There have been some attempts to solve the problem, for example, to determine the IRs of the chloroplast genomes of Fragaria × ananassa and Prunus persica with universal primers (Dhingra and Folta 2005). Similar to the logic of genome sequencing using NGS, the plastid genome de novo sequencing using either long-range or short-range PCR-based methods is theoretically reliable. The three identical genomes of L. formosana, determined by the three different methods used in this study, demonstrate the reliability of the three methods.
Reliability of Genomes Determined Using the Short-Range PCR-Based Method
The reliability of the assemblage is a major concern for the determination of a complete plastid genome through PCR direct sequencing. Although there is only approximately 2× coverage, the Sanger sequencing method is the most robust technique in use, and an overlap of 50–100 bp is sufficient to exclude paralogs from assembling into contigs. The concerted evolution of the two copies of IR is complete, which makes direct sequencing possible without cloning. Conventional practice allows for the copying and pasting of one IR sequence into the correct position. The IR regions can also be sequenced separately by amplifying each of them using the long-range PCR method, as used in Wu, Lai, et al. (2009).
The transitions of LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC (fig. 1) may be of concern. Although obtaining results is often not a problem, verifications are sometimes necessary because the IRa is usually based on IRb (Jansen et al. 2005). The primers for amplifying the single-copy IR boundary region (Supplementary table S7, Supplementary Material online) sometimes do not work because of genome substructuring or too variable flanking sequences of the IR regions, for instance, within Araceae (Ahmed et al. 2012). Taxon-specific primers are preferred after the genome has been assembled. The direction of SSC is potentially erroneous if the ycf1 and the ycf1 pseudogene are misplaced. To ensure the correct orientation of SSC, amplification of the entire IR region is necessary, preferably amplifying the IRb for shorter length and more specific priming.
One concern in the use of this method is the possibility that exogenous fragments would be incorporated into the plastid genome or that the fragments of different loci would be assembled into the wrong loci. The alien can hardly be assembled into a plastid genome because it is usually very different or has unreadable peaks when the copies are different. To solve this problem, PCR using adjacent primer combinations would finally eliminate the nonplastid copies. Although repeats have been reported in several groups of plants (Jansen et al. 2005 and the references there in), long repeats outside the IR regions are very rare. Exceptions are found on the biparentally inherited plastid genomes, for instance, Trachelium caeruleum (Haberle et al. 2008) and Geraniaceae (Guisinger et al. 2011), which have extensive structural reorganizations. To sequence such genomes, the short-range PCR method will provide sequences for designing taxa-specific primers for long-range PCR.
PCR-based plastid genome sequencing was previously considered labor-intensive and time consuming. With this set of primers, only one and a half PCR plates are required for the whole genome. The efficiency is identical to that of conventional PCR direct sequencing.
Usefulness of the Universal Primer Set
The primers given in supplementary table S5, Supplementary Material online, have been tested across all major groups of angiosperms (supplementary fig. S1, Supplementary Material online) and have proven to be efficient for most cases. In addition to that of L. formosana, the chloroplast genomes of Nelumbo lutea (Xue, Dong, et al. 2012), Penthorum chinense, S. sarmentosum, and Haloxylon spp. (unpublished) have been determined largely using the primer set. The primers can also be very useful for studies such as Soltis et al. (2011) and Jian et al. (2008), in which large data sets are required but the exact plastid genomes are of less consequence.
The universal primer set significantly eases the determination of individual genomes for investigators who have specific interests in certain species. The sequences generated using the universal primer set serve as references for designing taxon-specific primers. The primer set is also useful for those interested in the genomes of remotely related species. In this case, the PCR products can be sequenced using a long-reading NGS machine (such as Roche 454) directly without tag labels. Sequencing many species simultaneously in one run is very convenient; the primers significantly reduce the cost and quicken the accumulation of genomes in GenBank.
Utility of the Primer Set in the Phylogenetic Reconstruction of Saxifragales
Recent phylogenetic studies based on multiple genes (Moore et al. 2011; Soltis et al. 2011) suggested a close relationship among the families of Saxifragales and the families formerly belonging to Hamamelidales. The morphology-based circumscriptions of the two orders and their systematic positions are questionable from a molecular viewpoint. The systematic positions of several families formerly grouped in Hamamelidales and Saxifragales were no longer associated with these groups (fig. 3). Gunneraceae and Trochodendraceae are positioned among the basal eudicots rather than as close relatives of Haloragaceae and Hamamelidales, respectively. Philadelphus and Parnassia—formerly in Saxifragaceae—nest with asterids and rosids, respectively. Saxifragales can be treated as an order of the rosids. Considering short branches and relatively low bootstrap supports, it seems more natural to merge 1) Altingiaceae, Cercidiphyllaceae, and Daphniphyllaceae into Hamamelidaceae, 2) Grossulariaceae, Iteaceae, and Pterostemonaceae into Saxifragaceae, and 3) Aphanopetalaceae, Myriophyllaceae, Penthoraceae, and Tetracarpaeaceae into Haloragaceae.
Gaps of the Primer Set and Working Tips
Most of the primers used in this study are predicted to work for a majority of angiosperms. However, difficulties might arise from some primers located at rapidly evolving regions such as accD, matK, ndhF-rpl32, rpl20/clpP, rpoB, rps4, rps8, and ycf1. The ycf1 gene would require further analysis due to its long length and rapid evolution. For such regions, taxon-specific primers (Neubig and Abbott 2010; Franck et al. 2012) and long-range PCR are needed. Some PCR failures could result from the loss of genetic regions rather than primer problems. For example, ycf1 and ycf2 are absent in Poaceae. New combinations of forward and reverse primers at adjacent sites could rectify such problems. It is also strongly recommended to verify the IR regions with fragment sequences amplified with taxon-specific primers after the genome has been assembled.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S10 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Shan Yuan and Junjie Guo for their help obtaining samples. This work was supported in part by the National Natural Science Foundation of China (31270239 and 30930010) and the Ministry of Science and Technology (2011FY120200, 2012BAC01B05-6, and 2012AA021602) to S.Z.
Literature Cited
- Ahmed I, et al. Mutational dynamics of aroid chloroplast genomes. Genome Biol Evol. 2012;4:1316–1323. doi: 10.1093/gbe/evs110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton RA, et al. Whole genome sequencing of enriched chloroplast DNA using the Illumina GAII platform. Plant Methods. 2010;6:22. doi: 10.1186/1746-4811-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattolico RA, et al. Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics. 2008;9:211. doi: 10.1186/1471-2164-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SM, Gordon VS, Staub JE. Sequencing cucumber (Cucumis sativus L.) chloroplast genomes identifies differences between chilling-tolerant and -susceptible cucumber lines. Genome. 2007;50:215–225. doi: 10.1139/g07-003. [DOI] [PubMed] [Google Scholar]
- Dhingra A, Folta KM. ASAP: amplification, sequencing & annotation of plastomes. BMC Genomics. 2005;6:176. doi: 10.1186/1471-2164-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin L, et al. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AR, Cochrane BJ, Garey JR. Low-copy nuclear primers and ycf1 primers in Cactaceae. Am J Bot. 2012;99:e405–e407. doi: 10.3732/ajb.1200128. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. The chloroplast genome of the “basal” angiosperm Calycanthus fertilis—structural and phylogenetic analyses. Plant Syst Evol. 2003a;242:119–135. [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that amborella is not a basal angiosperm. Mol Biol Evol. 2003b;20:1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol. 2004;21:1445–1454. doi: 10.1093/molbev/msh147. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol. 2005;22:1813–1822. doi: 10.1093/molbev/msi173. [DOI] [PubMed] [Google Scholar]
- Grivet D, Heinze B, Vendramin GG, Petit RJ. Genome walking with consensus primers: application to the large single copy region of chloroplast DNA. Mol Ecol Notes. 2001;1:345–349. [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66:350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Heinze B. A database of PCR primers for the chloroplast genomes of higher plants. Plant Methods. 2007;3:4. doi: 10.1186/1746-4811-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim RI, Azuma J, Sakamoto M. Complete nucleotide sequence of the cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes Genet Syst. 2006;81:311–321. doi: 10.1266/ggs.81.311. [DOI] [PubMed] [Google Scholar]
- Jansen RK, et al. Methods for obtaining and analyzing whole chloroplast genome sequences. In: Zimmer EA, Roalson E, editors. Methods in enzymology. Boston: Academic Press; 2005. pp. 348–384. [DOI] [PubMed] [Google Scholar]
- Jansen RK, et al. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 2006;6:32. doi: 10.1186/1471-2148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian SG, et al. Resolving an ancient, rapid radiation in Saxifragales. Syst Biol. 2008;57:38–57. doi: 10.1080/10635150801888871. [DOI] [PubMed] [Google Scholar]
- Kumar S, et al. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hahn F, McMahan C, Cornish K, Whalen M. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009;9:131. doi: 10.1186/1471-2229-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leseberg CH, Duvall MR. The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J Mol Evol. 2009;69:311–318. doi: 10.1007/s00239-009-9275-9. [DOI] [PubMed] [Google Scholar]
- Li J, Wang S, Jing Y, Wang L, Zhou S. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 2013;48:72–78. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lin CP, Huang JP, Wu CS, Hsu CY, Chaw SM. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol. 2010;2:504–517. doi: 10.1093/gbe/evq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Wu CS, Huang YY, Chaw SM. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 2012;4:374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardanov AV, et al. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 2008;66:555–564. doi: 10.1007/s00239-008-9091-7. [DOI] [PubMed] [Google Scholar]
- Masood MS, et al. The complete nucleotide sequence of wild rice (Oryza nivara) chloroplast genome: first genome wide comparative sequence analysis of wild and cultivated rice. Gene. 2004;340:133–139. doi: 10.1016/j.gene.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Moore MJ, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, et al. Phylogenetic analysis of the plastid inverted repeat for 244 species: insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region. Int J Plant Sci. 2011;172:541–558. [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Nat Acad Sci U S A. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. How many species are there on earth and in the ocean? PLoS Biol. 2011;9:e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubig KM, Abbott JR. Primer development for the plastid region ycf1 in Annonaceae and other Magnoliids. Am J Bot. 2010;97:E52–E55. doi: 10.3732/ajb.1000128. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Stanyon R. Phylogenomics—ancestral primate viewed. Nature. 1999;402:365–366. doi: 10.1038/46450. [DOI] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A. Oxford: University of Oxford, Department of Zoology; 1996. Se-Al: sequence alignment editor, version 2.0. [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli N, et al. A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. PLoS One. 2011;6:e19954. doi: 10.1371/journal.pone.0019954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, et al. Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford D. 2002. PAUP*. Phylogenetic analysis using parsimony (and other methods) Version 4.0 beta. Sunderland (MA): Sinauer. [Google Scholar]
- Wortley AH, Rudall PJ, Harris DJ, Scotland RW. How much data are needed to resolve a difficult phylogeny? Case study in Lamiales. Syst Biol. 2005;54:697–709. doi: 10.1080/10635150500221028. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lai YT, Lin CP, Wang YN, Chaw SM. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: selection toward a lower-cost strategy. Mol Phylogenet Evol. 2009;52:115–124. doi: 10.1016/j.ympev.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 2011;3:309–319. doi: 10.1093/gbe/evr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 2011;3:1284–1295. doi: 10.1093/gbe/evr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Liu SM, Chaw SM. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol. 2007;24:1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- Wu FH, et al. Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol. 2009;29:847–856. doi: 10.1093/treephys/tpp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xue JH, Dong WP, Cheng T, Zhou SL. Nelumbonaceae: systematic position and species diversification revealed by the complete chloroplast genome. J Syst Evol. 2012;50:477–487. [Google Scholar]
- Xue JH, Wang S, Zhou SL. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae) Am J Bot. 2012;99:E240–E244. doi: 10.3732/ajb.1100547. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.