Abstract
Mitochondrial processing peptidase (MPP) consists of α and β subunits that catalyze the cleavage of N-terminal mitochondrial-targeting sequences (N-MTSs) and deliver preproteins to the mitochondria. In plants, both MPP subunits are associated with the respiratory complex bc1, which has been proposed to represent an ancestral form. Subsequent duplication of MPP subunits resulted in separate sets of genes encoding soluble MPP in the matrix and core proteins (cp1 and cp2) of the membrane-embedded bc1 complex. As only α-MPP was duplicated in Neurospora, its single β–MPP functions in both MPP and bc1 complexes. Herein, we investigated the MPP/core protein family and N-MTSs in the kinetoplastid Trypanosoma brucei, which is often considered one of the most ancient eukaryotes. Analysis of N-MTSs predicted in 336 mitochondrial proteins showed that trypanosomal N-MTSs were comparable with N-MTSs from other organisms. N-MTS cleavage is mediated by a standard heterodimeric MPP, which is present in the matrix of procyclic and bloodstream trypanosomes, and its expression is essential for the parasite. Distinct Genes encode cp1 and cp2, and in the bloodstream forms the expression of cp1 is downregulated along with the bc1 complex. Phylogenetic analysis revealed that all eukaryotic lineages include members with a Neurospora-type MPP/core protein family, whereas cp1 evolved independently in metazoans, some fungi and kinetoplastids. Evolution of cp1 allowed the independent regulation of respiration and protein import, which is essential for the procyclic and bloodstream forms of T. brucei. These results indicate that T. brucei possesses a highly derived MPP/core protein family that likely evolved in response to its complex life cycle and does not appear to have an ancient character proposed earlier for this eukaryote.
Keywords: mitochondrial processing peptidase, bc1 complex, mitochondrial targeting sequence, trypanosome, evolution
Introduction
Parasitic protists of the Trypanosoma brucei family are causative agents of African sleeping sickness in humans and nagana of domestic animals. Trypanosomes undergo a complex life cycle, with the procyclic form (PF) inhabiting the gut of a tsetse fly and the bloodstream form (BF) being pathogenic in vertebrate hosts. The cyclic changes between the BF and PF are accompanied by dramatic changes in the parasite’s metabolism, particularly by the activation of the citric acid cycle and respiratory chain in the mitochondrion of the PF and their downregulation, followed by a switch to glycolysis in the BF (Clayton and Michels 1996; Besteiro et al. 2005).
Trypanosoma brucei belongs to the class Kinetoplastea, which commonly have a large network of mitochondrial DNA termed the kinetoplast. Together with related euglenids and diplonemids, kinetoplastid flagellates form the taxon Euglenozoa within the eukaryotic supergroup Excavata (Adl et al. 2012). Several lineages of excavates were previously considered to be among the most ancient eukaryotes based on the ostensible absence of mitochondria (Cavalier-Smith 1987). However, the “amitochondriate” hypothesis eroded when hydrogenosomes of Trichomonas and mitosomes of Giardia were recognized as anaerobic reduced forms of mitochondria (Embley and Martin 2006). More recently, kinetoplastids, such as T. brucei, were proposed to be the most ancient eukaryotes based on some unique features of their mitochondria, particularly mitochondrial protein import (Cavalier-Smith 2010).
During eukaryogenesis, the majority of genes encoded in the genome of the proteobacterial ancestor of mitochondria were lost or transferred to the nucleus. Consequently, more than 95% of proteins in the contemporary mitochondria are encoded in the nucleus, synthesized in the cytosol and imported into the mitochondrial membranes or matrix (Lill and Neupert 1996; Neupert and Herrmann 2007). Correct protein targeting and translocation is assured mainly by targeting sequences at the amino termini (N-MTS) of imported mitochondrial preproteins (Chacinska et al. 2009). The N-MTS is initially recognized by a receptor in the outer membrane translocase (TOM) complex. The preprotein is then translocated across the β-barrel-shaped Tom40 channel and interacts with the inner membrane translocase machinery (TIM), which mediates its translocation through the inner membrane. Upon translocation, the N-MTS is cleaved by the mitochondrial processing peptidase (MPP). Trypanosoma brucei possesses a considerably divergent Tom40 that had not been recognized in initial bioinformatic searches of its genome, and a structurally similar voltage-dependent anion channel (VDAC) has been found as the only mitochondrial porin (Pusnik et al. 2009). More recent studies have revealed the presence of another import channel called the archaic translocase of the outer mitochondrial membrane (ATOM) (Pusnik et al. 2011). ATOM has been suggested to represent an ortholog of the bacterial Omp85 protein family, which is distinct from Tom40 and may represent an evolutionarily ancestral protein transport system. However, later this view has been challenged by Zarsky et al. (2012) who found that ATOM is most likely a divergent ortholog of a mitochondrial TOM40.
A typical mitochondrial system of inner membrane translocases includes the TIM23 complex, which is dedicated to the import of matrix proteins, and the TIM22 complex, which mediates the insertion of proteins into the inner membrane (Bauer et al. 2000). However, another unique feature of the T. brucei import machinery is that only a single member of the Tim17/22/23 protein family has been found in its genome, suggesting that a single TIM complex facilitates the functions of both TIM23 and TIM22 in the single mitochondrion of this unicellular parasite (Schneider et al. 2008). Finally, analysis of the N-MTSs of five mitochondrial preproteins revealed that they are considerably shorter in T. brucei (usually 8–20 amino acid residues [AA]) (Priest and Hajduk 1995; Hauser et al. 1996; Häusler et al. 1997; Bertrand and Hajduk 2000) compared with the average length of N-MTSs in ophistokonts and plants (20–80 AA in yeast/mammals; 20–70 AA in plants) (Burri and Keeling 2007; Huang et al. 2009). Although long N-MTSs were rarely documented in T. brucei (Long et al. 2008), the generally short nature of its N-MTSs was interpreted as a primitive feature of the import system (Schneider et al. 2008; Pusnik et al. 2009).
Primary sequences of AA residues in N-MTSs are usually rich in hydrophobic and basic residues, such as arginine (R), which allow for the formation of amphipathic α-helices and are generally not conserved (Gakh et al. 2002). The proximal arginine is either at position −2 (R-2) or −3 (R-3) from the cleavage site, whereas the distal arginines or other basic residues contribute to the overall positive net charge of N-MTS (Moriwaki et al. 1999; Taylor et al. 2001; Huang et al. 2009). The site of N-MTSs cleavage contains a loosely conserved motif, with aromatic and bulky hydrophobic AA at the +1 position and hydrophilic AA at the +2 and +3 positions (Shimokata et al. 1997; Song et al. 1998). A recent proteomic analysis of the amino termini of the mature mitochondrial proteins in yeast revealed that N-MTSs with the R-2 motif are processed by MPP, whereas those with R-3 motif undergo two-step processing by MPP and Icp55, with the latter enzyme removing just a single AA, such as tyrosine, leucine, or phenylalanine (Vogtle et al. 2009).
MPP is a zinc-dependent metallopeptidase that consists of two homologous subunits, α and β, which are both required for its processing activity (Schneider et al. 1990; Geli 1993). The α-MPP subunit possesses a highly conserved glycine-rich loop required for the recognition of N-MTSs (Nagao et al. 2000; Dvorakova-Hola et al. 2010), whereas the metal-binding motif HXXEHXnE of β-MPP is the catalytic site responsible for cleavage of the peptide bond (Kitada et al. 1995; Striebel et al. 1996). In various eukaryotes, such as metazoans and Saccharomyces cerevisiae, MPP is present in the mitochondrial matrix as a general presequence processing enzyme. In addition, these organisms contain core I (cp1) and core II (cp2) proteins, which are homologous to β-MPP and α-MPP, respectively (Braun and Schmitz 1995). The cp1/cp2 proteins are components of the mitochondrial ubiquinol–cytochrome c oxidoreductase (bc1) complex of the respiratory chain (Deng et al. 2001; Zara et al. 2004). The core proteins process a precursor of the Rieske protein, and the cleaved presequence is retained as subunit IX of the bc1 complex (Deng et al. 2001). Unlike α-MPP, cp2 does not contain a glycine-rich loop, and compared with β-MPP, the Zn-binding motif of mammalian cp1 is incomplete (Gencic et al. 1991). None of the Zn-binding ligands are present in cp1/cp2 of S. cerevisiae (Braun and Schmitz 1995). Interestingly, in plants, the core proteins are identical to the α- and β-MPP subunits, and the majority of the MPP activity is associated with the bc1 complex (Eriksson et al. 1996). Thus, in addition to its core function in the respiratory chain, the bc1 complex of plants is involved in general N-MTS processing. To distinguish plant MPP subunits from cp1/cp2, we designate herein the former as cb1-α-MPP and cb1-β-MPP. It has been proposed that MPP evolved from a single subunit of a bacterial protease, followed by gene duplication and specialization that gave rise to a heterodimeric enzyme in mitochondria (Kitada et al. 2007), although more recent studies suggested that the heterodimeric protease was formed already in bacteria (Maruyama et al. 2011). Subsequently, another gene duplication resulted in the appearance of the membrane-associated core proteins and soluble MPP subunits. The association between MPP and the bc1 complex in plants suggests that a heterodimeric MPP was part of this complex before the second gene duplication occurred, with the complete set of cp1/cp2 and soluble heterodimeric MPP representing a more recent phylogenetic event (Braun and Schmitz 1995).
The proposed ancient character of the T. brucei import system prompted us to re-evaluate features of N-MTSs in this flagellate, the enzyme responsible for their processing and the character of the core proteins. We predicted 336 N-MTSs that appeared to have standard properties when compared with other eukaryotes. We identified a complete set of genes coding for α/β-MPP subunits as well as for cp1/cp2 proteins. Both MPP subunits were expressed in PF and BF, and we demonstrated an ability of the recombinant MPP to catalyze processing of N-MTS in the predicted cleavage site. The cp1 protein was expressed only in PF, which is consistent with the independent regulation of respiration and protein import in T. brucei. To obtain insight into the evolutionary history of this interesting family of essential eukaryotic proteins, we subjected the T. brucei α/β-MPP and cp1/cp2 proteins to an extensive phylogenetic analysis that included members of all known eukaryotic supergroups.
Materials and Methods
Organisms
Trypanosoma brucei procyclic (PF) strain 29–13 with a T7 RNA polymerase under neomycin resistance and tetracycline (TET) repressor under hygromycin resistance was grown in SDM-79 medium with 10% fetal calf serum at 27 °C (Wirtz and Clayton 1995). BF strain STIB 920 of the same species was used for infection of rats and isolated from their blood using diethyl amino-ethyl cellulose chromatography (Chaudhuri et al. 1995). The BF single marker strain 427 (Wirtz et al. 1999) with a T7 RNA polymerase and TET repressor under neomycin resistance was grown in HMI-9 medium (Hirumi and Hirumi 1984) under 5% CO2 at 37 °C.
Preparation of Recombinant Proteins and Antibodies
Genes for the T. brucei Tbα-MPP (Tb927.2.4110) and Tbβ-MPP (Tb09.160.3110), Tbcp1 (Tb927.5.1060) and iron–sulfur cluster assembly protein (TbIscU) were amplified from genomic DNA of the 29–13 strain, cloned into pET42b (Novagen, Germany) and expressed in Escherichia coli strain BL21-DE3 with a C-terminal 6×histidine tag. The recombinant proteins were isolated using nickel column chromatography (HiTrap Chelating, Qiagen, Netherlands) under denaturing conditions. Tbα-MPP, Tbβ-MPP, and Tbcp1 were further purified by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and used to raise polyclonal antibodies in rats. TbIscU was used for the cleavage assay (discussed later). Polyclonal antibodies against Tb-Cpn60, Tb-enolase, Tb-porin, and the Leishmania tarentolae Rieske protein were kindly provided by S.L. Hajduk (University of Georgia, USA), P. Michels (University of Edinburgh, Scotland), M. Chaudhuri (Meharry Medical College, USA), and L. Simpson (University of California, USA), respectively.
Isolation of Native Recombinant TbMPP
To obtain native recombinant TbMPP, both subunits were co-expressed in E. coli Rosetta (DE3) pLysS with the chaperonins GroEL/ES (Du Pont) in the presence of 1 mM ZnCl2 and 0.1 mM IPTG at 20 °C for 16 h. Tbα-MPP with a carboxy-terminal 6× histidine tag was expressed using a pET28a plasmid (Novagen, Germany) with ampicillin resistance. Tbβ-MPP without a tag was produced using the pET42b plasmid (Novagen, Germany) with kanamycin resistance. The heterodimeric MPP was isolated under native conditions using a nickel column (Ni-NTA Agarose, Qiagen, Netherlands) that binds the polyhistidine tag of the Tbα-MPP subunit. Cells were resuspended in lysis buffer (50 mM HEPES, 20 mM NaCl, 10 mM imidazole, 10% glycerol, pH 8) supplemented with complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail (Roche, Switzerland) and disrupted twice by a French press (1,400 bar; Thermo Scientific, USA). The lysate was centrifuged at 200,000 × g for 10 min at 4 °C, and the resulting supernatant (cleared lysate) was loaded onto the nickel column. Unbound proteins were removed by two washes with 5 ml of lysis buffer containing 40 mM imidazole. Proteins retained on the column were stepwise eluted with lysis buffer containing increasing concentrations of imidazole (100–250 mM). Alternatively, both proteins were expressed separately in E. coli Rosetta (DE3) pLysS with the chaperonins GroEL/ES (Novagen, Germany). Histidine-tagged Tbα-MPP was isolated under native conditions on a nickel column (Ni-NTA Agarose, Qiagen, Netherlands) following the manufacturer’s instructions. Isolated Tbα-MPP (∼30 µg) was added to the cleared lysate (20 ml) from the E. coli cells expressing Tbβ-MPP and incubated in dialysis tubing (Membra-Cel; MWCO 3500; Serva, Germany) for 16 h at 4 °C in 5 l of the lysis buffer supplemented with 1 mM ZnCl2. The heterodimer of Tbα-MPP and Tbβ-MPP was then isolated on the nickel column as described earlier.
Cloning and Expression of TbMPP in T. brucei
Genes for Tbα-MPP and Tbβ-MPP subunits were cloned into the pJH54 plasmid (kindly provided by Christine Clayton, University of Heidelberg, Germany), which allows for the expression of recombinant preproteins with a carboxy-terminal HA-tag (Wirtz and Clayton 1995). The plasmids were linearized by the NotI restriction enzyme and electroporated into the PF T. brucei by Gene Pulser Xcell (Bio-Rad, USA). The protein expressing cells were selected using 2.5 μg/ml phleomycin.
Plasmid pT7-3V5-PAC (Flaspohler et al. 2010) was used for the expression of V5-tagged Tbα-MPP and Tbβ-MPP in BF T. brucei. Genes were subcloned, and the NotI-linearized plasmids were electroporated by Amaxa Nucleofector (Lonza, Switzerland) and transformed cells were selected using 0.1 μg/ml puromycin. Protein expression was triggered by adding 1 μg/ml of tetracycline to the medium.
Immunofluorescent Microscopy
Approximately 1×107 PF and BS trypanosomes were incubated with 0.5 µM Mitotracker Red CMXRos (Molecular Probes, USA) for 10 min in SDM-79 and HMI-9, respectively, washed with phosphate-buffered saline (PBS), incubated in corresponding media for another 20 min and washed with PBS. The PF trypanosomes were fixed and permeabilized on slides in −20 °C cold methanol for 5 min, followed by a 5-min incubation in −20 °C cold acetone. BF trypanosomes were fixed on slides in 4% formaldehyde for 5 min at room temperature, washed with PBS and permeabilized in methanol for 15 min at −20 °C. The slides were incubated in blocking solution (0.25% bovine serum albumin, 0.25% gelatin, and 0.05% Tween 20 in PBS) for 1 h at room temperature. Recombinant Tbα-MPP and Tbβ-MPP were visualized using the mouse monoclonal anti HA-tag (Sigma-Aldrich, USA) and anti V5-tag (Sigma-Aldrich, USA) antibodies, respectively, and donkey anti-rat Alexa Fluor 488 antibody (Invitrogen, USA). The cells were mounted in Vectashield with DAPI (Vector Laboratories, USA) and observed using an Olympus IX81 microscope. Images were captured using a Hamamatsu Orca-AG digital camera and processed using cell^R imaging software (Olympus, Japan).
Isolation and Analysis of Mitochondria
The mitochondrion-enriched fraction and cytosolic fraction from T. brucei BF and PF were isolated by digitonin fractionation (Smid et al. 2006). The purity of fractions was assessed by measuring the enzymatic activity of mitochondrial and cytosolic marker enzymes threonine dehydrogenase and pyruvate kinase, respectively (Smid et al. 2006). In addition, the purity of the fractions was evaluated by Western blot analysis using antibodies against mitochondrial (Cpn60 and porin) and cytosolic (enolase) marker proteins. Enzymatic assays indicated less than 2% cross-contamination between the mitochondrial and cytosolic fractions.
To separate mitochondrial membrane and matrix fractions, 2 mg of the mitochondrion-enriched fraction was treated with 1 mg of digitonin in 400 μl of SHE buffer (250 mM sucrose, 25 mM HEPES, and 1 mM EDTA, pH 7.4) on ice for 30 min, followed by centrifugation (200,000 × g for 20 min). The supernatant (mitochondrial matrix fraction) and washed pellet (mitochondrial membrane fraction) were resuspended in SDS-PAGE sample buffer and boiled for 5 min.
ImageJ software (http://rsbweb.nih.gov/ij/, last accessed February 19, 2013) was used for quantification and comparison of protein signals acquired by Western blot analysis.
TbMPP RNA Interference Knock-Down
The Tbβ-MPP protein was ablated in T. brucei using RNA interference (RNAi). Part of the gene for Tbβ-MPP (480 nt) was subcloned into the p2T7-177 plasmid containing a T7 promoter, TET operator and the gene for phleomycin resistance (Wickstead et al. 2002) and electroporated into PF 29–13 cells. Upon the addition of TET to the medium, the extent of RNAi-targeting Tbβ-MPP was followed by northern and Western blot analyses using a labeled probe and rat polyclonal anti-Tbβ-MPP antibody, respectively. Cell viability was observed for 14 days and recorded as a growth curve. Cells were counted by a Z2 Coulter Particle Counter (Beckman Coulter, USA).
MPP Cleavage Assay
The reaction mixture contained 0.5 µg of recombinant TbIscU as substrate and 0.2 µg of recombinant TbMPP heterodimer or 2 µg of mitochondrial lysate in the reaction buffer (50 mM HEPES, 20 mM NaCl, and 1 mM MnCl2, pH 8.6) in a total volume of 20 µl. The reaction was conducted for 30 min at 27 °C and stopped by boiling for 5 min in SDS-PAGE sample buffer. The cleavage products were separated by SDS-PAGE and stained with Coomassie brilliant blue. The mitochondrial lysate was prepared by treating the mitochondrion-enriched fraction for 5 min with 0.1% Triton X-100, followed by centrifugation for 5 min at 16,000 × g. The supernatant was used for the cleavage assays.
In Silico Analyses
The N-MTSs and cleavage sites were predicted using Gavel’s consensus patterns search in the PSORT II program (http://psort.hgc.jp/form2.html, last accessed February 19, 2013) (Gavel and von 1990; Braun and Schmitz 1995). The net charge of presequences was estimated as described elsewhere (Smid et al. 2008). Motifs of N-MTSs were visualized with Weblogo software (Crooks et al. 2004).
Phylogenetic analysis was conducted using 80 sequences of core/MPP homologs aligned with the program Muscle 3.8.31 (default parameters) (Edgar 2010) and trimmed with BMGE 1.1 (−b 1 -m BLOSUM30) (Hordijk and Gascuel 2005; Criscuolo and Gribaldo 2010) to a final length of 314 AA. Phylogenetic analysis was performed with PhyML 2.1.0 (topology search: best of NNIs and SPRs, initial tree: BioNJ, Substitution model: WAG, proportion of invariable sites: fixed (0), gamma distribution parameter: estimated; number of categories: 4; bootstrap replicates: 500) (Guindon and Gascuel 2003) and MrBayes 2.0.6 (rate matrix: WAG; rate variation: gamma; gamma categories: 4; chain length: 11,00,000; heated chains: 4; heated chain temp: 0.2; burn-in length: 100,000) (Huelsenbeck and Ronquist 2001).
Results
Predictions of Mitochondrial-Targeting Sequences
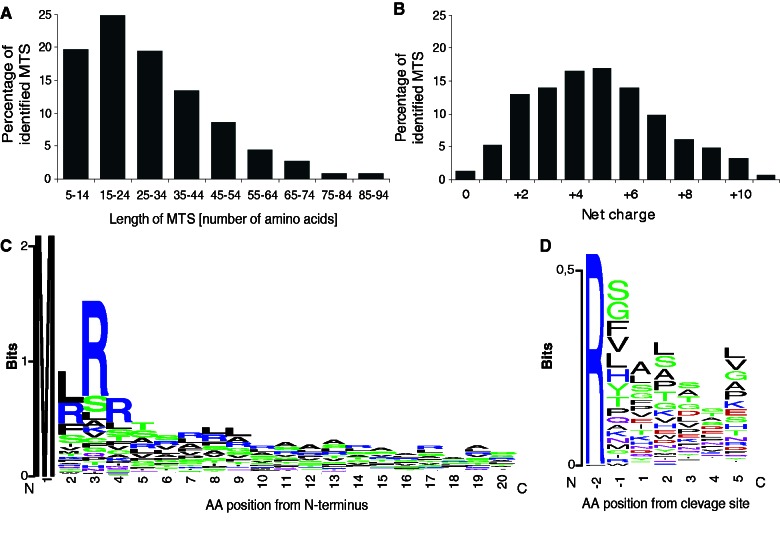
Data set of 402 proteins, identified in the T. brucei mitochondrial proteome and assigned to the organelle with the highest confidence (Panigrahi et al. 2009), were used for the amino-terminal mitochondrial targeting sequences (N-MTS) prediction using PSORT II software. In this set, 336 putative N-MTSs were identified to contain the predicted cleavage site according to the Gavel algorithm (Nakai and Horton 1999). Predicted N-MTSs were 3 to 147 AA in length, with most N-MTSs ranging between 8 and 37 AA (64%), with a median of 26 AA (fig. 1A). The majority of N-MTSs displayed a net charge between +2 and +8, with a median of +5 (fig. 1B) (supplementary table S1, Supplementary Material online). The net charge reflected the number of arginines distal from the cleavage site that were frequently present as doublets or triplets following the initial methionine. The double arginine motifs were found in 114 N-MTSs within the first 5 AA residues (fig. 1C). Longer N-MTS sequences (>16 AA) usually contained glycine or proline between distal and proximal arginines to allow for the flexibility of N-MTSs (Taylor et al. 2001). Negatively charged residues were rarely present in short (3–31 AA) N-MTSs. The analysis of predicted MPP cleavage sites with a conserved arginine residue at position −2 revealed a loosely conserved cleavage site motif RX↓(A/L/S)(L/S/A/T/P)(S/A/T) (fig. 1D). Altogether, the properties of the T. brucei N-MTSs in terms of length, net charge, and cleavage site motif were comparable with those of other organisms.
Fig. 1.—
Characteristics of amino-terminal MTSs identified in 336 proteins of Trypanosoma brucei mitochondria. (A) Length of N-MTSs predicted by PSORT II and (B) calculated net charge. (C) Sequence logo analysis of the N-MTS revealed the presence of a double arginine motif in 181 N-MTSs within the first 20 AA. (C) Sequence logo analysis of the MPP cleavage site in the 366 predicted N-MTSs.
Genes-Encoding MPP Subunits and Core Proteins in Genomes of T. brucei
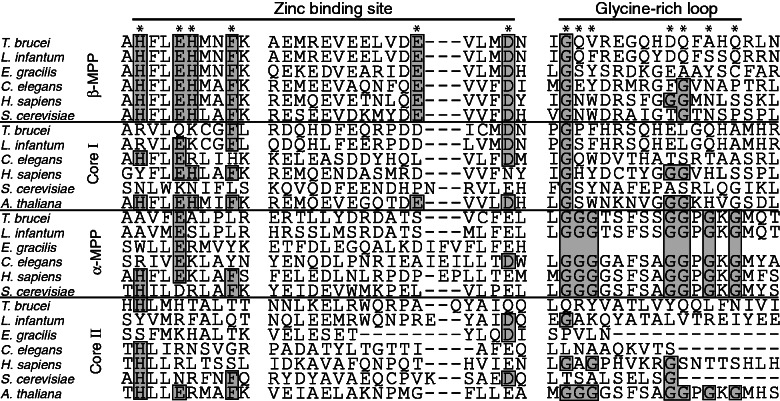
Processing of identified N-MTSs requires an active MPP in the mitochondrion. Thus, the T. brucei orthologs of MPP subunits and core proteins were searched for in the TriTryp database (http://tritrypdb.org, last accessed February 19, 2013) using the protein sequences of Euglena cp1 (P43264) and cp2 (P43265) as queries. Altogether, 5 genes were retrieved with an e value of 0.62–2.5e−16, including a single putative pseudogene (Tb927.10.2140). Sequence alignment of the retrieved sequences with eukaryotic orthologs revealed the presence of a glycine-rich loop in the Tb927.2.4110 gene that corresponded with an α-MPP subunit (fig. 2) and a catalytic zinc-binding motif in the Tb09.160.3110 gene that encoded a putative β-MPP subunit (fig. 2). Products of the two other genes possessed none of these MPP-specific motifs; however, Tb927.5.1060 and Tb11.02.1480 displayed 29.5% and 15.8% sequence similarity to β-MPP and α-MPP, respectively. Therefore, we predicted Tb927.5.1060 and Tb11.02.1480 to represent the cp1 and cp2 subunits of the bc1 complex, respectively. These analyses suggest that T. brucei contains a complete set of genes coding for a heterodimeric MPP and two distinct core proteins.
Fig. 2.—
Protein sequence alignment of MPP domains responsible for catalytic activity (zinc-binding site) and substrate recognition (glycine-rich loop). Positions of functionally important AA are marked by asterisks; conserved AA are in boxes. Accession numbers for protein sequences of α-MPP (Euglena gracilis, P43265; Trypanosoma brucei, XP_951618; Leishmania major, XP_001682967; Saccharomyces cerevisiae, NP_011889; Homo sapiens, NP_055975; Caenorhabditis elegans, NP_490888), β-MPP (E. gracilis, P43264; T. brucei, XP_803756; L. major, XP_003721587; S. cerevisiae, NP_013264; H. sapiens, NP_004270.2; C. elegans, NP_501576), cp1 (T. brucei, XP_844780; L. major, XP_001686023; S. cerevisiae, NP_009508; H. sapiens, NP_003356; Arabidopsis thaliana, NP_186858; C. elegans, NP_498202), and cp2 (E. gracilis, EC675176; T. brucei, XP_828477; L. major, XP_001681831; S. cerevisiae, NP_015517; H. sapiens, NP_003357; A. thaliana, NP_175610; C. elegans, NP_510011).
Cellular Localization
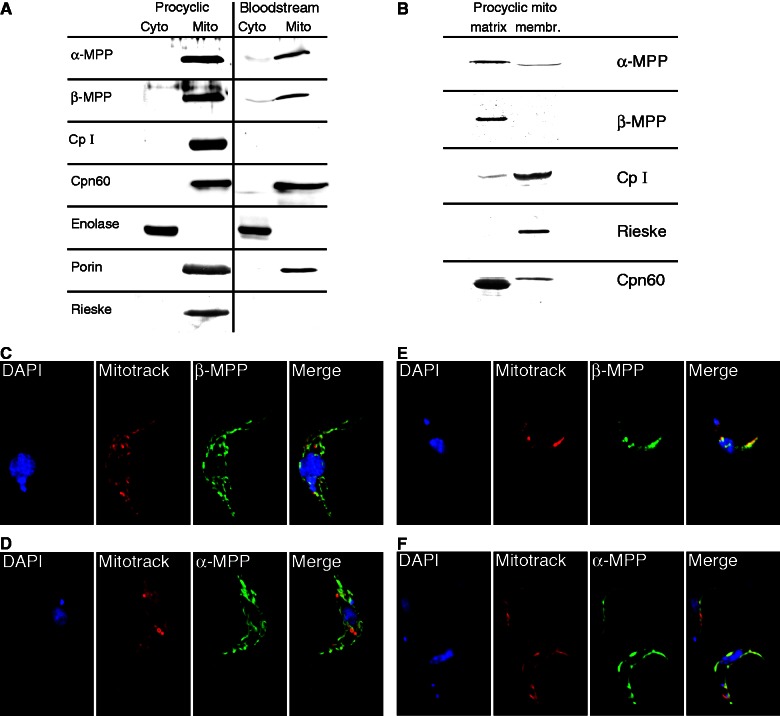
Comparison of the T. brucei MPP subunits and core proteins with Euglena orthologs revealed that Euglena cp1 possesses a conserved zinc-binding motif same as the T. brucei β-MPP (Tb09.160.3110) and plant cp1-β-MPP, whereas T. brucei Tb11.02.1480 and Euglena cp2 correspond to typical cp2 orthologs because they lack the glycine-rich loop present in the α-MPP from both organisms (fig. 2). This finding prompted us to establish the cellular localization of the MPP subunits and core proteins in T. brucei. Because key mitochondrial functions, such as the expression of respiratory complexes, dramatically change between the PF and BF cells, we also compared the cellular distribution of the MPP subunits and core proteins between both stages. Polyclonal antibodies were raised against recombinant T. brucei α-MPP, β-MPP, and cp1 and used in a Western blot analysis of the mitochondrial and cytosolic fractions obtained by digitonin fractionation, confirming the association of all these proteins with the organelle (fig. 3A). However, signals for both MPP subunits were approximately 40% lower in BF cells (fig. 3A). The cp1 protein was detected in the PF mitochondria, whereas no signal was observed in the BF mitochondria, which is consistent with the lack of the bc1 complex in this life cycle stage (Opperdoes and Michels 2008). The mitochondrial outer membrane porin, matrix Cpn60, and cytosolic enolase were used as controls to determine the purity of the cellular fractions. Mitochondrial localization of MPP was further confirmed by immunofluorescence microscopy of the PF and BF trypanosomes expressing the tagged Tbα-MPP and Tbβ-MPP. As shown in figures 3C–F, in both life stages, α and β-MPP subunits co-localized with MitoTracker, which was used here as a mitochondrial marker.
Fig. 3.—
Cellular localization of MPP subunits and core proteins in Trypanosoma brucei. (A) Immunoblot detection of MPP subunits and cp1 in cytosolic (Cyto) and mitochondrial (Mito) fractions isolated from T. brucei procyclic and bloodstream cells. The Rieske protein was used as a bc1 complex marker. Cpn60, porin, and enolase were used as marker proteins of the mitochondrial matrix, mitochondrial membrane, and cytosol, respectively. (B) Immunoblot analysis of membrane and matrix fractions of T. brucei mitochondria isolated from procyclic trypanosomes. (C–F) Detection of α- and β-MPP subunits in transformed T. brucei. procyclic (C, D) and bloodstream (E, F) forms. The proteins were expressed with carboxy terminal hemagglutinin tag in procyclic cells (C, D) or V5 tag in bloodstream cells (E, F) and detected using mouse monoclonal anti-hemagglutinin and anti-V5 antibodies, respectively. Mitotracker was used as a control for the visualization of mitochondria.
To investigate the mitochondrial topology of the MPP subunits and core proteins, soluble (matrix) and sedimentable (membrane) fractions were prepared from the T. brucei PF mitochondria. As expected, both MPP subunits together with matrix marker Cpn60 were associated predominantly with the matrix (fig. 3B), whereas cp1 and the Rieske protein (associated with the bc1 complex from the intermembrane space-facing side, used here as a control) were both present predominantly in the membrane fraction (fig. 3B).
Enzymatic Activity of the MPP Heterodimer and Complex Substrate Processing by the Mitochondrial Lysate
To investigate the function of the identified MPP subunits, recombinant histidine-tagged Tbα-MPP and non-tagged Tbβ-MPP were co-expressed in E. coli and isolated from the bacterial lysate using a nickel column via the tagged subunit. Affinity purification of the TbMPP complex from bacteria cultivated in standard LB media led to the isolation of a tagged Tbα-MPP subunit, whereas Tbβ-MPP was conspicuously absent (data not shown). However, when E. coli were cultivated in a medium supplemented with Zn2+, which is known to be required for formation of a functional MPP heterodimer (Luciano et al. 1998), MPP was indeed purified in an active form. The same results were obtained when isolated tagged Tbα–MPP was mixed with an E. coli lysate expressing only nontagged Tbβ–MPP in the buffer supplemented with Zn2+. Presence of both subunits in the MPP heterodimer was confirmed by Western blot analysis (supplementary fig. S1, Supplementary Material online) and mass spectrometry (data not shown).
The processing activity of the purified TbMPP heterodimer was tested on a substrate represented by a recombinant TbIscU that has been shown to reside in the T. brucei mitochondrion (Smid et al. 2006). According to PSORT II predictions, this protein possesses a 20 AA-long N-MTS with arginine in the -2 position from the putative MPP cleavage site. Incubation of TbIscU with the recombinant MPP heterodimer purified from E. coli resulted in a partial cleavage of the substrate. Determination of the cleavage site by Edman degradation of the processed product confirmed that TbIscU was cleaved exactly at the site RS/LY predicted by PSORT II (figs. 4A and B).
Fig. 4.—
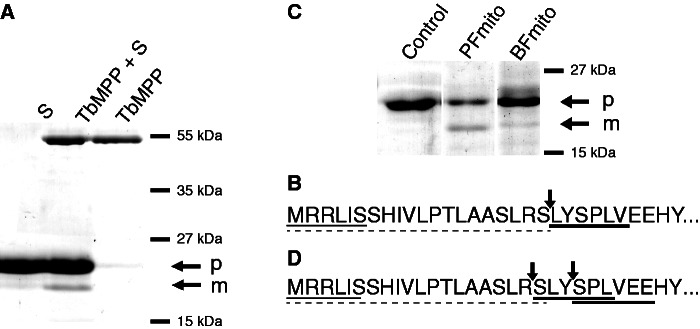
Processing of TbIscU preproteins by Trypanosoma brucei recombinant MPP activity and mitochondrial lysate isolated from PF and BF trypanosomes. (A) The TbIscU preprotein was treated with recombinant TbMPP and analyzed by SDS-PAGE. S, substrate (TbIscU); p, preprotein; m, mature protein. (B) Amino-terminal sequences of the preprotein and mature proteins were determined by Edman degradation. The peptide sequences of preprotein and mature protein are underlined by thin line and heavy line, respectively. Arrows indicates the TbIscU cleavage site. N-MTS was predicted by the PSORT II program (broken line). (C) The TbIscU preprotein was treated with mitochondrial lysates and analyzed by SDS-PAGE. Control, recombinant TbIscU without mitochondrial lysate. (D) Edman sequencing of mature TbIscU produced by PF and BF mitochondrial lysates revealed the presence of two amino-terminal peptides under both conditions (heavy line).
Next, we tested the processing of TbIscU in the mitochondrial fractions isolated from PF and BF cells. As in the case of recombinant MPP, incubation of recombinant TbIscU with the mitochondrial lysate resulted in the appearance of a single band of lower molecular weight (fig. 4C). Comparison of the processivity of the BF and PF mitochondrial lysate revealed an approximately 3-fold lower cleavage activity in the former lysate. However, Edman degradation of the cleavage product revealed the presence of two peptides. The first peptide commenced with serine, corresponding with the LR/SL cleavage, which is unlikely to be catalyzed by MPP (fig. 4D). The second peptide started with the second serine next to the proximal arginine, indicating cleavage between LY and SP (fig. 4D). In this case, TbIscU was most likely cleaved by MPP between RS and LY as observed with recombinant MPP and subsequently two following amino acid residues L and Y were removed by thus far unknown protease. These results were reproducible using organelles isolated from both life cycle stages and indicate that in addition to MPP, other proteases are involved in protein processing in the T. brucei mitochondria.
MPP Is Essential for T. brucei
To test whether MPP is an indispensable protein for T. brucei, we silenced the gene for Tbβ-MPP by RNAi. The Tbβ-MPP subunit became virtually undetectable within 4 days following RNAi induction, arresting the growth of the cells (fig. 5A and B). Western blot analysis of the mitochondria isolated from the PF cells ablated for Tbβ-MPP revealed the accumulation of unprocessed TbIscU at the expense of its fully processed form, which is consistent with Tbβ-MPP-dependent processing (fig. 5B). Taken together, these results demonstrate that MPP activity is essential for T. brucei and that both soluble MPP subunits are required for the processing activity within its mitochondria.
Fig. 5.—
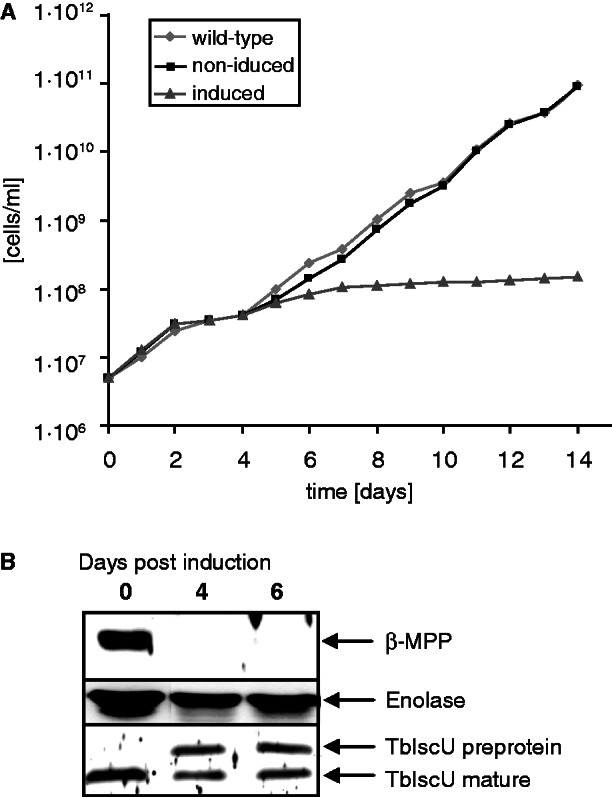
Effect of β–MPP silencing on Trypanosoma brucei growth and TbIscU maturation. (A) Growth curve of wild–type cells (29–13) strain (filled diamonds) and strain with induced (filled triangles) and noninduced (filled squares) RNAi against β-MPP. (B) Immunoblot analysis of mitochondrial fractions isolated from transformed T. brucei before and after induction of RNAi against β-MPP. The β-MPP depletion after a 4-day induction corresponded with an accumulation of TbIscU preprotein. Enolase was used as a loading control.
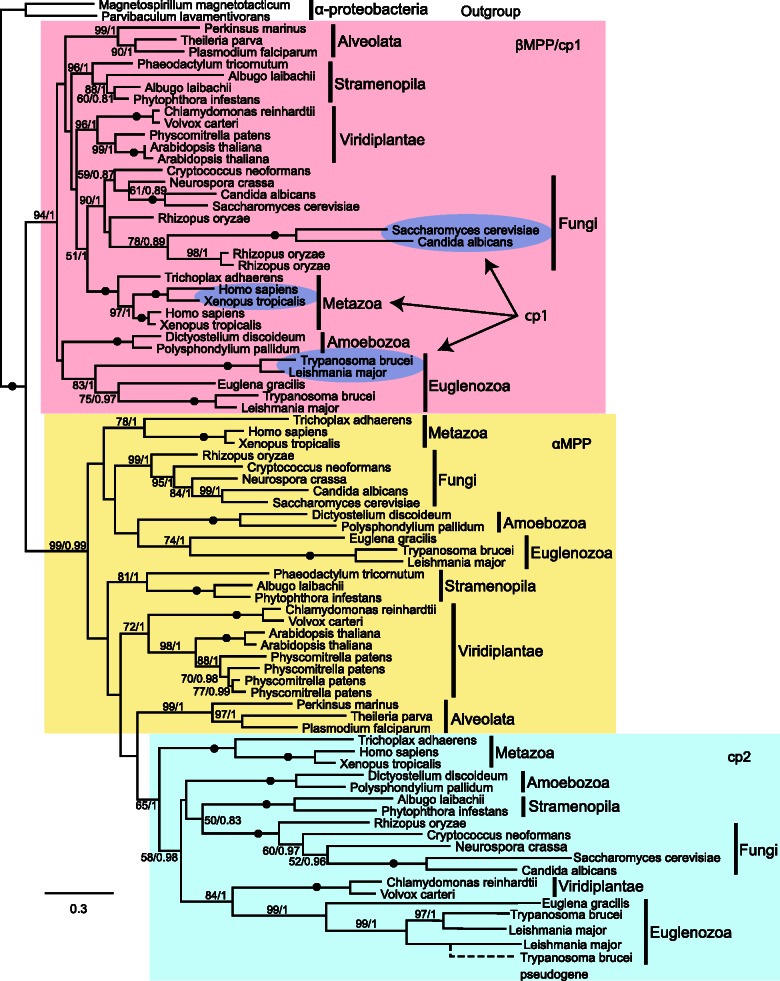
Distribution and Phylogenetic Analysis of MPP and Core Proteins in Eukaryotes
To perform phylogenetic analysis, we searched for the T. brucei α/β-MPP and the cp1/cp2 orthologs across main eukaryotic groups. Interestingly, we were able to identify the complete set of α/β-MPP subunits and cp1/cp2, present in T. brucei and closely related Leishmania spp., only in metazoans and some fungi (table 1). In all major eukaryotic groups, we found lineages that possess only three components, namely α-MPP, β-MPP, and cp2. Only two components (bc1-α-MPP and bc1-β-MPP) were present in the majority of plants; however, the green algae Chlamydomonas and Volvox also possess cp2, in addition to both MPP subunits. The same as in higher plants, the cp1/cp2 pair is absent from the apicomplexans. Protists with functionally and/or morphologically reduced mitochondria that lack the bc1 complex either retain only the soluble MPP (e.g., Trichomonas, Giardia) or lost both MPP and the core proteins (e.g., Entamoeba, Encephalitozoon) (table 1).
Table 1.
Presence of the α/β-MPP and the cp1/cp2 Orthologs across Main Eukaryotic Groups
| α-MPP | β-MPP | bc1-α- MPP | bc1-β- MPP | Cp2 | Cp1 | bc1 Complex | |
|---|---|---|---|---|---|---|---|
| Homo | ✓ | ✓ | — | — | ✓ | ✓ | ✓ |
| Saccharomyces | ✓ | ✓ | — | — | ✓ | ✓ | ✓ |
| Neurospora | ✓ | ✓a | — | ✓a | ✓ | — | ✓ |
| Encephalitozoon | — | — | — | — | — | — | — |
| Dictyostelium | ✓ | ✓b | — | ✓b | ✓ | — | ✓ |
| Mastigamoeba | ✓ | ✓ | — | — | — | — | — |
| Entamoeba | — | — | — | — | — | — | — |
| Arabidopsis | — | — | ✓ | ✓ | — | — | ✓ |
| Chlamydomonas | ✓ | ✓b | — | ✓b | ✓ | — | ✓ |
| Volvox | ✓ | ✓b | — | ✓b | ✓ | — | ✓ |
| Phytophthora | ✓ | ✓b | — | ✓b | ✓ | — | ✓ |
| Thalassiosira | — | — | ✓c | ✓c | — | — | ✓ |
| Blastocystis | ✓d | ✓d | — | — | — | — | — |
| Plasmodium | — | — | ✓c | ✓c | — | — | ✓ |
| Toxoplasma | — | — | ✓c | ✓c | — | — | ✓ |
| Cryptosporidium | ✓d | ✓d | — | — | — | — | — |
| Tetrahymena | — | — | ✓c | ✓c | — | — | ✓ |
| Trypanosoma | ✓ | ✓ | — | — | ✓ | ✓ | ✓ |
| Leishmania | ✓ | ✓ | — | — | ✓ | ✓ | ✓ |
| Euglenaf | ✓e | — | — | ✓ | ✓ | — | ✓ |
| Naegleria | ✓ | ✓b | — | ✓b | ✓ | — | ✓ |
| Andalucia | ✓ | ✓b | — | ✓b | ✓ | — | ✓ |
| Trichomonas | ✓ | ✓ | — | — | — | — | — |
| Giardia | — | ✓ | — | — | — | — | — |
aA single gene encodes β-MPP and cp1 protein.
bDual function of a single β-MPP/cp1 protein was inferred based on presence of cp2, and bc1 complex together with absence of cp1 genes.
cPresence of soluble α/β-MPP was inferred based on absence of bc1 complex.
dIncorporation of α/β-MPP within bc complex was inferred based on presence of bc complex and absence of cp1 and cp2 proteins as in plants.
ePartial sequence.
fIncomplete genome.
Phylogenetic analysis was performed with 80 selected eukaryotic orthologs of MPP and cp1/cp2 proteins (fig. 6). As expected, the analysis revealed 2 major, well-supported clades corresponding to α-MPP/cp2 and β-MPP/cp1. The α-MPP subunits and cp2 proteins were separated into two subclades, revealing a moderately supported monophyletic origin of cp2. The α-MPP group includes the bc1-α-MPP subunits of plants. Surprisingly, the topology of cp1 suggests that this β-MPP-related core protein evolved independently at least three times, namely in the kinetoplastid, fungal, and metazoan lineages (fig. 6). In each of these three groups, the cp1 protein is closely related to the corresponding β-MPP from the same taxon. Multiple origins of cp1 in kinetoplastids, fungi and metazoans, together with the conspicuous absence of cp1 in other lineages, suggests that the 3-component system (α-MPP, β-MPP, and cp2) represents an ancestral stage, whereas the acquisition of the fourth component, as in the case of T. brucei and other organisms, represent a more evolutionarily derived stage.
Fig. 6.—
Phylogeny of MPP subunits and bc1 core proteins. Phylogenetic analysis was performed with PhyML 2.1.0 and MrBayes 2.0.6. The numbers at the nodes represent the bootstrap values for 500 replicates (PhyML) and Bayesian posterior probability. Branches with bootstrap support lower than 50 are shown as polytomy. The scale bar indicates the substitution rate. Accession numbers for protein sequences that were used in phylogenetic analyses are shown in supplementary table S2, Supplementary Material online.
Discussion
Here, we investigated a MPP/core protein family in T. brucei to contribute to the understanding of the evolutionary history of trypanosomatid mitochondria, which is in many features highly divergent from the mitochondria of other eukaryotes (Madison-Antenucci et al. 2002; Besteiro et al. 2005; Allen et al. 2008). These features include a substantially simplified mitochondrial protein import machinery that might be considered either as a primitive feature of an ancient mitochondrion (Cavalier-Smith 2010; Pusnik et al. 2011) or the result of specific adaptation, as was proposed for the reduced import machinery in the mitochondria-derived organelles of Giardia and Trichomonas (Smid et al. 2008; Dagley et al. 2009; Rada et al. 2011). Our investigation revealed that T. brucei possesses a complete set of proteins consisting of soluble α- and β-MPP subunits and cp1 and cp2 subunits associated with the inner membrane respiratory complex bc1. We demonstrated that the matrix-located heterodimeric MPP is expressed in both stages of T. brucei. It apparently catalyzed processing of the mitochondrial preprotein TbIscU in vitro, and the substrate processing was impaired when Tbβ-MPP expression was silenced by RNAi in vivo. During preparation of this manuscript, another group reported the presence of canonical MPP in T. brucei (Desy et al. 2012). In addition to MPP, we identified cp1 and cp2 genes that are paralogs of β-MPP and α-MPP, respectively. We were able to show that cp1 is associated with the mitochondrial membrane fraction and that in BF cells, its expression is downregulated together with the bc1 complex. Accordingly, recent proteomic analysis of the T. brucei mitochondrial respirasome revealed the presence of the Tb927.5.1060 and Tb11.02.1480 gene products that correspond to the cp1 and cp2 proteins in the bc1 complex (Acestor et al. 2011). Unlike the MPP subunits, the T. brucei cp1/cp2 proteins lack the domains required for the processing activity. It is therefore likely that in T. brucei, these proteins are proteolytically inactive and serve as structural bc1 components, a situation reminiscent of that described for S. cerevisiae (Zara et al. 2004).
It has been proposed that plants represent an ancient stage of MPP evolution, with active α- and β-MPP subunits peripherally associated with the bc1 complex, whereas metazoans and yeast represent a more recent evolutionary stage in which MPP subunits were duplicated and formed membrane-associated cp1/cp2 proteins and soluble α/β-MPP (Braun and Schmitz 1995, 1997; Glaser and Dessi 1999). Neurospora was considered to represent an intermediate stage in which a single gene encodes a bi-functional β-MPP that is shared between α-MPP to constitute a soluble MPP heterodimer and cp2 in the bc1 complex (Neurospora-type system) (Schulte et al. 1989). A similar system was later observed also in Dictyostelium (Nagayama et al. 2008). Our search across major eukaryotic groups revealed that the combined presence of α/β-MPP subunits together with cp2 known from Neurospora and Dictyostelium thus far is not an exception but that this Neurospora-type system is present in at least some members of all major eukaryotic groups. The α-MPP and cp2 proteins differ in the confinement of the glycine-rich loop, which is important for substrate recognition, to the former protein (Nagao et al. 2000). However, it has been shown that bovine cp2 forms with cp1 a proteolytically active heterodimer capable of processing substrates such as the Rieske protein (Deng et al. 2001). Therefore, we can hypothesize that the original processing machinery consisted of an enzymatically active cp2-like/β-MPP enzyme associated with the bc1 complex that processed a limited number of imported proteins (fig. 7). As a consequence of growing mitochondrial complexity, the gene encoding a cp2-like protein was duplicated: One of the copies gained the glycine-rich loop (α-MPP) and formed a soluble heterodimer with β-MPP. It has been shown that the association of α-MPP with β-MPP is essential for the enzyme’s stability in solution (Janata et al. 2004). At this stage, β-MPP has acquired a dual function, as observed in Neurospora, while in plants, cp2 was likely replaced with the α-MPP to increase substrate specificity of the membrane-associated complex. Indeed, a more stringent recognition system was needed in plants to distinguish preproteins targeted into the mitochondria from those aimed for the plastids, as both systems used a similar targeting mechanism (Teixeira and Glaser 2012). In support of the secondary loss of cp2 in plants, we identified cp2 orthologs in the green algae Chlamydomonas and Volvox that displayed monophyly with other eukaryotic cp2 proteins. Similar to plants, apicomplexans also possess exclusively α- and β-MPP subunits. Although their cellular localization remains to be established, the presence of the bc1 complex and a remnant plastid suggests that the situation in these parasitic protists might be similar to plants. In several organisms lacking bc1, the MPP/core protein family is limited to a soluble MPP (Giardia and Trichomonas) (Smid et al. 2008), or MPP is completely absent as a result of reductive evolution, as is the case of Encephalitozoon and Entamoeba (Burri et al. 2006; Burri and Keeling 2007). On the other side of the spectrum, the Neurospora-type 3-component MPP/core family gained another member, cp1, by gene duplication and specialization of β-MPP. According to our phylogenetic analysis, cp1 emerged independently in metazoans, fungi, and higher kinetoplastids, indicating significant evolutionary pressure for this event. The cp1 proteins either possess a degenerate zinc-binding motif and retain enzymatic activity, such as in metazoans, or this motif has been lost, as is the case of yeasts and kinetoplastid flagellates. Evolution of the cp1 subunit allowed for separate respiration and mitochondrial biogenesis; for example, in yeast, the acquisition of the cp1 system allowed for the downregulation of the respiratory chain during anaerobic fermentation when mitochondrial protein import still occurs. Similarly, the cytochrome c-dependent respiration is silenced in the BF T. brucei; however, the respective mitochondrion still needs to import proteins to maintain processes such as Fe–S cluster assembly and fatty acid synthesis. Indeed, we demonstrated that cp1, along with the bc1 complex, is absent from the BF trypanosomes, whereas a soluble MPP heterodimer remains active. Interestingly, isolation and characterization of the bc1 complex in Euglena identified cp2 and β-MPP as its genuine subunits (Cui et al. 1994). Although the complete genome sequence for Euglena is not yet available, identification of β-MPP in the bc1 complex suggests that unlike T. brucei, Euglena contains the Neurospora-type system. Altogether, identification, functional characterization, and phylogenetic analysis of the MPP/core protein family in T. brucei did not support the ancient character of its mitochondrial biogenesis.
Fig. 7.—
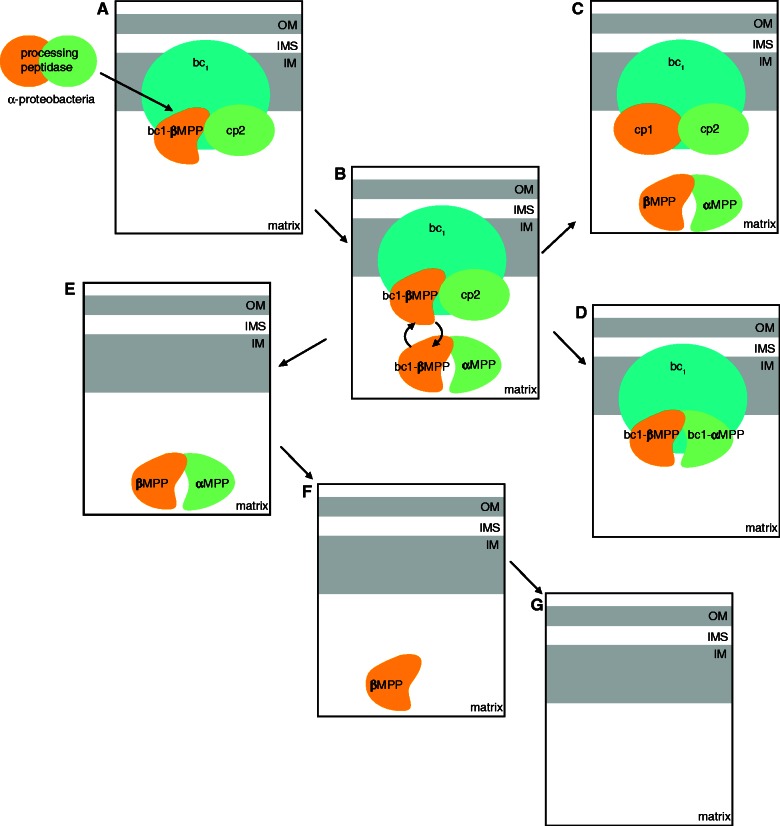
Model of MPP/core protein family evolution. MPP/core proteins most likely evolved from an α-proteobacterial protease present in the endosymbiotic ancestor of mitochondria. (A) The genes coding for the original bacterial protease were inherited from the mitochondrial ancestor, and the active cp2-like/β-MPP enzyme became associated with a respiratory bc1 complex in the inner mitochondrial membrane. (B) Subsequently, the gene encoding the cp2-like protein was duplicated. One paralog (cp2) was retained as the bc1 complex subunit, and the second paralog (α-MPP) gained the glycine-rich loop and formed a soluble heterodimer with β-MPP. At this stage, a single β-MPP has dual function as a subunit of soluble MPP and bc1 complex. (C) In three eukaryotic lineages (metazoa, fungi, and kinetoplastea), the β-MPP was also duplicated, forming core protein cp1 and soluble β-MPP. This event allows for the functional separation of respiration and protein processing. (D) In plants, we propose that cp2 was replaced by α-MPP. Consequently, enzymatically active α-MPP and β-MPP subunits were associated with the bc1 complex (bc1-αMPP, and bc1-βMPP). In this case, core proteins and MPP subunits are identical, and the bc1 complex facilitates a dual function: respiration and protein processing. Similar systems might be present in apicomplexans. In some anaerobic organisms and intracellular parasites with reduced mitochondria, the core proteins were lost together with the bc1 complex, and they possess only soluble MPP heterodimers (e.g., Trichomonas) (E), a single subunit MPP (e.g., Giardia) (F), or the MPP is absent (e.g., Entamoeba) (G). IM, inner mitochondrial membrane; IMS, intermembrane space; OM, outer mitochondrial membrane.
We further analyzed the T. brucei N-MTSs, which are substrates for MPP. It has been noticed that T. brucei MTSs are often shorter compared with those in metazoans and plants (Clayton et al. 1995). Until recently, only a limited number of T. brucei proteins were available with experimentally confirmed mitochondrial localization. Since the T. brucei mitochondrial proteome is now available (Panigrahi et al. 2009), we were able to analyze a set of 402 proteins assigned to the organelle with high confidence, in which 336 N-MTSs were predicted by PSORTII. The length of in silico predicted that T. brucei N-MTSs widely vary from 3 to 147 AA, with a median length of 26 AA. Similar results were recently obtained by a bioinformatic analysis of 38 putative T. brucei mitochondrial proteins using two other programs, MITOPROT and TargetP, which predicted N-MTSs with median lengths of 25 and 30 AA, respectively (Krnacova et al. 2012). Taken together, these analyses determined that mitochondrial proteins of this model flagellate are furnished with N-MTSs comparable in size with those in yeast and mammals (Schneider et al. 1998). Furthermore, other features of the T. brucei N-MTS, such as net charge, AA composition and cleavage site motif did not show any remarkable deviation from the usual pattern in other eukaryotic lineages, with the sole exception of a frequent occurrence of double and triple arginines close to the amino terminus. Therefore, the analyzed N-MTSs do not share the simplicity of the Giardia and Trichomonas presequences as suggested previously (Pusnik et al. 2011; Schneider et al. 2008). On the contrary, T. brucei is able to import and process mitochondrial preproteins from a large spectrum of evolutionary distant organisms, including the excavate Trichomonas, the model plant Arabidopsis, the stramenopile Blastocystis and humans (Long, Jirku, et al. 2008; Long et al. 2008; Tsaousis et al. 2012). Moreover, it has been shown that the expression of the human frataxin preprotein with a 55 AA long N-MTS in T. brucei resulted in its correct import and processing as in human mitochondria (Long, Jirku, et al. 2008).
Production of the active recombinant MPP allowed us to demonstrate in vitro that T. brucei MPP recognizes the typical cleavage site in N-MTS of TbIscU with arginine in the −2 position, as it was predicted by the PSORT II program. However, the protein processing within mitochondria seems to be a more complex process (Teixeira and Glaser 2012). The treatment of TbIscU with mitochondrial lysate resulted in the production of two peptides with arginine in −1 and −4 positions from the cleavage sites, respectively. It is not clear which process is responsible for the peptide cleavage in the R-1 position. The second (R-4) peptide is most likely a product of multistep processing in which the first step is carried out by MPP, which catalyses the cleavage between RS and LYS. Subsequently, L and Y might be removed by another mitochondrial protease, such as Icp55 (Teixeira and Glaser 2012). Indeed, the Icp55 typically removes L, Y, or F from the cleavage motif (F/L/Y)↓(S/A), and we found several candidates for Icp55-like peptidase in T. brucei genome (e.g., Tb09.211.4330). However, Icp55 is known to cleave off a single amino acid residue at the R-3 position (Naamati et al. 2009), while two amino acids were removed following MPP cleavage site in TbIscU. Similar R-4 cleavage was also observed in the case of hydrogenosomal flavodiiron protein in T. vaginalis (Smutna et al. 2009). We can speculate that the observed R-4 trimming of TbIscU is performed either in a single step in which LY dipeptide is removed or L and Y are removed in two successive steps. Future studies of putative N-MTS trimming enzymes in T. brucei will be needed to understand their possible role in the processing of N-MTSs.
Recently, important support for an ancient character of the trypanosomatid import system came from the identification of a putative ATOM as the main gate of the T. brucei outer mitochondrial membrane, whereas Tom40 was considered to be absent (Pusnik et al. 2011). It has been proposed that unlike Tom40, ATOM is related to the YtfM subgroup of the bacterial β barrel Omp85 protein family. The ATOM-like translocase was suggested to mediate mitochondrial protein import in the last common eukaryotic ancestor that was in the course of eukaryotic evolution replaced by Tom40 in and is present in all extant eukaryotes except trypanosomes (Pusnik et al. 2011). In addition, several other scenarios were considered, including a possibility that ATOM replaced Tom40 in trypanosomatids during excavate evolution by a lateral gene transfer from bacteria or that Tom40 might have evolved from the ATOM. However, later bioinformatic analysis questioned the relationship between the ATOM and bacterial Omp85-like proteins and showed that the former protein shares a Porin_3 domain with Tom40 and VDAC sequences and thus more likely represents a divergent ortholog of Tom40 (Zarsky et al. 2012).
Taken together, we believe that the features of the T. brucei MPP/core protein family described herein, together with the known features of its mitochondrial import machinery, can be best explained as adaptations to a complex life cycle. The T. brucei MPP/core proteins seem to represent a highly evolved form that allows a separate regulation of respiration and protein import in insect- and vertebrate-dwelling stages of the parasite. Our extensive analysis of N-MTSs revealed standard properties encountered in mitochondrial preproteins across main eukaryotic lineages. Although the TOM and TIM complexes of T. brucei show apparent simplicity compared with S. cerevisiae and other eukaryotes, currently there is insufficient evidence to distinguish whether this simplicity reflects a primitive evolutionary trait or resulted from an extensive reductive evolution, which occurred independently in anaerobic Giardia and Trichomonas that belong to the same eukaryotic supergroup.
Supplementary Material
Supplementary tables S1 and S2 and figure S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Czech Ministry of Education grant MSM0021620858 and Czech Science Foundation grant P305/11/1061 to J.T.; Grant Agency of the Czech Republic grant P305/11/2179 and a Praemium Academiae award to J.L., who is also a Fellow of the Canadian Institute for Advanced Research; Grant Agency of Charles University grant GAUK62209 to J.M.; and the Czech Ministry of Education grant OP VK CZ.1.07/2.3.00/20.0055 to J.J. The authors thank Eva Kutějová for assistance in the preparation of recombinant proteins.
Literature Cited
- Acestor N, et al. Trypanosoma brucei mitochondrial respiratome: composition and organization in procyclic form. Mol Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JW, et al. Order within a mosaic distribution of mitochondrial c-type cytochrome biogenesis systems? FEBS J. 2008;275:2385–2402. doi: 10.1111/j.1742-4658.2008.06380.x. [DOI] [PubMed] [Google Scholar]
- Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- Bertrand KI, Hajduk SL. Import of a constitutively expressed protein into mitochondria from procyclic and bloodstream forms of Trypanosoma brucei. Mol Biochem Parasit. 2000;106:249–260. doi: 10.1016/s0166-6851(99)00218-2. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Barrett MP, Riviere L, Bringaud F. Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol. 2005;21:185–191. doi: 10.1016/j.pt.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK. Are the ‘core' proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease. Trends Biochem Sci. 1995;20:171–175. doi: 10.1016/s0968-0004(00)88999-9. [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK. The mitochondrial processing peptidase. Int J Biochem Cell B. 1997;29:1043–1045. doi: 10.1016/s1357-2725(97)00032-0. [DOI] [PubMed] [Google Scholar]
- Burri L, Keeling PJ. Protein targeting in parasites with cryptic mitochondria. Int J Parasitol. 2007;37:265–272. doi: 10.1016/j.ijpara.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Burri L, Williams BAP, Bursac D, Lithgow T, Keeling PJ. Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc Natl Acad Sci U S A. 2006;103:15916–15920. doi: 10.1073/pnas.0604109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of eukaryotic and archaebacterial cells. Ann N Y Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri M, Ajayi W, Temple S, Hill GC. Identification and partial purification of a stage-specific 33 kDa mitochondrial protein as the alternative oxidase of the Trypanosoma brucei brucei bloodstream trypomastigotes. J Eukaryot Microbiol. 1995;42:467–472. doi: 10.1111/j.1550-7408.1995.tb05892.x. [DOI] [PubMed] [Google Scholar]
- Clayton CE, Häusler T, Blattner J. Protein trafficking in kinetoplastid protozoa. Microbiol Rev. 1995;59:325–344. doi: 10.1128/mr.59.3.325-344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton CE, Michels P. Metabolic compartmentation in African trypanosomes. Parasitol Today. 1996;12:465–471. doi: 10.1016/s0169-4758(96)10073-9. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Mukai K, Saeki K, Matsubara H. Molecular cloning and nucleotide sequences of cDNAs encoding subunits I, II, and IX of Euglena gracilis mitochondrial complex III. J Biochem. 1994;115:98–107. doi: 10.1093/oxfordjournals.jbchem.a124312. [DOI] [PubMed] [Google Scholar]
- Dagley MJ, et al. The protein import channel in the outer mitosomal membrane of Giardia intestinalis. Mol Biol Evol. 2009;26:1941–1947. doi: 10.1093/molbev/msp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, Shenoy SK, Tso SC, Yu L, Yu CA. Reconstitution of mitochondrial processing peptidase from the core proteins (subunits I and II) of bovine heart mitochondrial cytochrome bc(1) complex. J Biol Chem. 2001;276:6499–6505. doi: 10.1074/jbc.M007128200. [DOI] [PubMed] [Google Scholar]
- Desy S, Schneider A, Mani J. Trypanosoma brucei has a canonical mitochondrial processing peptidase. Mol Biochem Parasit. 2012;185:161–164. doi: 10.1016/j.molbiopara.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Dvorakova-Hola K, et al. Glycine-rich loop of mitochondrial processing peptidase alpha-subunit is responsible for substrate recognition by a mechanism analogous to mitochondrial receptor Tom20. J Mol Biol. 2010;396:1197–1210. doi: 10.1016/j.jmb.2009.12.054. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Quality measures for protein alignment benchmarks. Nucleic Acids Res. 2010;38:2145–2153. doi: 10.1093/nar/gkp1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Eriksson A, Sjoling S, Glaser E. Characterization of the bifunctional mitochondrial processing peptidase (MPP)/bc1 complex in Spinacia oleracea. J Bioenerg Biomembr. 1996;28:285–292. doi: 10.1007/BF02110702. [DOI] [PubMed] [Google Scholar]
- Flaspohler JA, Jensen BC, Saveria T, Kifer CT, Parsons M. A novel protein kinase localized to lipid droplets is required for droplet biogenesis in trypanosomes. Eukaryot Cell. 2010;9:1702–1710. doi: 10.1128/EC.00106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh E, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- Gavel Y, von HG. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990;4:33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Geli V. Functional reconstitution in Escherichia coli of the yeast mitochondrial matrix peptidase from its 2 inactive subunits. Proc Natl Acad Sci U S A. 1993;90:6247–6251. doi: 10.1073/pnas.90.13.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencic S, Schagger H, Vonjagow G. Core I protein of bovine ubiquinol-cytochrome-c Reductase; an additional member of the mitochondrial-protein-processing family. Cloning of bovine core I and core II cDNAs and primary structure of the proteins. Eur J Biochem. 1991;199:123–131. doi: 10.1111/j.1432-1033.1991.tb16099.x. [DOI] [PubMed] [Google Scholar]
- Glaser E, Dessi P. Integration of the mitochondrial-processing peptidase into the cytochrome bc1 complex in plants. J Bioenerg Biomembr. 1999;31:259–274. doi: 10.1023/a:1005475930477. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hauser R, Pypaert M, Hausler T, Horn EK, Schneider A. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Sci. 1996;109:517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- Häusler T, Stierhof YD, Blattner J, Clayton C. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur J Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of animal-infective bloodstream forms of an East African Trypanosoma congolense stock. Ann Trop Med Parasitol. 1984;78:327–330. doi: 10.1080/00034983.1984.11811824. [DOI] [PubMed] [Google Scholar]
- Hordijk W, Gascuel O. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics. 2005;21:4338–4347. doi: 10.1093/bioinformatics/bti713. [DOI] [PubMed] [Google Scholar]
- Huang SB, Taylor NL, Whelan J, Millar AH. Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol. 2009;150:1272–1285. doi: 10.1104/pp.109.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Janata J, et al. Substrate evokes translocation of both domains in the mitochondrial processing peptidase alpha-subunit during which the C-terminus acts as a stabilizing element. Biochem Biophys Res Commun. 2004;316:211–217. doi: 10.1016/j.bbrc.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Kitada S, et al. A protein from a parasitic microorganism, Rickettsia prowazekii, can cleave the signal sequences of proteins targeting mitochondria. J Bacteriol. 2007;189:844–850. doi: 10.1128/JB.01261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada S, Shimokata K, Niidome T, Ogishima T, Ito A. A putative metal-binding site in the beta subunit of rat mitochondrial processing peptidase is essential for its catalytic activity. J Biochem. 1995;117:1148–1150. doi: 10.1093/oxfordjournals.jbchem.a124836. [DOI] [PubMed] [Google Scholar]
- Krnacova K, Vesteg M, Hampl V, Vlcek C, Horvath A. Euglena gracilis and trypanosomatids possess common patterns in predicted mitochondrial targeting presequences. J Mol Evol. 2012;75:119–129. doi: 10.1007/s00239-012-9523-2. [DOI] [PubMed] [Google Scholar]
- Lill R, Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- Long SJ, et al. Ancestral roles of eukaryotic frataxin: mitochondrial frataxin function and heterologous expression of hydrogenosomal Trichomonas homologues in trypanosomes. Mol Microbiol. 2008;69:94–109. doi: 10.1111/j.1365-2958.2008.06260.x. [DOI] [PubMed] [Google Scholar]
- Long SJ, Jirku M, Ayala FJ, Lukes J. Mitochondrial localization of human frataxin is necessary but processing is not for rescuing frataxin deficiency in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2008;105:13468–13473. doi: 10.1073/pnas.0806762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P, Tokatlidis K, Chambre I, Germanique JC, Geli V. The mitochondrial processing peptidase behaves as a zinc-metallopeptidase. J Mol Biol. 1998;280:193–199. doi: 10.1006/jmbi.1998.1858. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S, Grams J, Hajduk SL. Editing machines: the complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- Maruyama Y, Chuma A, Mikami B, Hashimoto W, Murata K. Heterosubunit composition and crystal structures of a novel bacterial M16B metallopeptidase. J Mol Biol. 2011;407:180–192. doi: 10.1016/j.jmb.2011.01.038. [DOI] [PubMed] [Google Scholar]
- Moriwaki K, Ogishima T, Ito A. Analysis of recognition elements for mitochondrial processing peptidase using artificial amino acids: roles of the intervening portion and proximal arginine. J Biochem. 1999;126:874–878. doi: 10.1093/oxfordjournals.jbchem.a022529. [DOI] [PubMed] [Google Scholar]
- Naamati A, Regev-Rudzki N, Galperin S, Lill R, Pines O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J Biol Chem. 2009;284:30200–30208. doi: 10.1074/jbc.M109.034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao Y, et al. Glycine-rich region of mitochondrial processing peptidase alpha-subunit is essential for binding and cleavage of the precursor proteins. J Biol Chem. 2000;275:34552–34556. doi: 10.1074/jbc.M003110200. [DOI] [PubMed] [Google Scholar]
- Nagayama K, et al. Antisense RNA inhibition of the beta subunit of the Dictyostelium discoideum mitochondrial processing peptidase induces the expression of mitochondrial proteins. Biosci Biotechnol Biochem. 2008;72:1836–1846. doi: 10.1271/bbb.80106. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Opperdoes FR, Michels PAM. Complex I of trypanosomatidae: does it exist? Trends Parasitol. 2008;24:310–317. doi: 10.1016/j.pt.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, et al. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. The trypanosomatid Rieske iron-sulfur proteins have a cleaved presequence that may direct mitochondrial import. Biochim Biophys Acta. 1995;1269:201–204. doi: 10.1016/0167-4889(95)00154-6. [DOI] [PubMed] [Google Scholar]
- Pusnik M, et al. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- Pusnik M, et al. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol. 2011;21:1738–1743. doi: 10.1016/j.cub.2011.08.060. [DOI] [PubMed] [Google Scholar]
- Rada P, et al. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS One. 2011;6:e24428. doi: 10.1371/journal.pone.0024428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Bursac D, Lithgow T. The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol. 2008;18:12–18. doi: 10.1016/j.tcb.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Schneider G, et al. Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins. 1998;30:49–60. [PubMed] [Google Scholar]
- Schneider H, Arretz M, Wachter E, Neupert W. Matrix processing peptidase of mitochondria. Structure-function relationships. J Biol Chem. 1990;265:9881–9887. [PubMed] [Google Scholar]
- Schulte U, et al. A family of mitochondrial proteins involved in bioenergetics and biogenesis. Nature. 1989;339:147–149. doi: 10.1038/339147a0. [DOI] [PubMed] [Google Scholar]
- Shimokata K, et al. Substrate recognition by mitochondrial processing peptidase toward the malate dehydrogenase precursor. J Biochem. 1997;122:1019–1023. doi: 10.1093/oxfordjournals.jbchem.a021841. [DOI] [PubMed] [Google Scholar]
- Smid O, et al. Knock-downs of iron-sulfur cluster assembly proteins IscS and IscU down-regulate the active mitochondrion of procyclic Trypanosoma brucei. J Biol Chem. 2006;281:28679–28686. doi: 10.1074/jbc.M513781200. [DOI] [PubMed] [Google Scholar]
- Smid O, et al. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites Trichomonas vaginalis and Giardia intestinalis. PLoS Pathog. 2008;4: e1000243. doi: 10.1371/journal.ppat.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutna T, et al. Flavodiiron protein from Trichomonas vaginalis hydrogenosomes: the terminal oxygen reductase. Eukaryot Cell. 2009;8:47–55. doi: 10.1128/EC.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MC, Ogishima T, Ito A. Importance of residues carboxyl terminal relative to the cleavage site in substrates of mitochondrial processing peptidase for their specific recognition and cleavage. J Biochem. 1998;124:1045–1049. doi: 10.1093/oxfordjournals.jbchem.a022198. [DOI] [PubMed] [Google Scholar]
- Striebel HM, Rysavy P, Adamec J, Spizek J, Kalousek F. Mutational analysis of both subunits from rat mitochondrial processing peptidase. Arch Biochem Biophys. 1996;335:211–218. doi: 10.1006/abbi.1996.0500. [DOI] [PubMed] [Google Scholar]
- Taylor AB, et al. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9:615–625. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta. 2012;1833:360–370. doi: 10.1016/j.bbamcr.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, et al. Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc Natl Acad Sci U S A. 2012;109:10426–10431. doi: 10.1073/pnas.1116067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtle F, et al. Global analysis of the mitochondrial N-Proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasit. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Clayton C. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science. 1995;268:1179–1183. doi: 10.1126/science.7761835. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasit. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Zara V, Palmisano I, Conte L, Trumpower BL. Further insights into the assembly of the yeast cytochrome bc1 complex based on analysis of single and double deletion mutants lacking supernumerary subunits and cytochrome b. Eur J Biochem. 2004;271:1209–1218. doi: 10.1111/j.1432-1033.2004.04024.x. [DOI] [PubMed] [Google Scholar]
- Zarsky V, Tachezy J, Dolezal P. Tom40 is likely common to all mitochondria. Curr Biol. 2012;22:R479–R481. doi: 10.1016/j.cub.2012.03.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.