Abstract
Widespread structural alterations of cancer genomes are increasingly observed in a broad spectrum of tumors. In a recent issue of Cell, Baca and colleagues describe large chains of rearrangements that coordinately affect multiple chromosomes in prostate cancer. This phenomenon of chromoplexy may define cancer subtypes and drive punctuated tumor evolution.
The application of next-generation sequencing technologies has resulted in systematic efforts to characterize the mutational spectrum, genomic alterations, and clonal evolution of a wide range of tumors. In particular, whole-genome sequencing approaches can reveal extensive structural rearrangements throughout the tumor genome, which can be difficult to detect using more limited exome sequencing approaches. The whole-genome sequencing analysis of prostate tumors by Baca et al. represents the most comprehensive such analysis to date for one of the most common human malignancies, resulting in a surprising new insight into cancer genomes (Baca et al., 2013).
Prostate cancer was the first solid tumor shown to have frequent large-scale chromosomal rearrangements, as originally demonstrated by the discovery of the TMPRSS2-ERG fusion (Tomlins et al., 2005). Subsequent studies have shown that chromosomal rearrangements and extensive copy number alterations are prevalent in prostate cancer whereas point mutations are relatively infrequent, suggesting that structural alterations in tumor genomes represent the primary drivers of prostate cancer progression (Barbieri et al., 2012; Rubin et al., 2011; Taylor et al., 2010). Furthermore, previous whole-genome sequencing of 7 prostate tumors showed frequent occurrence of complex chains of balanced rearrangements, involving both intra-chromosomal and inter-chromosomal events (Berger et al., 2011).
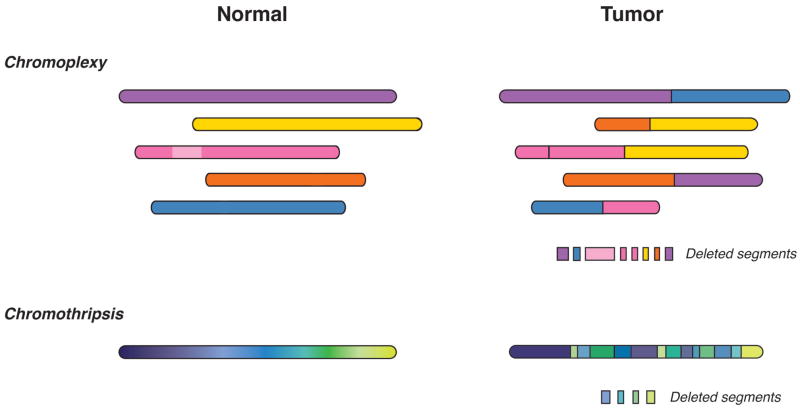
In the current study, Baca et al. performed whole-genome sequencing analysis of 57 prostate tumors and identify 5596 somatic rearrangements. Notably, almost 40% of the detected rearrangements were components of complex and lengthy series of rearrangements, often occurring as closed chains. Such chained rearrangements can display precise or nearly precise joins at their breakpoints, or alternatively are associated with large DNA deletions at their junctions, corresponding to “deletion bridges”. The number of rearrangements within a chain was highly variable, ranging from three to over forty, with six or more chromosomes possibly involved. Nearly 90% of the tumors contained chains with five or more rearrangements, and more than 60% of the tumors contained more than one such chain. Importantly, statistical analyses indicated that such rearrangements are unlikely to arise independently, and instead may form in a coordinated and simultaneous fashion. Thus, Baca et al. coin the term “chromoplexy” to describe this phenomenon of intricately weaved genomic rearrangements occurring in concert (Figure 1).
Figure 1. Comparison of chromoplexy and chromothripsis.
Shown are schematic representations of chromosomal rearrangements that occur in tumor genomes as a consequence of chromoplexy (top) or chromothripsis (bottom).
Chromoplexy can account for many of the known genomic alterations found in prostate cancer by generation of oncogenic fusion genes as well as by disruption or deletion of genes located near rearrangement breakpoints (Baca et al., 2013). Although no novel recurrent rearrangements were discovered in this analysis, the TMPRSS2-ERG fusion was often found as part of chromoplectic rearrangement chains. Moreover, putative oncogenic fusions involving BRAF and MAPK1 were uniquely identified in individual tumors, while loss-of-function alterations due to chromoplexy for the putative tumor suppressor genes PTEN, NKX3.1, TP53, and CDKN1B were observed in multiple tumors. In particular, several tumors contained evidence for multiple loss- and gain-of-function alterations occurring in the context of a single chromoplectic event.
The coordinated structural rearrangements characteristic of chromoplexy display features similar to but distinct from the phenomenon of chromothripsis that has been observed in a diverse range of malignancies (Figure 1) (Forment et al., 2012; Jones and Jallepalli, 2012). Both chromothripsis and chromoplexy display random breakage and fusion of genomic segments with low copy number states, most likely mediated by non-homologous end-joining. However, the genomic breakpoints associated with chromothripsis typically number in the hundreds and are locally clustered within one or two chromosomes, whereas the chained rearrangements characteristic of chromoplexy are unclustered, usually number in the tens, and include multiple chromosomes. Furthermore, chromothripsis appears to occur as a single clonal event early in tumor progression, while chromoplexy can occur more than once in prostate cancer evolution, with sequential events detected at clonal or subclonal frequencies. Finally, chromothripsis represents a relatively infrequent event for all tumor types analyzed to date, whereas chromoplexy is a common event in prostate cancer. Nonetheless, the distinction between chromothripsis and chromoplexy is not well defined, and it is conceivable that some coordinated structural rearrangements may have intermediate properties.
Distinct molecular mechanisms are likely to underlie chromothripsis versus chromoplexy. Although several models have been advanced to explain the mechanistic basis for chromothripsis, major causes are likely to be replication stress and mitotic errors, perhaps in association with the formation of micronuclei and premature chromosome compaction (Forment et al., 2012; Jones and Jallepalli, 2012). Notably, micronucleus formation is a characteristic feature of genomic instability, and loss of p53 appears to result in increased frequency of chromothripsis. In contrast, the mechanistic basis of chromoplexy is less well understood at present, but may be causally related to DNA damage induced by transcription factor binding. Previous studies have shown that formation of the TMPRSS2-ERG fusion in tumor cell lines is mediated by double-strand breaks induced by binding of androgen receptor (AR), and can be facilitated by genotoxic stress (Haffner et al., 2010; Rubin et al., 2011). Consistent with such a model, chromoplectic rearrangement breakpoints are associated with active transcription and open chromatin configurations (Baca et al. 2013).
The study by Baca et al. also sheds light on potential molecular subtypes of prostate adenocarcinoma. Despite the existence of categories of prostate cancer patients with markedly different survival outcomes, previous attempts to discern distinct histopathological or molecular subtypes of prostate cancer have met with limited success (Shen and Abate-Shen, 2010). Baca et al. now suggest the existence of at least two distinct molecular subtypes defined by the presence or absence of the TMPRSS2-ERG fusion (ETS+) and by the mutational status of CHD1, which encodes a chromodomain helicase involved in chromatin remodeling (Baca et al., 2013). Whereas rearrangements occurring in ETS+ CHD1WT tumors are predominantly inter-chromosomal and display features of chromoplexy, ETS− CHD1del tumors display a higher frequency of intra-chromosomal rearrangements, which more closely resemble chromothripsis. Moreover, the rearrangement breakpoints in ETS+ CHD1WT tumors tend to occur near highly expressed loci, consistent with a transcription-associated mechanism for chromoplexy, but the ETS− CHD1del tumors instead contain rearrangements that are often associated with heterochromatin. Whether these molecularly distinct groups correspond to distinct patient outcomes and/or treatment responses will undoubtedly represent a major issue for future studies.
Although chromoplexy appears to be a cardinal feature of many prostate tumor genomes, it is currently unclear whether it plays a key role in other cancer types. Interestingly, Baca et al. detect chained rearrangements in a significant proportion of non-small cell lung cancers, head and neck cancers, and melanomas, suggesting that chromoplexy can also occur in a broad spectrum of tumors. Given the apparent central role of AR and possibly ETS transcription factors in promoting chromoplexy in prostate tumors, it will be interesting to determine whether the chromoplectic events found in other cancers are qualitatively similar or distinct.
Overall, chromoplexy and chromothripsis undoubtedly represent key drivers of tumor evolution. In contrast to traditional “gradualist” views of sequential accumulation of cancer-promoting mutations, the large-scale structural alterations of both chromoplexy and chromothripsis are likely to promote discontinuous tumor evolution in a form of “punctuated equilibrium” (Baca et al., 2013). Moreover, further discrete steps in tumor evolution can potentially arise through the sequential occurrence of chromothripsis followed by chromoplexy, or by successive rounds of chromoplexy. Modeling the causes and consequences of such genomic alterations will be a serious challenge for cancer biologists, while their impact on treatment response and resistance will pose significant clinical challenges.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baca, et al. 2013 [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MJ, Jallepalli PV. Chromothripsis: chromosomes in crisis. Dev Cell. 2012;23:908–917. doi: 10.1016/j.devcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]