Abstract
The Ser/Thr-specific IκB kinase (IKK), which comprises IKKα or IKKβ and the regulatory protein NEMO, is at the bottleneck for NF-κB activation. IKK activity relies on interaction between NEMO and IKKα or IKKβ. A conserved region in the C-terminal tail of IKKβ or IKKα (NEMO-binding domain, NBD, residues 734–745 of IKKβ) is important for interaction with NEMO. Here we show that the NBD peptide of IKKβ is not sufficient for interaction with NEMO. Instead, a longer region of the IKKβ C-terminal region provides high affinity for NEMO. Quantitative measurements using surface plasmon resonance and isothermal titration calorimetry confirm the differential affinities of these interactions and provide insight into the kinetic and thermodynamic behaviors of the interactions. Biochemical characterization using multiangle light scattering (MALS) coupled with refractive index shows that the longer IKKβ C-terminal region forms a 2:2 stoichiometirc complex with NEMO.
NF-κB proteins (NF-κBs) are evolutionarily conserved master regulators of immune and inflammatory responses (1, 2). They play critical roles in a wide array of biological processes, including innate and adaptive immunity, oncogenesis, and development. They are activated in response to ligation of many receptors, including T-cell receptors, B-cell receptors, members of the tumor necrosis factor (TNF) receptor superfamily, and the Toll-like receptor/interleukin-1 receptor (TLR/IL-1R) superfamily.
NF-κBs share a highly conserved DNA-binding/dimerization domain called the Rel homology domain (RHD) (2, 3). The mammalian NF-κB family consists of p65 (RelA), RelB, c-Rel, p50/p105 (NF-κB1), and p52/p100 (NF-κB2). While p65, RelB, and c-Rel contain C-terminal transactivation domains, p105 and p100 contain long C-terminal domains that contain multiple ankyrin repeats and act to inhibit these proteins. The activity of NF-κB family members p65, RelB, and c-Rel is tightly regulated by interaction with the inhibitor of κB (IκB) proteins, which also contain ankyrin repeats, like the C-terminal domain of p105 and p100. Thus, in most cells, NF-κBs are held captive in the cytoplasm from translocating to the nucleus by the IκB proteins or IκB-like domains.
The Ser/Thr-specific IκB kinase (IKK) is at the bottleneck for NF-κB activation (4). Activated IKK phosphorylates NF-κB-bound IκBs and the IκB-like domains of p100 and p105. This leads to Lys48-linked polyubiquitination and subsequent degradation of IκBs and processing of p100 and p105, respectively, by the proteasome. The freed or processed NF-κB dimers translocate to the nucleus to mediate specific target gene transcription. Recent studies have revealed that IKK activation by cytokines such as TNF and IL-1 and by Toll-like receptors is dependent on Lys63-linked nondegradative polyubiquitination (5). Using biochemical purification and in vitro reconstitution, it was shown that together with a ubiquitin activating enzyme (E1) and a specific dimeric ubiquitin conjugating enzyme Ubc13–Uev1A complex (E2), the RING domain containing protein TRAF6 acts as a specific ubiquitin ligase (E3) in this Lys63-linked polyubiquitination (6–8). A MAP kinase kinase kinase complex (MAP3K) known as the TAK1 complex containing the kinase TAK1 and two adapter proteins TAB1 and TAB2 is the intermediary in IKK activation. Formation of a signaling complex containing TRAF6, the TAK1 complex, and the IKK complex leads to IKK activation (9–16).
The IKK holo complex contains the kinase, IKKα or IKKβ, and the regulatory protein NEMO (also known as IKKγ or FIP-3) (17–21). IKK activity relies on the interaction between the kinase and the regulatory protein NEMO (21–23). IKK activation is accompanied by phosphorylation of the activation loop Ser residues in the canonical MEK (MAK kinase kinase) consensus motif, SxxxS, in the kinase domain (S177 and S181 in human IKKβ) (17–21, 24). In cells lacking NEMO, IKKα and IKKβ cannot be activated by any of the classical NF-κB inducers (21). Mutational analysis of the activation loop Ser residues revealed that IKKβ is the primary target of proinflammatory stimuli (25). In vitro, IKKβ has a higher catalytic activity toward IκB than does IKKα, which in turn is a more proficient kinase for p100.
IKKβ (residues 1–756) and IKKα (residues 1–745) contain the following recognizable domains: a kinase domain (KD), a leucine zipper domain (LZ), a helix–loop–helix domain (HLH), and a C-terminal NEMO-binding domain (NBD). NEMO (residues 1–419) is highly conserved, and sequence analysis indicates a high helical content with an N-terminal kinase-binding domain (KBD), three coiled coil regions (CC1–3), and a zinc finger domain. In NEMO, the kinase binding domain (KBD) has been mapped to the N-terminal region (24, 26, 27).
A growing body of evidence suggests that the NF-κB signaling pathways play important roles in both inflammation and tumor development (1, 28–37). Because of its importance in NF-κB activation, IKK has become a potential therapeutic target for both inflammation and cancer (1, 38–42). Given the essential role of NEMO for the activation of IKK (2), any disruption of its interaction with the kinase can impair IKK function. A conserved region in the C-terminal tail of IKKβ or IKKα [NEMO-binding domain (NBD)] has been shown to be important for the interaction with NEMO (26). The NBD peptide containing only amino acid residues 735–745 of IKKβ, when fused to the antennapedia or HIV-Tat protein cell transduction domains, blocks IKK and NF-κB activation in transformed tumor cells, in primary human cells, and in mice (26, 43, 44).
Here we show that the short conserved C-terminal NBD peptide of IKKβ is not sufficient for interaction with NEMO. Instead, we mapped a longer region of the IKKβ C-terminal region (residues 705–745) that provides high-affinity interaction with NEMO. Characterization of the interaction using multiangle light scattering (MALS) coupled with refractive index shows that NEMO and the IKKβ C-terminal region mostly form a 2:2 stoichiometric complex. Quantitative measurements of the interactions using surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) further confirm the differential affinities of these interactions and provide additional mechanistic insights into the kinetic and thermodynamic behaviors of the interactions.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human NEMO and IKKβ were expressed in Escherichia coli. The NEMO cDNA corresponding to amino acid residues 1–196 or residues 38–196 was amplified by PCR using gene-specific primers containing NdeI (5′) and NotI (3′) sites. The PCR fragment was subsequently digested and ligated into the pET-28a expression vector (Novagen) containing hexahistidine tags (His tags). The IKKβ cDNA corresponding to amino acid residues 640–756, residues 680–756, and residues 705–745 was amplified by PCR using gene-specific primers containing NdeI and NotI sites. The PCR fragments were subsequently digested and ligated into the pET-28a expression vector (Novagen) containing His tags.
After transformation with the plasmids, E. coli BL21(DE3) cells were grown in Luria broth supplemented with 50 μg/mL kanamycin at 37 °C to an A600 of 1.0. Protein expression was induced by adding 1 mM isopropyl thio-β-d-galactopyranoside (IPTG), and the cells were grown overnight at 20 °C. Cells were pelleted by centrifugation at 5000 rpm and resuspended in buffer A containing 50 mM sodium phosphate buffer (pH 7.4), 500 mM KCl, 1 mM DTT, 20 mM imidazole, and a protease inhibitor mixture (Sigma). The cells were lysed by sonication, and the lysate was centrifuged at 16000 rpm for 1 h. The supernatant was decanted and incubated with Ni-NTA beads (Qiagen) for 45 min. The beads were loaded onto a column and washed with buffer B containing 50 mM sodium phosphate buffer (pH 7.4), 250 mM KCl, 1 mM DTT, 20 mM imidazole, and a protease inhibitor mixture (Sigma). The protein was eluted with buffer C containing 50 mM sodium phosphate buffer (pH 7.4), 250 mM KCl, 1 mM DTT, and 200 mM imidazole.
Peptide Synthesis
The IKKβ NBD peptide (residues 735–745) used in this study was chemically synthesized at the University of Maryland Biopolymer Core Facility with amino-terminal acetylation and carboxy-terminal amidation to mimic the intact protein. They were purified by reverse phase HPLC using a C18 column (Vydac, Hesperia, CA) and lyophilized. The molecular mass of each peptide was verified by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry.
Native PAGE and Gel Filtration Chromatography
Interactions between IKKβ (residues 640–756, residues 680–756, residues 735–745, or residues 705–745) and NEMO (residues 1–196 or residues 38–196) were first assayed using native PAGE on a PhastSystem (GE Healthcare). For gel filtration analyses, purified NEMO was mixed with a molar excess of IKKβ and applied to a gel filtration column (Superdex 200 HR10/30, GE Healthcare) which was pre-equilibrated with a solution of 50 mM sodium phosphate buffer (pH 7.4) and 1 mM tris(2-carboxyethyl)phosphine hydrochloride. The fractions were collected and subjected to SDS–PAGE analysis. Similarly, the short IKKβ NBD peptide (residues 735–745) was mixed in molar excess with NEMO (residues 1–196) and applied to the gel filtration column (Superdex 200 HR10/30, GE Healthcare). The fractions were subjected to mass spectrometry for the identification of formation of a complex between NEMO and the NBD of IKKβ.
Isothermal Titration Calorimetry (ITC) and Data Analysis
Prior to analysis, affinity-purified His-tagged NEMO (residues 38–196) and IKKβ (residues 680–756) were further purified by gel filtration using the Superdex 200 HR10/30 column (GE Healthcare). Following overnight dialysis against identical buffer [50 mM sodium phosphate buffer (pH 7.4) and 1 mM tris(2-carboxyethyl)phosphine hydrochloride], the proteins were concentrated in Amicon Ultra-4 tubes (Millipore). ITC measurements were performed at 25 °C using a VP-ITC machine that was connected to a computer with ORIGIN (Microcal Inc., Northampton, MA). Prior to titration, the protein and NBD peptide samples were centrifuged at 10000 rpm and 4 °C for 10 min to remove any debris and degassed by vacuum aspiration for ~10 min. The calorimeter cell and titration syringe were extensively rinsed with dialysis buffer [50 mM sodium phosphate buffer (pH 7.4) and 1 mM tris(2-carboxyethyl)phosphine hydrochloride]. The calorimetric titration was carried out at 25 °C either with 20 injections of 10 μL of 0.8 mM IKKβ NBD peptide (residues 735–745), spaced 120 s apart, into the sample cell containing a solution of 1.334 mL of 40 μM NEMO or with 15 injections of 10 μL of 0.2 mM IKKβ, spaced 120 s apart, into the sample cell containing a solution of 1.334 mL of 13 μM NEMO. Similarly, the same IKKβ NBD peptide or IKKβ protein was titrated into the sample cell of buffer alone to obtain the heat of dilution. After subtraction of the heat of dilution, the association constant (KA), enthalpy change (ΔH), and stoichiometry (N) were obtained by fitting the thermograms to a single-binding site model using ORIGIN. The remaining thermodynamic parameters, the dissociation constant (KD), the free energy change (ΔG), and the entropy change (ΔS), were calculated from the relationships
Surface Plasmon Resonance (SPR)
Binding studies were performed at 25 °C using a Biacore S51 optical biosensor equipped with a streptavidin-coated CM5 research-grade sensor chip and equilibrated with running buffer [10 mM HEPES, 150 mM NaCl, 5 mM DTT, and 0.005% P20 (pH 7.4)]. NEMO was incubated with EZ-link sulfo-NHS-LCLC-biotin for 2 h at 4 °C and then passed over a fast-desalting column to remove free biotin. The minimally biotinylated protein was captured at different densities (~500 and 3500 RU) at two spots within a streptavidin-coated flow cell. A concentration series (0, 0.617, 1.85, 5.56, 16.7, and 50.0 nM) of IKKβ (residues 680–756) and a concentration series (0, 18.8, 37.5, 75.0, 150, and 300 μM) of IKKβ NBD were tested in duplicate for binding to surface-tethered NEMO. Between binding cycles, the NEMO surfaces were regenerated with 1/3000 phosphoric acid. The data sets for the IKKβ-NEMO interaction were fit to a 1:1 interaction model (45). The binding responses were concentration-dependent and the duplicate analyses of each IKKβ concentration overlaid, indicating the assay was reproducible. Since IKKβ NBD and NEMO associated and dissociated so quickly, reliable kinetic parameters could not be determined. Instead, fitting the responses at equilibrium plotted as a function of peptide concentration to a binding isotherm yielded an estimate of the affinity constant.
Multiangle Light Scattering (MALS)
The molar masses of IKKβ (residues 680–756), NEMO (residues 38–196), and the IKKβ-NEMO complex were determined by MALS. The mixture of IKKβ and NEMO with IKKβ in excess was injected into a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) equilibrated in a buffer containing 20 mM Tris (pH 8.0) and 150 mM NaCl. The chromatography system was coupled to a three-angle light scattering detector (mini-DAWN TRISTAR) and a refractive index detector (Optilab DSP) (Wyatt Technology). Data were collected every 0.5 s at a flow rate of 0.2 mL/min. Data analysis was carried out using ASTRA.
RESULTS
The C-Terminal NBD Peptide of IKKβ Is Not Sufficient for NEMO Interaction, while a Longer C-Terminal Region Confers High-Affinity Interaction on NEMO
Previously published data have shown that a short region at the C-terminal tail of IKKβ (residues 735–745) is important for NEMO interaction [NEMO-binding domain (NBD)], and the cell permeable peptide comprising this region can inhibit cytokine-induced NF-κB activation and associated biological functions (26, 46–49).
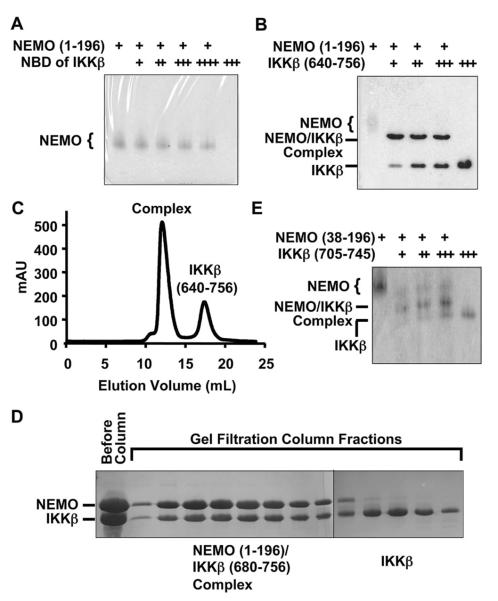
To determine whether the 11-residue NBD sequence is sufficient for NEMO interaction in vitro, we chemically synthesized the NBD peptide and used it to assess complex formation. Previous deletion studies have shown that NEMO (residues 1–196) is sufficient for IKKβ interaction (26). On native PAGE, addition of the NBD peptide did not cause a position shift of the purified NEMO protein (residues 1–196) (Figure 1A), suggesting that the NBD peptide does not form a stable complex with NEMO. To further confirm this, we performed gel filtration chromatography on the mixture of NEMO and a molar excess of NBD peptide. Mass spectrometry analysis of the gel filtration fractions showed that the NDB peptide did not comigrate with NEMO. These data on the apparent weak affinity between NEMO and the NBD peptide may be consistent with the high concentration of the NBD peptide required for inhibiting the NEMO–IKKβ interaction and their associated biological effects (26, 46, 47).
Figure 1.
Qualitative characterization of the interaction between NEMO and IKKβ. (A) Native PAGE analysis for the interaction between NEMO (residues 1–196) and the NEMO-binding domain (NBD) peptide of IKKβ (residues 735–745): lane 1, 690 pmol of NEMO; lane 2, 276 pmol of NEMO and 600 pmol of IKKβ; lane 3, 276 pmol of NEMO and 1.2 nmol of IKKβ; lane 4, 276 pmol of NEMO and 1.7 nmol of IKKβ; lane 5, 276 pmol of NEMO and 2.8 nmol of IKKβ; and lane 6, 2.8 nmol of IKKβ. (B) Native PAGE analysis for the interaction between NEMO (residues 1–196) and the C-terminal region of IKKβ (residues 640–756): lane 1, 690 pmol of NEMO; lane 2, 276 pmol of NEMO and 264 pmol of IKKβ; lane 3, 276 pmol of NEMO and 554 pmol of IKKβ; lane 4, 276 pmol of NEMO and 818 pmol of IKKβ; and lane 5, 2.0 nmol of IKKβ. (C) Gel filtration profile showing formation of the complex between NEMO (residues 1–196) and the C-terminal region of IKKβ (residues 640–756). (D) SDS–PAGE of gel filtration fractions showing formation of the complex between NEMO (residues 1–196) and the C-terminal region of IKKβ (residues 680–756). (E) Native PAGE analysis of the interaction between NEMO (residues 38–196) and the C-terminal region of IKKβ (residues 705–745): lane 1, 123 pmol of NEMO; lane 2, 123 pmol of NEMO and 124 pmol of IKKβ; lane 3, 123 pmol of NEMO and 248 pmol of IKKβ; lane 4, 123 pmol of NEMO and 372 pmol of IKKβ; and lane 5, 498 pmol of IKKβ.
To map the NEMO–IKKβ interaction more precisely to facilitate structural studies, we expressed and purified an IKKβ construct (residues 640–756), which contains the entire C-terminal region immediately after the HLH domain. In contrast to the 11-residue IKKβ NBD peptide, IKKβ (residues 640–756) interacted well with NEMO, as shown by complex formation on native PAGE (Figure 1B) and on gel filtration chromatography (Figure 1C). The purified complex was then subjected to limited proteolysis by the protease subtilisin followed by N-terminal sequencing and mass spectrometry analyses. The experiment showed that the proteolysis removed the first 37 residues in the NEMO construct and the first 40 residues in the IKKβ construct, suggesting that residues 38–196 of NEMO and residues 680–756 of IKKβ are mutually sufficient for the interaction. Indeed, IKKβ (residues 680–756) formed a stable complex with NEMO (residues 1–196) or NEMO (residues 38–196) on a gel filtration column (Figure 1D and Table 1).
Table 1.
Qualitative Assessment of the NEMO–IKKβ Interaction
| NEMO | IKKβ | native PAGE | gel filtration |
|---|---|---|---|
| 1–196 | 735–745 | − | − |
| 1–196 | 640–756 | + | + |
| 1–196 | 680–756 | + | + |
| 38–196 | 680–756 | + | + |
| 38–196 | 705–745 | + | NDa |
Not determined.
Sequence analysis of IKKβ as well as IKKα showed that the region around residues 705–732 of IKKβ has a high propensity for forming α-helix and coiled-coil structures as predicted using PHD and Multicoil (50, 51) (Figure 2). Because NEMO appears to be an oligomer (27, 52), we wondered whether the predicted α-helical and coiled-coil region is sufficient for enhancing the affinity of the longer construct of IKKβ for NEMO. Since the C-terminal tail (residues 746–756) of IKKβ is not present in IKKα and may not be important for IKKβ interaction, we made a new construct of IKKβ comprising just the predicted α-helical and coiled-coil region and the NBD peptide region (residues 705–745). Indeed, native PAGE showed that IKKβ (residues 705–745) formed a complex with NEMO (residues 38–196). This region in IKKβ is significantly longer than the previously defined NBD of IKKβ (residues 735–745) (26). Because of the higher expression levels of IKKβ (residues 680–756), further quantitative characterizations were performed with this construct.
Figure 2.
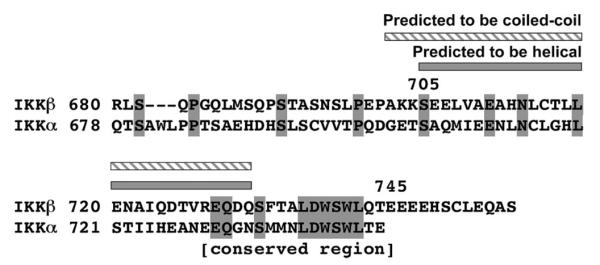
Sequence alignment of human IKKβ with human IKKα at the C-terminal region. Identical residues are highlighted in gray. The predicted α-helical and coiled-coil region is shown.
Characterization of the Interaction between IKKβ and NEMO Using Isothermal Titration Calorimetry (ITC)
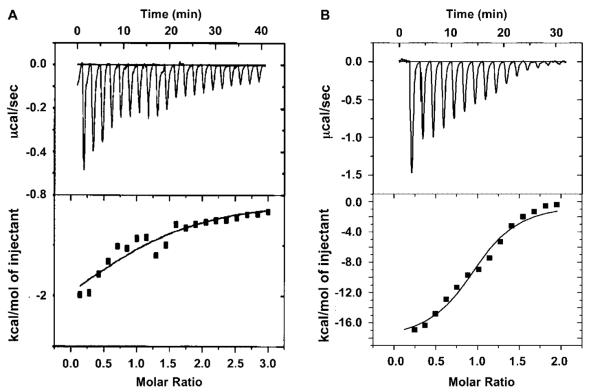
Because we could not detect formation of a stable complex between the NBD peptide and NEMO using either native PAGE or gel filtration chromatography, we attempted to use ITC to assess this interaction quantitatively (Figure 3A and Table 2). ITC measurements require large amounts of proteins and are sensitive for detection of weak interactions. The NBD peptide was titrated into the NEMO solution (residues 38–196). The release of heat during the titration was in excellent agreement with ideal binding, indicating the presence of a single type of binding site and the lack of cooperativity in the interaction. The heat of peptide dilution was measured by titrating the peptide into buffer alone. This was small and subtracted from the binding titration curve. Consistent with the lack of stable complex formation, the association constant of the interaction is 2.8 × 104 M−1 and the dissociation constant is 35 μM. The ITC measurements showed that favorable enthalpy is a minor component while favorable entropy contributes the most to the free energy of the interaction. This dominance of entropy is consistent with the importance of hydrophobic residues in the NEMO–IKKβ interaction (26).
Figure 3.
Isothermal titration calorimetry (ITC) used in assessing the IKKβ–NEMO interaction. (A) Titration of the IKKβ NBD peptide (residues 735–745) into a NEMO solution (residues 38–196). The top panel shows the calorimetric titration, which was carried out at 25 °C with 20 injections of 10 μL of 0.8 mM IKKβ NBD peptide, spaced 120 s apart, into the sample cell containing a solution of 1.334 mL of 40 μM NEMO. The bottom panel shows the fitting of the data to a single-site interaction model. (B) Titration of the IKKβ C-terminal region (residues 680–756) into a NEMO solution (residues 38–196). The top panel shows the calorimetric titration, which was carried out at 25 °C with 15 injections of 10 μL of 0.2 mM IKKμ, spaced 120 s apart, into the sample cell containing a solution of 1.334 mL of 13 μM NEMO. The bottom panel shows the fitting of the data to a single-site interaction model.
Table 2.
Thermodynamic Parameters for the IKKβ NBD (residues 735–745) and IKKβ C-Terminal Region (residues 680–756) Binding to NEMO (residues 38–196) As Measured by Isothermal Titration Calorimetrya
| IKKβ residues | N | KA (M−1) | KD (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|---|---|
| 735–745 | 1.16 ± 0.30 | 2.84 ± 0.23 × 104 | 35 | −6.07 | −1.15 ± 0.40 | −5.57 |
| 680–756 | 0.98 ± 0.04 | 9.38 ± 2.26 × 105 | 1 | −8.17 | −18.47 ± 1.07 | 10.30 |
Stoichiometry (N), association constant (KA), and enthalpy change (ΔH) are the mean values obtained from three independent ITC experiments. KD was determined using the relationship KD = KA−1. ΔG and −TΔS were determined from the relationship ΔG = −RT ln KA = ΔH − TΔS.
In parallel, we performed ITC studies on the interaction of NEMO with the longer IKKβ C-terminal region (residues 680–756) (Figure 3B and Table 2). Similarly, the IKKβ protein was titrated into the NEMO solution (residues 38–196) to obtain a binding isotherm as well as into buffer alone to obtain the heat of dilution. The measured association constant of the interaction is 0.94 × 105 M−1, and the dissociation constant is 1 μM. This is much stronger than the NEMO–IKKβ NBD peptide interaction with an affinity difference of approximately 35-fold. Interestingly, this interaction between NEMO and the longer IKKβ C-terminal region is dominated by favorable enthalpy rather than favorable entropy, suggesting a thermodynamic difference with the IKKβ NBD.
Characterization of the Interaction between IKKβ and NEMO Using Surface Plasmon Resonance (SPR)
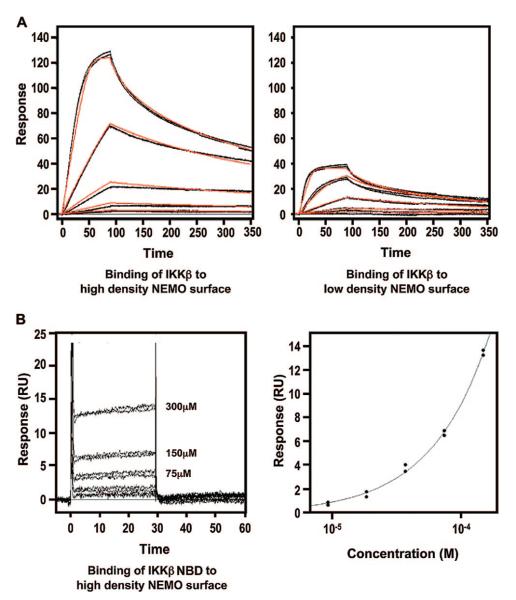
To quantitatively characterize the stable interaction between the IKKβ C-terminal domain and NEMO, we used surface plasmon resonance (SPR) (Figure 4A and Table 3). Purified NEMO (residues 38–196) was coupled to sensor chips via the biotin–streptavidin system to a high density and a low density. A concentration series of IKKβ (residues 680–756) was then assayed for their binding to NEMO surfaces. Both density level NEMO surfaces resulted in significant concentration-dependent interactions between NEMO and IKKβ. The obtained kinetic and affinity parameters were similar for both measurements and were averaged between the two surfaces. The interaction exhibits an association rate similar to that of diffusion-controlled rigid body macromolecular interactions, which are on the order of 106 M−1 s−1 for many different systems (53). This suggests that the interactions do not involve large-scale conformational changes. The obtained dissociation constant of the interaction is 3.4 nM, much lower than that for the interaction between the NBD peptide and NEMO.
Figure 4.
Surface plasmon resonance (SPR) characterization of the IKKβ–NEMO interaction. (A) Binding of the IKKβ C-terminal domain (residues 680–756) to a NEMO (residues 38–196)-coupled sensor chip. The left and right panels show the binding of IKKβ (0.617–50.0 nM) to the high-density (left) and low-density (right) NEMO surfaces, respectively. Each of these data sets was fit to a 1:1 interaction model (red lines), and a KD of 3.4 nM was obtained. (B) Binding of the IKKβ NBD (residues 735–745) to a NEMO (residues 38–196)-coupled sensor chip. The left panel shows the binding of IKKβ (18.8–300.0 μM) to the high-density NEMO surface. The right panel shows the fitting of the responses at equilibrium against peptide concentration which yields a KD of 3.6 mM.
Table 3.
Kinetic and Affinity Parameters for IKKβ (residues 680–756) and IKKβ NBD (residues 735–745) Binding to NEMO (residues 38–196) As Measured by Surface Plasmon Resonancea
| ka (× 106 M−1 s−1) | kd (s−1) | K D | |
|---|---|---|---|
| IKKβ to high-density NEMO surface | 6.3 ± 0.1 | 0.0216 ± 0.0003 | 3.43 ± 0.06 nM |
| IKKβ to low-density NEMO surface | 7.6 ± 0.2 | 0.0263 ± 0.0005 | 3.33 ± 0.01 nM |
| average parameters for binding of IKKβ to NEMO | 7.0 ± 0.7 | 0.023 ± 0.002 | 3.38 ± 0.06 nM |
| IKKβ NBD to high-density NEMO surface | NDb | NDb | 3.6 ± 0.4 mM |
ka is the kinetic association rate, kd the kinetic dissociation rate, and KD the equilibrium dissociation constant.
Cannot be determined.
Similar SPR measurements were also performed using the same NEMO surface, but with the IKKβ NBD peptide as the analyte. Unlike the IKKβ C-terminal proteins (residues 680–756), the NBD peptide only exhibited appreciable binding to the high-density NEMO surface with no detectable binding to the low-density NEMO surface. Even with the high-density NEMO surface, the complex associated and dissociated so quickly that reliable kinetic parameters could not be determined. Instead, fitting the responses at equilibrium plotted against peptide concentration to a binding isotherm yielded a dissociation constant of 3.6 mM.
Characterization of the Interaction between IKKβ and NEMO Using Multiangle Light Scattering (MALS)
To elucidate the stoichiometry of the IKKβ–NEMO interaction, we determined the accurate molecular masses of IKKβ, NEMO, and its complex (Figure 5). We used static multiangle light scattering (MALS) in conjunction with refractive index because the measurements are not functions of the shape of the molecules. The calculated molecular masses of NEMO (residues 38–196) and IKKβ (residues 680–756) are 21764.3 and 10776.7 Da, respectively. MALS measurements of NEMO (residues 38–196) alone showed that it existed in several different oligomeric states, high-order oligomer (~80 monomers), pentamer, trimer, dimer, and monomer, with measured molecular masses of 1722 kDa (0.4% fitting error), 109 kDa (0.3% error), 62.9 kDa (0.2% error), 44 kDa (0.2% error), and 22 kDa (0.4% error), respectively (Figure 5A). When the mixture of NEMO with excess IKKβ (residues 680–756) was subjected to gel filtration coupled with MALS, the majority of the NEMO–IKKβ complex was in a 2:2 stoichimetric complex with a measured molecular mass of 69.1 kDa (0.0% fitting error) (Figure 5B). In contrast, the IKKβ peak from gel filtration chromatography exhibited a molecular mass of 10.9 kDa (1% error), consistent with IKKβ being monomers in solution. A small portion of the NEMO–IKKβ complex appears to exist in a 4:4 complex with a measured molecular mass of 136 kDa (0.2% error).
Figure 5.
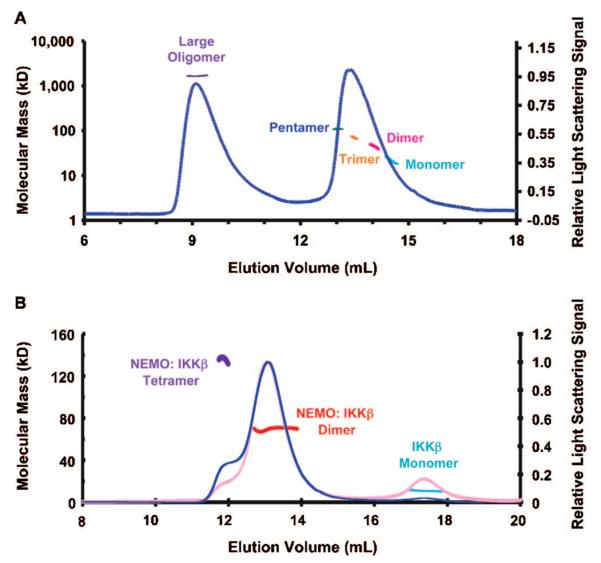
Multiangle light scattering (MALS) measurements of the IKKβ–NEMO interaction. (A) MALS measurement of NEMO alone, showing the relative light scattering signal as a function of elution volume. The derived molecular masses of peaks or peak locations are shown as lines that match the readings on the left axis. Several NEMO species are shown, such as large oligomer (purple), pentamer (blue), trimer (orange), dimer (dark red), and monomer (cyan). (B) MALS measurement of the mixture of NEMO with excess IKKβ, showing the relative light scattering signal as a function of elution volume. The derived molecular masses of peaks are shown as lines that match the readings on the left axis. NEMO–IKKβ complexes are mostly dimeric (red) with some tetrameric species (purple). Excess IKKβ alone exists as monomer (cyan). The refractive index reading of the chromatographic trace is colored pink.
DISCUSSION
In this report, we used both qualitative methods such as native PAGE and gel filtration chromatography and quantitative measurements such as SPR ad ITC in dissecting the NEMO–IKKβ interaction. While ITC measures the heat change during titration of one binding partner to the other partner and derives the corresponding dissociation constants, SPR measures the kinetics of the interactions and derives dissociation constants from the ratios of dissociation rates to association rates. ITC experiments are performed entirely in solution without surface tethering, while SPR requires the coupling of one binding partner to the surface of a sensor chip. Dissociation constants of 35 and 1 μM were obtained using ITC for the interactions of NEMO with the NBD peptide of IKKβ (residues 735–745) and with the longer C-terminal region of IKKβ (residues 680–756), respectively. These measurements show a difference of 35-fold in affinity between IKKβ NBD and the longer IKKβ C-terminal region. The difference in the apparent affinity is much larger in the SPR measurements with dissociation constants of 3.6 mM and 3.4 nM for the interactions of NEMO with the IKKβ NBD and with the longer C-terminal region of IKKβ (residues 680–756), respectively. Generally, ITC and SPR give consistent results (54). In this report, both ITC and SPR show a significantly higher affinity for the longer IKKβ C-terminal region. The much larger difference shown in the SPR measurements could be due to avidity effects. As shown by the MALS measurements, the NEMO–IKKβ interaction is mostly 2:2 but with some 4:4 components. The surface tethering may have promoted the higher-order interaction. The importance of the predicted IKKβ α-helical and coiled-coil region preceding the NBD region in the NEMO interaction suggests that although the IKKβ C-terminal region is monomeric in solution, it may form a coiled-coil structure upon contacting NEMO and facilitate the 2:2 or 4:4 interactions between the two proteins.
In any case, the data reported here defined a much more robust NEMO interaction domain in IKKβ, which includes additional sequences beyond the core conserved sequences of residues 735–745. Sequence alignment of IKKα and IKKβ shows that the additional sequences beyond the highly conserved NBD peptide region (residues 735–745) are not conserved between IKKα and IKKβ (Figure 4). The lack of sequence conservation suggests that these additional sequences may be specific for the NEMO–IKKβ interaction only, not for the homologous NEMO–IKKα interaction. It is possible that these sequences are distinguishing features between NEMO–IKKα and NEMO–IKKβ interactions. Therefore, they may be exploited for the development of specific peptide inhibitors against either the NEMO–IKKβ or the NEMO–IKKα interaction.
ACKNOWLEDGMENT
We thank the generosity of the structural biology groups at the Memorial Sloan-Kettering Cancer Center for access to the Micro Calorimetry System instrument and Dr. Jung-Hyun Min and Dr. Miao Lu for help with the ITC experiments. Y.-C. Lo is a postdoctoral fellow of Irvington Institute Fellowship Program of the Cancer Research Institute.
REFERENCES
- 1.Greten FR, Karin M. The IKK/NF-κB activation pathway: A target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 3.Gilmore TD. Introduction to NF-κB: Players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 4.Scheidereit C. IκB kinase complexes: Gateways to NF-κB activation and transcription. Oncogene. 2006;25:6685–6705. doi: 10.1038/sj.onc.1209934. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 7.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 8.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 9.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 10.Holtmann H, Enninga J, Kalble S, Thiefes A, Dorrie A, Broemer M, Winzen R, Wilhelm A, Ninomiya-Tsuji J, Matsumoto K, Resch K, Kracht M. The MAPK kinase kinase TAK1 plays a central role in coupling the interleukin-1 receptor to both transcriptional and RNA-targeted mechanisms of gene regulation. J. Biol. Chem. 2001;276:3508–3516. doi: 10.1074/jbc.M004376200. [DOI] [PubMed] [Google Scholar]
- 11.Irie T, Muta T, Takeshige K. TAK1 mediates an activation signal from toll-like receptor(s) to nuclear factor-κB in lipopolysaccharide-stimulated macrophages. FEBS Lett. 2000;467:160–164. doi: 10.1016/s0014-5793(00)01146-7. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Mira-Arbibe L, Ulevitch RJ. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J. Leukocyte Biol. 2000;68:909–915. [PubMed] [Google Scholar]
- 13.Jiang Z, Zamanian-Daryoush M, Nie H, Silva AM, Williams BR, Li X. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFκB and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J. Biol. Chem. 2003;278:16713–16719. doi: 10.1074/jbc.M300562200. [DOI] [PubMed] [Google Scholar]
- 14.Mizukami J, Takaesu G, Akatsuka H, Sakurai H, Ninomiya-Tsuji J, Matsumoto K, Sakurai N. Receptor activator of NF-κB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol. Cell. Biol. 2002;22:992–1000. doi: 10.1128/MCB.22.4.992-1000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besse A, Lamothe B, Campos AD, Webster WK, Maddineni U, Lin SC, Wu H, Darnay BG. TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J. Biol. Chem. 2007;282:3918–3928. doi: 10.1074/jbc.M608867200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J. Biol. Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 18.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 19.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 20.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 21.Yamaoka S, Courtois G, Bessia C, Whiteside ST, Weil R, Agou F, Kirk HE, Kay RJ, Israel A. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 22.Rothwarf DM, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 23.Mercurio F, Murray BW, Shevchenko A, Bennett BL, Young DB, Li JW, Pascual G, Motiwala A, Zhu H, Mann M, Manning AM. IκB kinase (IKK)-associated protein 1, a common component of the heterogeneous IKK complex. Mol. Cell. Biol. 1999;19:1526–1538. doi: 10.1128/mcb.19.2.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyet JL, Srinivasula SM, Lin JH, Fernandes-Alnemri T, Yamaoka S, Tsichlis PN, Alnemri ES. Activation of the IκB kinases by RIP via IKKγ/NEMO-mediated oligomerization. J. Biol. Chem. 2000;275:37966–37977. doi: 10.1074/jbc.M006643200. [DOI] [PubMed] [Google Scholar]
- 25.Delhase M, Hayakawa M, Chen Y, Karin M. Positive and negative regulation of IκB kinase activity through IKKβ subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 26.May MJ, D'Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-κB activation by a peptide that blocks the interaction of NEMO with the IκB kinase complex. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 27.Tegethoff S, Behlke J, Scheidereit C. Tetrameric oligomerization of IκB kinase γ (IKKγ) is obligatory for IKK complex activity and NF-κB activation. Mol. Cell. Biol. 2003;23:2029–2041. doi: 10.1128/MCB.23.6.2029-2041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, Iglehart JD. NF-κB activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Eppenberger-Castori S, Marx C, Yau C, Scott GK, Eppenberger U, Benz CC. Activation of nuclear factor-κB (NFκB) identifies a high-risk subset of hormone-dependent breast cancers. Int. J. Biochem. Cell Biol. 2005;37:1130–1144. doi: 10.1016/j.biocel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Andresen L, Jorgensen VL, Perner A, Hansen A, Eugen-Olsen J, Rask-Madsen J. Activation of nuclear factor κB in colonic mucosa from patients with collagenous and ulcerative colitis. Gut. 2005;54:503–509. doi: 10.1136/gut.2003.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla S, Maclennan GT, Marengo SR, Resnick MI, Gupta S. Constitutive activation of PI3K-Akt and NF-κB during prostate cancer progression in autochthonous transgenic mouse model. Prostate. 2005;YY:224–239. doi: 10.1002/pros.20217. [DOI] [PubMed] [Google Scholar]
- 34.Krappmann D, Emmerich F, Kordes U, Scharschmidt E, Dorken B, Scheidereit C. Molecular mechanisms of constitutive NF-κB/Rel activation in Hodgkin/Reed-Sternberg cells. Oncogene. 1999;18:943–953. doi: 10.1038/sj.onc.1202351. [DOI] [PubMed] [Google Scholar]
- 35.Aupperle K, Bennett B, Han Z, Boyle D, Manning A, Firestein G. NF-κB regulation by IκB kinase-2 in rheumatoid arthritis synoviocytes. J. Immunol. 2001;166:2705–2711. doi: 10.4049/jimmunol.166.4.2705. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig L, Kessler H, Wagner M, Hoang-Vu C, Dralle H, Adler G, Bohm BO, Schmid RM. Nuclear factor-κB is constitutively active in C-cell carcinoma and required for RET-induced transformation. Cancer Res. 2001;61:4526–4535. [PubMed] [Google Scholar]
- 37.Tamatani T, Azuma M, Aota K, Yamashita T, Bando T, Sato M. Enhanced IκB kinase activity is responsible for the augmented activity of NF-κB in human head and neck carcinoma cells. Cancer Lett. 2001;171:165–172. doi: 10.1016/s0304-3835(01)00611-5. [DOI] [PubMed] [Google Scholar]
- 38.Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: A treasure trove for drug development. Nat. Rev. Drug Discovery. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 39.Burke JR. Targeting IκB kinase for the treatment of inflammatory and other disorders. Curr. Opin. Drug Discovery Dev. 2003;6:720–728. [PubMed] [Google Scholar]
- 40.Yang J, Richmond A. Constitutive IκB kinase activity correlates with nuclear factor-κB activation in human melanoma cells. Cancer Res. 2001;61:4901–4909. [PubMed] [Google Scholar]
- 41.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, Munshi N, Dang L, Castro A, Palombella V, Adams J, Anderson KC. NF-κB as a therapeutic target in multiple myeloma. J. Biol. Chem. 2002;277:16639–16647. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 42.Jin SH, Kim TI, Han DS, Shin SK, Kim WH. Thalidomide suppresses the interleukin 1β-induced NFκB signaling pathway in colon cancer cells. Ann. N.Y. Acad. Sci. 2002;973:414–418. doi: 10.1111/j.1749-6632.2002.tb04674.x. [DOI] [PubMed] [Google Scholar]
- 43.Choi M, Rolle S, Wellner M, Cardoso MC, Scheidereit C, Luft FC, Kettritz R. Inhibition of NF-κB by a TAT-NEMO-binding domain peptide accelerates constitutive apoptosis and abrogates LPS-delayed neutrophil apoptosis. Blood. 2003;102:2259–2267. doi: 10.1182/blood-2002-09-2960. [DOI] [PubMed] [Google Scholar]
- 44.Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, Sudo T, Kojima T, Okamoto F, Fukushima H, Okabe K, Ohya K, Ghosh S. Selective inhibition of NF-κB blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat. Med. 2004;10:617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 45.Myszka DG. Improving biosensor analysis. J. Mol. Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 46.May MJ, Marienfeld RB, Ghosh S. Characterization of the IκB-kinase NEMO binding domain. J. Biol. Chem. 2002;277:45992–46000. doi: 10.1074/jbc.M206494200. [DOI] [PubMed] [Google Scholar]
- 47.Tas SW, de Jong EC, Hajji N, May MJ, Ghosh S, Vervoordeldonk MJ, Tak PP. Selective inhibition of NF-κB in dendritic cells by the NEMO-binding domain peptide blocks maturation and prevents T cell proliferation and polarization. Eur. J. Immunol. 2005;YY:1164–1174. doi: 10.1002/eji.200425956. [DOI] [PubMed] [Google Scholar]
- 48.Rehman KK, Bertera S, Bottino R, Balamurugan AN, Mai JC, Mi Z, Trucco M, Robbins PD. Protection of islets by in situ peptide-mediated transduction of the IκB kinase inhibitor Nemo-binding domain peptide. J. Biol. Chem. 2003;278:9862–9868. doi: 10.1074/jbc.M207700200. [DOI] [PubMed] [Google Scholar]
- 49.Dai S, Hirayama T, Abbas S, Abu-Amer Y. The IκB kinase (IKK) inhibitor, NEMO-binding domain peptide, blocks osteoclastogenesis and bone erosion in inflammatory arthritis. J. Biol. Chem. 2004;279:37219–37222. doi: 10.1074/jbc.C400258200. [DOI] [PubMed] [Google Scholar]
- 50.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf E, Kim PS, Berger B. MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agou F, Courtois G, Chiaravalli J, Baleux F, Coic YM, Traincard F, Israel A, Veron M. Inhibition of NF-κB activation by peptides targeting NF-κB essential modulator (nemo) oligomerization. J. Biol. Chem. 2004;279:54248–54257. doi: 10.1074/jbc.M406423200. [DOI] [PubMed] [Google Scholar]
- 53.Wu H, Myszka DG, Tendian SW, Brouillette CG, Sweet RW, Chaiken IM, Hendrickson HA. Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15030–15035. doi: 10.1073/pnas.93.26.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chesnokova LS, Slepenkov SV, Protasevich II, Sehorn MG, Brouillette CG, Witt SN. Deletion of DnaK's lid strengthens binding to the nucleotide exchange factor, GrpE: A kinetic and thermodynamic analysis. Biochemistry. 2003;42:9028–9040. doi: 10.1021/bi0346493. [DOI] [PubMed] [Google Scholar]