This study reports the highly efficient differentiation of human induced pluripotent stem cells (hiPSCs) toward a relatively homogeneous population of functional hepatocytes. A model was generated for the efficient generation of hepatocytes from induced pluripotent stem cells in vitro and in vivo, and it appears that the cells function similarly to primary human hepatocytes, including developing a complete metabolic function. These results represent a significant step toward using patient/disease-specific hepatocytes for cell-based therapeutics as well as for pharmacology and toxicology studies.
Keywords: Induced pluripotent stem cells, Hepatocyte differentiation, Stem cell transplantation, Liver regeneration
Abstract
Human induced pluripotent stem cells (hiPSCs) hold great potential for use in regenerative medicine, novel drug development, and disease progression/developmental studies. Here, we report highly efficient differentiation of hiPSCs toward a relatively homogeneous population of functional hepatocytes. hiPSC-derived hepatocytes (hiHs) not only showed a high expression of hepatocyte-specific proteins and liver-specific functions, but they also developed a functional biotransformation system including phase I and II metabolizing enzymes and phase III transporters. Nuclear receptors, which are critical for regulating the expression of metabolizing enzymes, were also expressed in hiHs. hiHs also responded to different compounds/inducers of cytochrome P450 as mature hepatocytes do. To follow up on this observation, we analyzed the drug metabolizing capacity of hiHs in real time using a novel ultraperformance liquid chromatography-tandem mass spectrometry. We found that, like freshly isolated primary human hepatocytes, the seven major metabolic pathways of the drug bufuralol were found in hiHs. In addition, transplanted hiHs engrafted, integrated, and proliferated in livers of an immune-deficient mouse model, and secreted human albumin, indicating that hiHs also function in vivo. In conclusion, we have generated a method for the efficient generation of hepatocytes from induced pluripotent stem cells in vitro and in vivo, and it appears that the cells function similarly to primary human hepatocytes, including developing a complete metabolic function. These results represent a significant step toward using patient/disease-specific hepatocytes for cell-based therapeutics as well as for pharmacology and toxicology studies.
Introduction
Liver dysfunction is a major health problem in the world, and liver transplantation is the only established successful treatment of end-stage liver failure. However, the source of human donor livers is limited. As an alternative to organ transplantation, hepatocyte transplantation and the use of hepatocytes with extracorporeal bioartificial liver devices may potentially provide effective treatment for many liver diseases. Therefore, it would be greatly beneficial if an unlimited supply of functional hepatocytes from other sources could be generated. It is postulated by many that pluripotent embryonic stem cells (ESCs) may be the most effective source of hepatocytes for regenerative medicine and for the pharmaceutical industry [1–6]. However, the ethical and social issues, and potential immune rejection after transplantation, have hindered ESC applications. Induced pluripotent stem cells (iPSCs) bypass some of these problems and have energized the fields of stem cell biology and regenerative medicine.
iPSCs are generated from somatic cells by forced expression of pluripotency factors [7–9], and they represent a pluripotent stem cell population that can differentiate into the same cell types as ESCs. Importantly, iPSCs provide the opportunity to generate patient/disease-specific cells. Therefore, iPSCs not only can be used to generate cells for autologous transplantation in regenerative medicine but also may be used to study disease mechanisms, and for drug development and toxicity studies.
The liver is a major location for the detoxification of drugs or other xenobiotics as well as endogenous substrates; therefore, metabolic function is a critical function of hepatocytes in vitro and in vivo. Although the differentiation of human iPSCs (hiPSCs) into hepatocytes has been reported in recent investigations [10–13], the cells generated in these reports showed limited hepatocyte-specific metabolic functions. In the present study, we report the generation of hiPSC-derived hepatocytes (hiHs) with a relatively homogeneous population, which exhibit properties of metabolically functional hepatocytes.
Materials and Methods
Human iPSC Line
hiPSC line IMR90-4 was purchased from WiCell Research Institute (Madison, WI, http://wicell.org), and cell culture was undertaken according to the instructions provided by WiCell.
Differentiation of hiPSCs Toward Hepatocytes
hiPSCs were induced to definitive endoderm (DE) as previously described [6]. The period of induction could be extended to 11 days or longer if hiPSCs were cultured with high confluence. The DE cells were treated with Accutase (Millipore, Billerica, MA, http://www.millipore.com) or trypsin (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) for a very short time and rapidly reseeded on collagen I-coated plates for hepatic differentiation using our previously described culture conditions [6]. The average splitting rate was determined by the seeded DE cells, which would grow to 80%–100% confluence within 7 days after differentiation. The DE cells with less than 50% confluence could be used to differentiate directly, without splitting. Fourteen days after differentiation, the cells were further differentiated and maintained in hepatocyte culture medium supplemented with growth factors and chemical reagents [6] for an additional 2–3 weeks, until use.
Gene Expression by hiHs
During the induction of DE and differentiation toward hepatocytes, the cells were analyzed for the expression of CXCR4, SOX17, and FOXA2, as well as for determining the percentage of the cells positive for albumin (ALB), α1-antitrypsin (α1-AT), and α-fetoprotein (AFP) by immunochemistry or/and flow cytometry as described previously [6].
Expressions of Metabolizing Enzymes, Transporters, and Nuclear Receptors in hiHs
The expression of metabolizing phase I and II enzymes, transporter proteins (phase III) and their regulators, and nuclear receptors in hiHs was determined by reverse transcription-polymerase chain reaction (PCR), immunohistochemistry, and Western blots. Total RNA and protein were extracted from undifferentiated hiPSCs, hiHs, human embryonic stem cell (hESC)-derived hepatocytes (hEHs), human primary hepatocytes (hPHs), and Hep G2 cells as described previously [6]. The primers and antibodies used are listed in supplemental online Tables 1 and 2.
Analysis of Liver Specific Functions in hiHs
Differentiated hiPSCs at day 18 were used to detect glycogen storage by using the periodic acid-Schiff's (PAS) staining [14], and cellular uptake and excretion of indocyanine green (ICG) (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) by ICG staining; [6] the excretion of ICG was examined by microscopy at 1 and 3.5 hours after removal of ICG. The secretion of ALB by hiHs was measured by enzyme-linked immunosorbent assay (ELISA) [6].
Induction of Cytochrome P450 Metabolic Activity by Inducers
hiHs were treated for 72 hours with rifampicin (final concentration, 25 μM) and phenobarbital (final concentration, 500 μM) at 22 days after differentiation. The supernatants were collected at 72 hours after treatment. Metabolism was assessed by the measurement of luciferase activity with the P450-Glo CYP3A4 assay kit (Promega, Madison, WI, http://www.promega.com) following the manufacturer's instructions.
Metabolic Profiling of Drug (+/−)-Bufuralol in hiHs
hiHs and freshly isolated hPHs were treated with (+/−)-bufuralol (BF) at a working concentration of 50 μM. The supernatants were collected at 24 and 48 hours after treatment, and the sample processing and ultraperformance liquid chromatography-tandem mass spectrometry (UPLC/MS/MS) were performed as described in the supplemental online data; the major change in the liquid chromatography (LC) condition was to shorten retention time (RT) from 15 minutes to 4 minutes [6]. Metabolite identification and validation are described in the supplemental online data. The amount of the metabolism products from each pathway at both phase I and phase II stages was determined as analyte peak area and normalized to the cell number used.
Transplantation of hiHs Into Mice
hiHs were injected into the spleens of NOD/SCID mice at approximately 5 × 105 cells per mouse. NOD/SCID mice were treated with retrorsine 2 weeks prior to transplantation [15]. Surgical procedures for transplantation and subsequent collection of mouse serum and liver tissue were approved by the Animal Care and Use Administrative Advisory Committees of the University of California, Davis.
Analysis of Human Liver Gene Expression in Mouse Liver and Serum
Mice were sacrificed at different time points after transplantation. The sample collection and processing, immunochemistry analysis, and ELISA were performed as described previously [14].
Human Primary Hepatocytes
Adult human primary hepatocytes freshly isolated from donor livers were provided by the Liver Tissue Cell Distribution System of the NIH (University of Pittsburg) and were used in the present studies with the approval of the institutional review board of the University of California, Davis.
Statistics
All data were summarized as means ± SEM from at least three independent measurements. An unpaired Student t test was used to analyze the data. p < .05 was considered statistically significant.
Results
Differentiation of hiPSCs Toward Hepatocytes
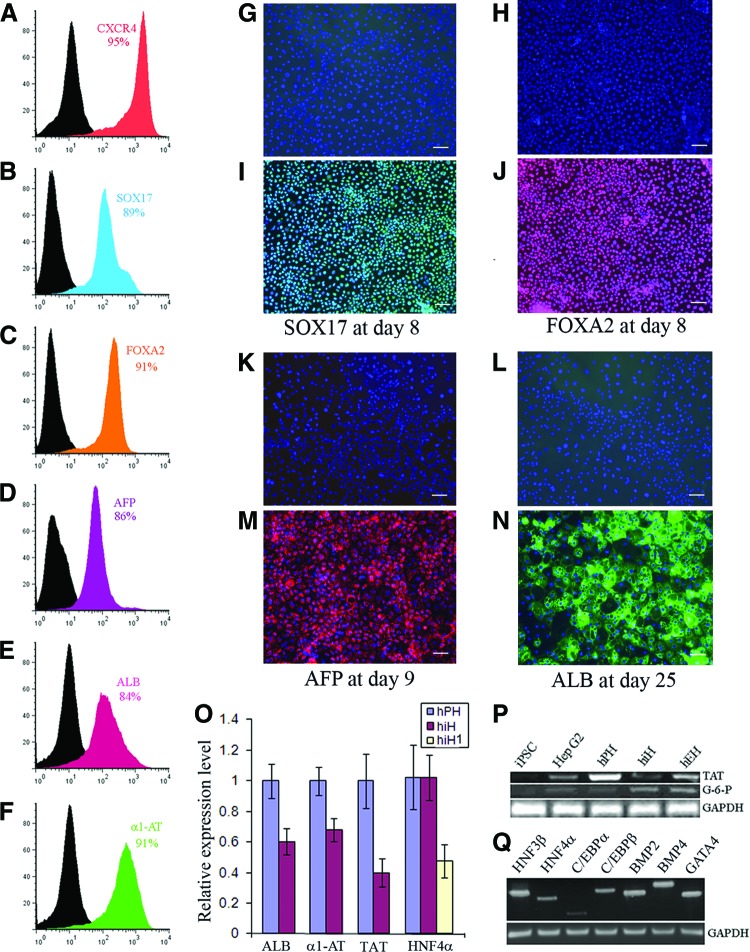
The differentiation protocol was illustrated as supplemental online Figure 1. hiPSCs were initially induced to contain a high percentage of DE cells as assessed by the expression of SOX17, FOXA2, and CXCR4 at levels of 89%, 91%, and 95%, respectively (Fig. 1A—1C, 1I, 1J). These DE cells showed a uniform morphology, indicating a relatively homogeneous population (supplemental online Fig. 1B). Under our differentiation culture condition, the DE cells were successfully differentiated into hepatic progenitor cells with 86% of the cells positive for AFP within 9 days, as determined by flow cytometry (FC) (Fig. 1D) and immunohistochemistry (Fig. 1M). The percentage of ALB-positive cells gradually increased to 84% at day 18 as determined by FC (Fig. 1E) and immunochemistry (Fig. 1N), and 91% of the cells were positive for α1-AT at day 18 (Fig. 1F). hiPSCs underwent a series of morphological changes during differentiation, and the hepatocyte morphology appeared from day 7 showing a polygonal shape and round single or double nuclei (Fig. 1M, 1N; supplemental online Fig. 1C, 1D).
Figure 1.
Differentiation of human induced pluripotent stem cells toward hepatocytes. (A–F): The cells were analyzed by flow cytometry for the percentage of positive cells for CXCR4 (A), SOX17 (B), and FOXA2 (C) at day 8 during induction of definitive endoderm; for AFP (D) at day 9 after differentiation; and for ALB (E) and α1-AT (F) at day 18 after differentiation. (G, N): Definitive endodermal cells were stained with the primary antibodies goat anti-SOX 17 (green) (I) and FOXA2 (red) (J) during induction of human induced pluripotent stem (iPS) cells to definitive endoderm; the differentiated cells were stained with the primary antibodies monoclonal antibody against AFP (red) (M) and goat anti-ALB (green) (N) during differentiation of human iPS cells toward hepatocytes. (G, H, K, L): Negative controls of SOX17 (G), FOXA2 (H), AFP (K), and ALB (L) stained with isotype antibodies. All immunohistochemistry analyses were merged with 4′,6′-diamidino-2-phenylindole nucleic acid staining (G–N). (O): Relative expression of ALB, α1-AT, TAT, and HNF4α in hiHs and hPHs determined by quantitative reverse transcription-polymerase chain reaction. (P, Q): Polymerase chain reaction was used to determine expression of the liver-associated genes G-6-P and TAT (P), and liver-associated transcriptional factors and BMP signaling (Q). Scale bars = 100 μm (G–N). Details of primers and antibodies can be found in the supplemental online tables. Abbreviations: α1-AT, α1-antitrypsin; AFP, α-fetoprotein; ALB, albumin; BMP, bone morphogenetic protein; G-6-P, glucose-6-phosphatase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; hiH, human induced pluripotent stem cell-derived hepatocytes at day 25; hiH1, human induced pluripotent stem cell-derived hepatocytes at day 10; hPH, human primary hepatocyte; HNF4α, hepatocyte nuclear factor 4α; iPSC, induced pluripotent stem cell; TAT, tyrosine aminotransferase.
Gene Expression by hiHs
The expression of liver-specific genes in our hiHs was determined at day 25 by quantitative reverse transcription-PCR. The results showed the average relative levels were 60 ± 8.8% for ALB, 68 ± 7.6% for α1-AT, 40 ± 9% for tyrosine aminotransferase (TAT), and 102 ± 15% and 48 ± 11% (at day 10) for hepatocyte nuclear factor 4α (HNF4α) compared with freshly isolated hPHs (Fig. 1O). Another indicator of more mature hepatocytes, glucose-6-phosphatase (G-6-P), was also expressed in our hiHs (Fig. 1P). In addition, the liver-associated transcription factors HNF3β, HNF4, GATA4, C/EBPα, C/EBPβ, and bone morphogenetic protein (BMP) signaling (BMP2 and BMP4) were all expressed, as determined by PCR (Fig. 1Q).
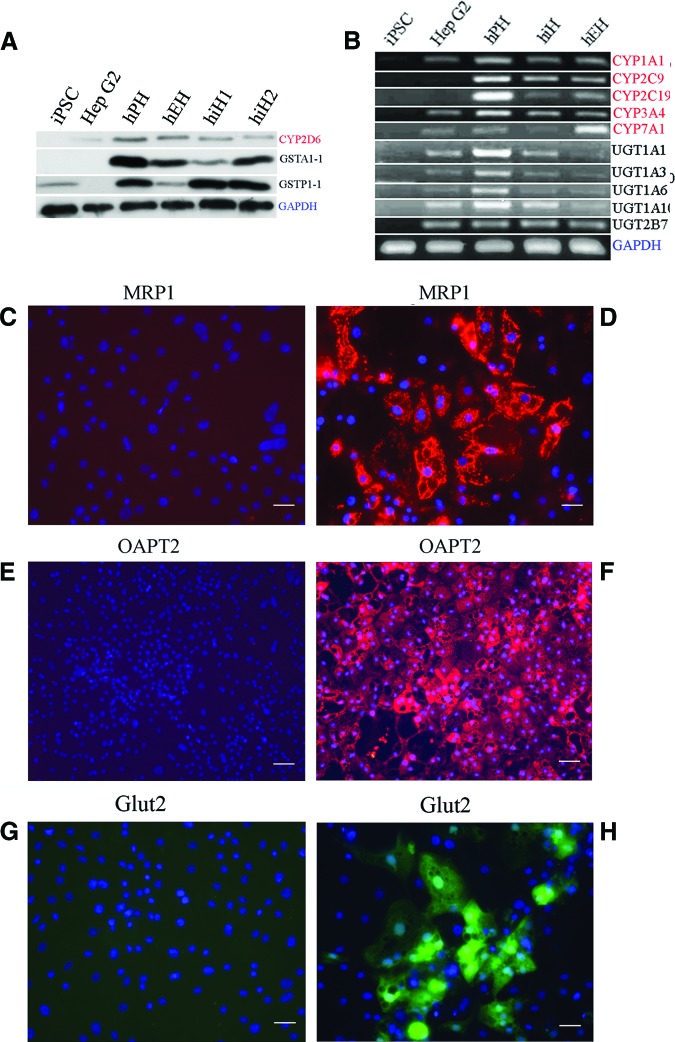
Metabolizing phase I enzymes (such as cytochrome P450 [CYP] 1A1, 2C9, 2C19, 2D6, 3A4, and 7A1) and phase II enzymes (such as glutathione S-transferase [GST] A1-1, GSTP1-1, UDP-glucuronosyl-S-transferase [UGT] 1A1, UGT1A3, UGT1A10, and UGT2B7) were expressed in hiHs, as determined by Western blot (Fig. 2A) and reverse transcription-PCR (Fig. 2B). Moreover, transporters and phase III proteins such as multidrug-resistant protein 1 (MRP1), organic anion transporting polypeptide 2 (OATP2), and glucose transporter 2 (Glut2) were also expressed in hiHs, as determined by immunohistochemistry (Fig. 2C–2H).
Figure 2.
Gene expression in hiHs. (A, B): Expression of phase I (red) and phase II (black) enzymes in hiHs determined by Western blot (A) and reverse transcription-polymerase chain reaction (B). hiH1 and hiH2 are the cells from different batches of differentiation. (D, F, H): Merge of immunostainings for expressions of MRP1 (D), OATP2 (F), and Glut2 (H) with nucleic acid staining by 4′,6′-diamidino-2-phenylindole (DAPI). (C, E, G): Negative controls of MRP1 (C), OAPT2 (E), and Glut2 (G) stained with isotype antibodies, respectively, in immunohistochemistry analysis merged with DAPI nucleic acid staining. Scale bars = 100 μm (C–F). Abbreviations: CYP, cytochrome P450; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Glut2, glucose transporter 2; GST, glutathione S-transferase; hEH, human embryonic stem cell-derived hepatocyte; hiH, human induced pluripotent stem cell-derived hepatocyte; hPH, human primary hepatocyte; iPSC, induced pluripotent stem cell; MRP1, multidrug-resistant protein 1; OATP2, organic anion transporting polypeptide 2; UGT, UDP-glucuronosyl-S-transferase.
Some important nuclear receptors, critical in regulating the expression of metabolizing enzymes, including the constitute androstane receptor (CAR), aryl hydrocarbon receptor (AhR), cytochrome P450 reductase (CPR), and pregnane X receptor (PXR), were also expressed in hiHs, as assessed by immunochemistry (Fig. 3).
Figure 3.
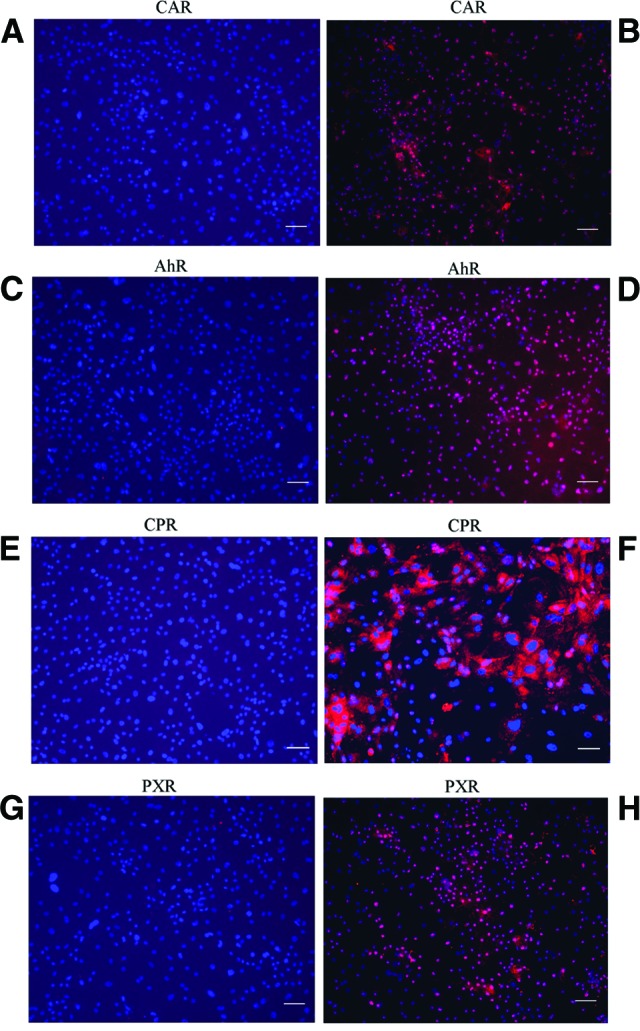
Expression of nuclear receptors in human induced pluripotent stem cell-derived hepatocytes (hiHs). (B, D, F, H): Merge of immunostainings for expressions of the nuclear receptors CAR, AhR, CPR, and PXR with nucleic acid by 4′,6′-diamidino-2-phenylindole (DAPI) in hiHs determined by immunohistochemistry. (A, C, E, G): Negative controls of CAR, AhR, CPR, and PXR stained with the corresponding isotype antibodies in immunohistochemistry analysis merged with nucleic acid staining by DAPI. Scale bars = 100 μm (A–H). Abbreviations: AhR, aryl hydrocarbon receptor; CAR, constitute androstane receptor; CPR, cytochrome P450 reductase; PXR, pregnane X receptor.
Liver-Specific Function in hiHs
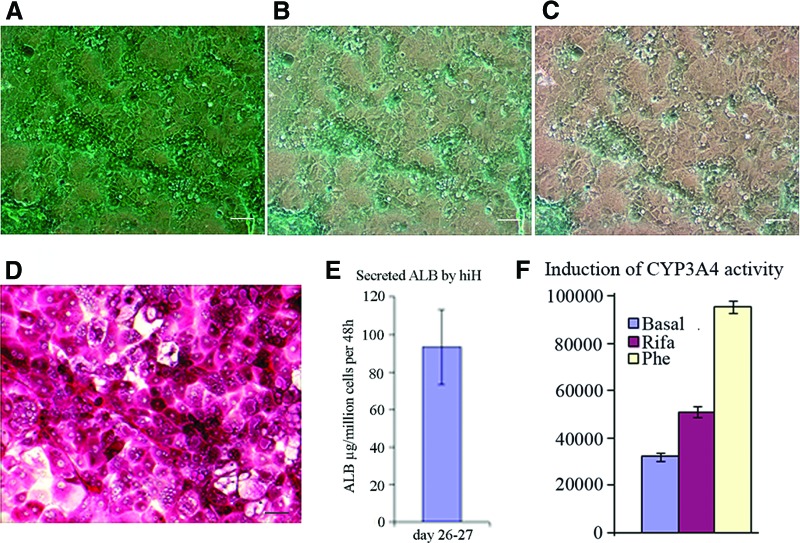
The cellular uptake and excretion of ICG is a unique characteristic of hepatocytes. The cellular uptake was observed in our hiHs in a very high percentage of cells (Fig. 4A), and the excretion of ICG appeared 1 hour after the removal of ICG (Fig. 4B), with most of the ICG excreted within 3.5 hours (Fig. 4C), and it almost completely disappeared by the next day (data not shown), indicating that a functional biotransforming system was generated by our hiHs. hiHs also showed glycogen accumulation by PAS staining, and these PAS-positive cells had a hepatocyte morphology (Fig. 4D). One of the important functions of hepatocytes is to secrete functioning proteins into the blood, such as albumin. ELISA showed that 93 ± 20 μg of ALB was secreted into the medium per million cells within 48 hours at days 26–27 of differentiation (Fig. 4E). An important property of hepatocyte function is the activity to respond to compounds/inducers. The activity of CYP3A4 was significantly increased 60% and 300% when cells were treated with either rifampicin or phenobarbital, respectively, compared with untreated hiHs (Fig. 4F).
Figure 4.
Liver function assays of hiHs. (A): hiHs were stained at 37°C for 60 minutes with indocyanine green (ICG) at day 18 after differentiation, and then the cellular uptake of ICG was examined by microscopy after the cells were washed with phosphate-buffered saline (PBS). (B, C): After PBS was replaced by the culture medium and the cells were incubated at 37°C again, the cellular excretion of ICG was examined by microscopy at 1 hour (B) and 3.5 hours (C) after the removal of ICG. (D): hiHs were analyzed by periodic acid-Schiff's staining for glycogen storage at day 20 after differentiation. Scale bars = 100 μm (A–D). (E): The supernatants were collected during the differentiation of human induced pluripotent stem cells, and albumin secretion by hiHs in supernatants was determined by enzyme-linked immunosorbent assay. The total secreted albumin was normalized to the cell numbers at days 26–27. (F): The supernatants were collected at 72 hours after hiHs were treated with the inducers Rifa and Phe at day 22 after differentiation. The increase of CYP3A4 activity was assessed by measurement of luciferase activity with the P450-Glo CYP3A4 assay kit. Abbreviations: ALB, albumin; hiH, human induced pluripotent stem cell-derived hepatocyte; Phe, phenobarbital; Rifa, rifampicin.
Metabolite Identification and Validation in hiHs
(+/−)-Bufuralol is a nonselective β-adrenoceptor blocking agent; its major metabolic pathways in humans are oxidation and glucuronidation [16, 17], and our recent study showed that there are seven major metabolic pathways in vitro [6]. Predictive multiple reaction monitoring (MRM) is the most sensitive approach in metabolite identification [18, 19]. The predictive MRM transitions of most of the metabolic pathways of BF are listed in supplemental online Table 3.
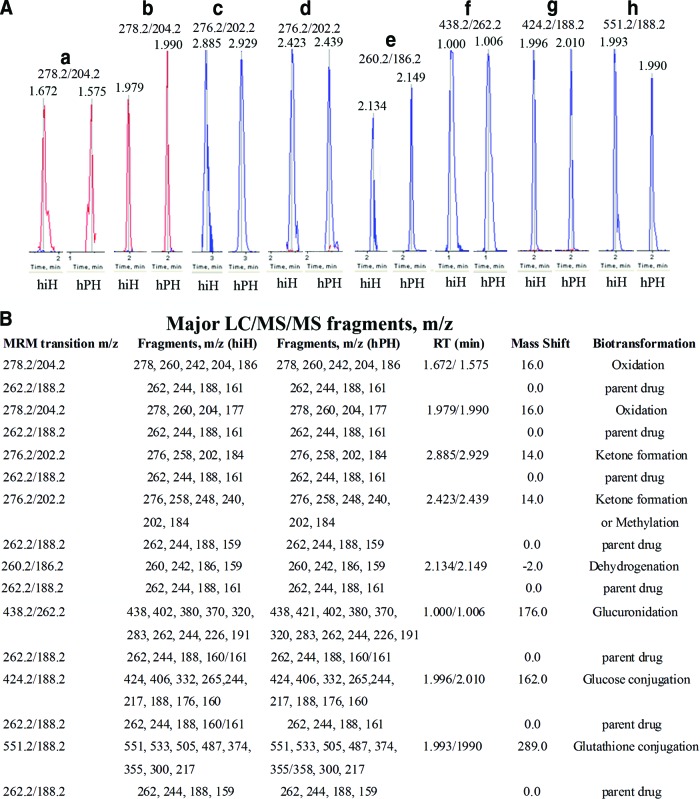
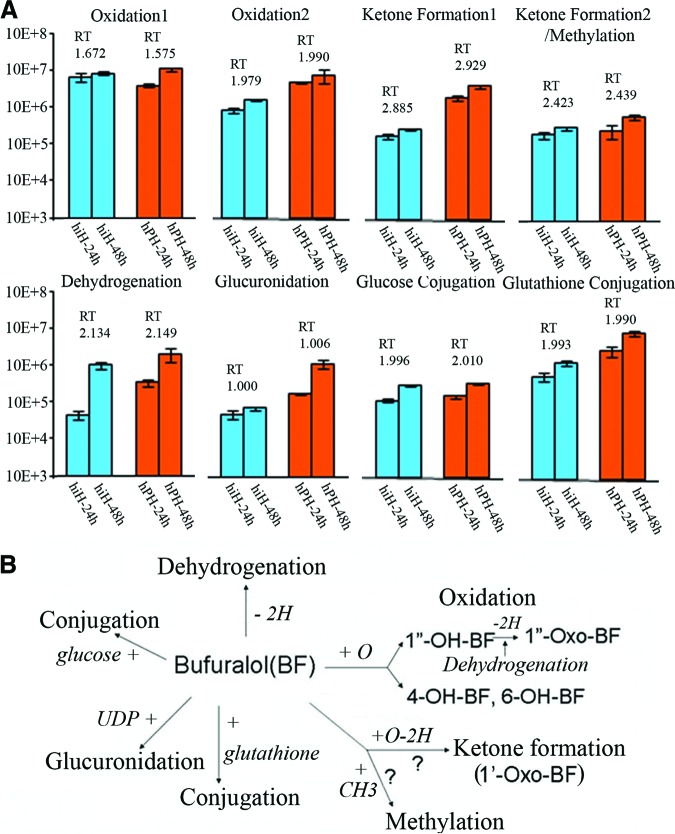
Four peaks with the same MRM transition of m/z 278.2/204.2 and RTs of 1.672 and 1.979 minutes (hiHs) and 1.575 and 1.990 minutes (hPHs) (Fig. 5A, peaks a and b) had time-course effects (Fig. 6A), suggesting a secondary metabolite of oxidation (supplemental online Table 3). Compared with the parent BF, the MS/MS fragmentation pattern showed an m/z shift of +16 on most of the major fragments in these secondary metabolites of the four peaks, confirming the biotransformation of oxidation (Fig. 5B; supplemental online Fig. 2). Because we shortened the LC RT in this study, we found that the metabolites with RTs of 1.672 and 1.575 minutes (oxidation 1) (supplemental online Fig. 2A, 2B) and 1.979 and 1.990 minutes (oxidation 2) (supplemental online Fig. 2C, 2D) are the same as those from oxidation 1 at RT of 8.8 minutes and oxidation 2 at RT 9.0 minutes, which were shown in our previous study [6]. We found that the oxidation of the metabolite with RTs 1.672/1.575 minutes occurred at the aromatic 7-position forming 1″-OH-BF, and the metabolites with RTs of 1.979/1.990 minutes are aromatic hydroxylations at the 4- and 6-positions (details are described in the legend of supplemental online Fig. 2).
Figure 5.
Metabolites (peaks) identified with MRM transition code and liquid chromatography (LC) RT and MS/MS fragment patterns. (A): Peaks a and b: Metabolites with MRM transition of m/z 278.2/204.2 and RTs of 1.672 and 1.979 minutes (from hiHs) and 1.575 and 1.990 minutes (from hPHs) were identified from oxidations/+16.0. Peaks c and d: Metabolites with MRM transition of m/z 276.2/202.2 and RTs of 2.885 and 2.423 minutes (from hiHs) and 2.929 and 2.438 minutes (from hPHs) were identified from ketone formation and/or methylation/+14.0. Peak e: Metabolites with MRM transition of m/z 260.2/186.2 and RTs of 2.134 minutes (from hiHs) and 2.149 minutes (from hPHs) were identified from dehydrogenation/−2.0. Peak f: Metabolites with MRM transition of m/z 438.2/262.2 and RTs of 1.000 minute (from hiHs) and 1.006 minutes (from in hPHs) were identified from glucuronidation/+289.0. Peak g: Metabolites with MRM transition of m/z 424.2/188.2 and RTs of 1.996 minutes (from in hiHs) and 2.010 minutes (from hPHs) were identified from conjugation of glucose/+162.0. Peak h: Metabolites with MRM transition of m/z 551.2/188.2 and RTs of 1.993 minutes (from hiHs) and 1.990 minutes (from hPHs) were identified from conjugation of glutathione/+289.0. (B): The major LC/MS/MS fragments of metabolites from each pathway are listed with those of parent bufuralol. Details of mass spectrometry methods, metabolite identification, and validation can be found in the supplemental online data. Abbreviations: hiH, human induced pluripotent stem cell-derived hepatocyte; hPH, human primary hepatocyte; LC/MS/MS, liquid chromatography tandem mass spectrometry; MRM, multiple reaction monitoring; RT, retention time.
Figure 6.
Time course effects of metabolites after treatment with BF and their metabolic pathways in hiHs and hPHs. (A): Supernatants from hiHs and hPHs treated with BF were collected at 24 and 48 hours after treatment, and the amount of metabolite products was determined as analyte peak area and normalized to cell number used. Metabolites identified by MRM transition codes and liquid chromatography RTs (shown in Fig. 5) from each pathway showed time course effects in both hiHs and hPHs. (B): Summary of in vitro metabolic pathways of bufuralol in hiHs and hPHs. The metabolic pathways of drug bufuralol in hiHs were shown to be the same as those in hPHs. Abbreviations: BF, bufuralol; hiH, human induced pluripotent stem cell-derived hepatocyte; hPH, human primary hepatocyte; RT, retention time.
Four peaks with the same MRM transition of m/z 276.2/202.2 and RTs of 2.885 and 2.423 minutes (hiHs) and 2.929 and 2.439 minutes (hPHs) (Fig. 5A, peaks c and d) showed a time-course effect (Fig. 6A), suggesting a secondary metabolite of methylation (supplemental online Table 3). Compared with the parent BF, the MS/MS fragmentation pattern showed an m/z shift of +14 on most of the major fragments in these secondary metabolites from these peaks (Fig. 5B; supplemental online Fig. 3). Similar to the situation in the oxidation pathway, we found that the metabolite with RTs of 2.885 and 2.929 minutes (ketone formation 1) (supplemental online Fig. 3A, 3B) and 2.423 and 2.439 minutes (supplemental online Fig. 3C, 3D) (ketone formation 2 or methylation) are the same as those with ketone formation 1 at RT of 8.8 minutes and ketone formation 2 or methylation at 8.4 minutes, which were shown in our previous study [6]. Therefore, the metabolites with RTs of 2.885/2.929 minutes showed a biotransformation of a ketone formation producing 1″-oxo-BF by dehydrogenation from the oxidation forming 1″-OH-BF, and the metabolites with RTs of 2.423/2.439 minutes might be from another ketone formation occurring at the 1′-position or from another possibility, which is that this metabolic attack is a methylation of BF.
The peaks with MRM transition of m/z 260.2/186.2 and RTs of 2.134 minutes (hiHs) and 2.149 minutes (hPHs) (Fig. 5A, peak e) showed a time-course effect (Fig. 6A), suggesting a secondary metabolite of dehydrogenation (supplemental online Table 3). Compared with parent BF, the MS/MS fragmentation pattern showed an m/z shift of −2 on most of the major fragments in the secondary metabolite, confirming the biotransformation of dehydrogenation (Fig. 5B; supplemental online Fig. 4A, 4B).
The peaks with MRM transition of m/z 438.2/262.2 and RTs of 1.000 minute (hiHs) and 1.006 minutes (hPHs) (Fig. 5A, peak f) had a time-course effect (Fig. 6A), indicating a secondary metabolite of glucuronidation (supplemental online Table 3). In the LC/MS/MS fragmentation pattern of the metabolite from this biotransformation, the major fragment of the metabolite was at m/z 438 (m/z shift of BF + m/z 176 from the uridine diphosphate [UDP] group), confirming that these metabolites are the results of BF glucuronidation (Fig. 5B; supplemental online Fig. 4C, 4D). We found that there are two major fragments, m/z 438 (m/z shift of + m/z 176 from the UDP group) and m/z 420 (m/z 244 [the remaining parent of BF after cleavage] shift + m/z 176), indicating that the UDP group is linked to the parent BF molecule during cleavage of the metabolite. Before removing the parent BF, there are some major fragments, m/z 380, m/z 370, and m/z 320 (parent BF with partial UDP), in its LC/MS/MS fragmentation (Fig. 5B; supplemental online Fig. 4C, 4D). After removal of the parent BF, its LC/MS/MS fragments of the remaining molecule consist of the major fragments from parent BF (m/z 262, m/z 244). The MS/MS fragmentation patterns in hPHs and hiHs were the same or similar (Fig. 5B; supplemental online Fig. 4C, 4D).
The peaks with MRM transition of m/z 424.2/188.2 and RTs of 1.996 minutes (hiHs) and 2.010 minutes (hPHs) (Fig. 5A, peak g) had a time-course effect (Fig. 6A), suggesting a secondary metabolite of conjugation with glucose (supplemental online Table 3). The LC/MS/MS fragmentation pattern showed that these metabolites are from BF conjugation with glucose (supplemental online Fig. 5A, 5B). There are two major fragments, m/z 424 (m/z shift of + m/z 162 from glucose) and m/z 406 (m/z 244 shift + m/z 162), indicating the glucose is linked to the BF molecule during cleavage of the metabolites. Therefore, the LC/MS/MS fragmentation patterns of BF glucose conjugates include the fragments from parent BF (m/z 262), conjugates (m/z 424), and/or glucose (m/z 162) during cleavage. The major fragments of the metabolites from BF glucose conjugates in hiHs and hPHs were the same or similar (Fig. 5B; supplemental online Fig. 5A, 5B).
The peaks with MRM transition of m/z 551.2/188.2 and RTs of 1.993 minutes (hiHs) and 1.990 minutes (hPHs) (Fig. 5A, peak h) had a time-course effect (Fig. 6A), suggesting a secondary metabolite of conjugation with glutathione (supplemental online Table 3). The LC/MS/MS fragmentation pattern showed that these metabolites are the results of BF glutathione conjugates (Fig. 5B; supplemental online Fig. 5C, 5D). Two major fragments exist in the LC/MS/MS fragmentation pattern of this metabolite: m/z 551 (m/z shift of +m/z 289 from glutathione) and m/z 533 (m/z 244 shift of +m/z 289), indicating that the glutathione is also linked to the BF molecule during the cleavage of the metabolites. Thus, the LC/MS/MS fragmentation pattern of BF glutathione conjugates consists of the fragments of parent BF (m/z 262), conjugate (m/z 551), and/or glutathione (m/z 289) during cleavage. The major fragments in hiHs and hPHs were the same or similar (Fig. 5B; supplemental online Fig. 5C, 5D).
Therefore, with the evidence for the same MRM transitions, LC RT, LC/MS/MS fragmentation patterns, and time-course effects of these secondary metabolites, we have successfully identified seven major metabolic pathways of BF (oxidation, dehydrogenation, ketone formation, and potential methylation in the phase I stage, and glucuronidation and conjugations of glucose and glutathione in the phase II stage) in hiHs, which are the same as those found in hPHs (Figs. 5, 6). This demonstrated for the first time that our hiHs had developed the full metabolic function of hPHs.
Analysis of Human Liver-Specific Gene Expression by hiHs in Mouse Liver and Serum
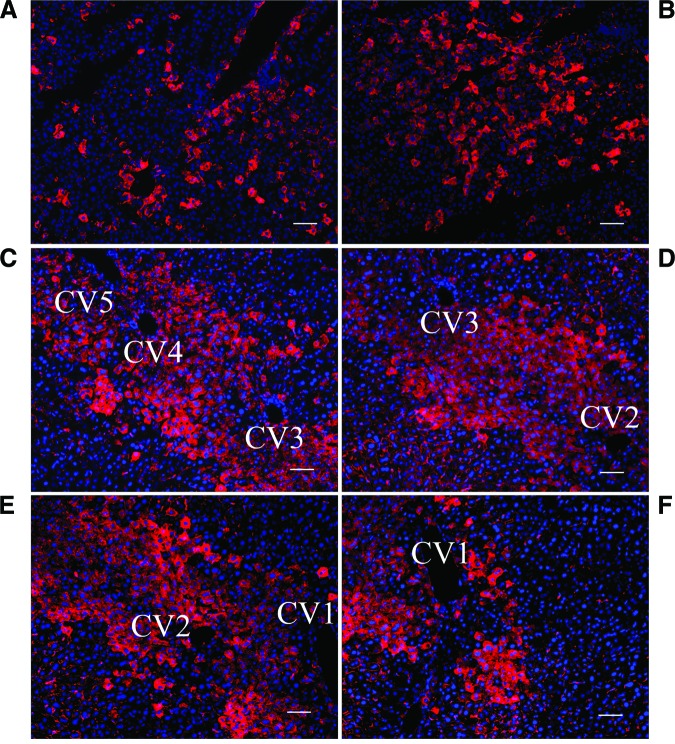
Liver tissue from the mice that were sacrificed at different time points after transplantation with hiHs through spleenic injection was immunostained with antibody against human ALB [14]. The results showed that a few single cells were positive for human ALB staining around the central vein and in the parenchyma in the livers of mice that were sacrificed 96 hours after injection, indicating that hiHs engrafted in the liver as early as day 4 after transplantation (data not show) and that a large number of cells around the central vein were positive for ALB in the livers of NOD/SCID mice. Also, a large number of positive cells were seen in the parenchyma at 4 weeks after transplantation (Fig. 7A, 7B), demonstrating that hiHs engrafted and integrated after transplantation. Interestingly, we also found that hiHs proliferated along at least five adjacent central veins as determined by human ALB staining in the livers of NOD/SCID mice that were treated with retrorsine 2 weeks prior to transplantation (Fig. 7C–7F). One function of hepatocytes is to secrete proteins into the blood stream, such as ALB. ELISA detected human ALB in the serum of 5 of 12 mice transplanted with hiHs at 4 weeks after transplantation; the values of human ALB were in the range of 5–48 ng/ml. Taken together, these results indicated that our hiHs also functioned in vivo.
Figure 7.
Analysis of human albumin expression by human induced pluripotent stem cell-derived hepatocytes (hiHs) in mouse livers after transplantation. (A–D): Liver sections from transplanted mice were immunostained with primary goat anti-human albumin antibody in livers of NOD/SCID mice sacrificed at 4 weeks after transplantation with hiHs. Large numbers of hiHs engrafted around central veins (A), integrated in the parenchyma of mouse livers (B), and proliferated along at least five adjacent central veins (CV1–CV5) in the livers of mice treated with retrorsine 2 weeks prior to transplantation (C–F). All immunostainings of liver sections were merged with nucleic acid staining by 4′,6-diamidino-2-phenylindole. Scale bars = 100 μm (A–F). Abbreviation: CV, central vein.
Discussion
The recent discovery of iPSCs has opened up new avenues in the field of regenerative medicine [7–9], including autologous cell therapies, and provides opportunities for drug development and studies of disease mechanisms. In the present study, hiPSCs were enriched to contain more than 90% DE cells, and DE cells were differentiated into cell populations with a high percentage of cells positive for AFP and ALB, similar to what we recently achieved in hESCs [6], suggesting that there was no difference in the hepatocyte differentiation capacity of hESCs and hiPSCs.
Liver transcription factors HNF3β, GATA4, C/EBPα, C/EBPβ, and BMP signaling (BMP2, and BMP4), which play important roles in hepatocyte differentiation and liver development [14, 20], were all expressed during differentiation; the expression of these factors in our hiHs is crucial to prime these cells for further hepatocyte differentiation. Our hiHs expressed a series of liver-specific genes such as ALB, α1-AT, TAT, and G-6-P, which are indicators of more mature hepatocytes and play important functions in the liver. Moreover, in functional assays, our hiHs accumulated glycogen and demonstrated uptake and excretion of ICG, and secretion of ALB into the medium. Interestingly, we found that it took a few more days (26–27 days after differentiation) for hiHs to secrete similar amounts of ALB compared with hESC-derived hepatocytes (20–22 days) [6]. This is the first difference we found in the differentiation between hESCs and hiPSCs in our present study. Thus, we extended the period of time for both the induction of DE and differentiation/maturation in our protocol for hiPSCs differentiation.
Metabolic activity is the most important function of hepatocytes in vitro and in vivo; this function is performed by the complex biotransforming system, which consists of phase I and II metabolizing enzymes and phase III transporters. The results demonstrated that our hiHs not only expressed phase I and II enzymes, as well as phase III proteins, but also appeared to express a functional biotransforming system, shown by the cellular uptake, conjunction, and excretion of ICG. Moreover, some nuclear receptors, key mediators regulating drug-metabolizing enzymes and transporters [21], were also expressed by our hiHs. Importantly, metabolic function of our hiHs was enhanced by two common inducers; responding to compounds/inducers is characteristic of functioning hepatocytes. Taken together, these results indicated that our hiHs appear to have developed a full biotransformation system.
To further assess this finding, we used UPLC/MS/MS technology for drug metabolic profiling and metabolism to determine the real-time functions of the biotransforming system, as described for differentiated hESCs in our previous study [6]. With the evidence of MRM transitions, LC RT, MS/MS fragmentation patterns, and time-course effects of the secondary metabolites, we identified seven major metabolic pathways of the drug bufuralol from phase I and II stages in our hiHs (Fig. 6B), which are the same as those in hPHs, thus indicating that our hiHs appear to have complete metabolic function that is comparable to those functions in primary liver cells. Interestingly, we found that the value of metabolites in oxidation 1 at RT of 1.672 minutes from hiHs was higher than or similar to those from hPHs (Fig. 6A) and that the values of metabolites in ketone formation 2 or methylation at RT of 2.423 minutes (Fig. 6A) and the conjugation of glucose were also very similar to those from hPHs (Fig. 6A). It appears that these three metabolic pathways are more active in these hiHs than in hEHs [6]. This is the first report to systematically evaluate the presence and distribution of the biotransforming enzymes/proteins and their regulators, the nuclear receptors, and to determine the real-time activities and functions of these biotransforming enzymes/proteins in hiPSC-derived hepatocytes, indicating that differentiated hiPSCs can function in vitro as differentiated hESCs do.
We also showed that hiHs can engraft similarly to hEHs in livers of NOD/SCID mice (Fig. 7) [14]. We noticed that hiHs engrafted in the liver as early as day 4 after transplantation, and we also found significant numbers of hiHs engrafted around central veins and integrated in the parenchyma of mouse livers. By using retrorsine to treat NOD/SCID mice 2 weeks prior to transplantation, we found that hiHs proliferated in the central vein area at 4 weeks after transplantation (Fig. 7C–7F). Human ALB was also detected in the mouse serum. The concentrations of human ALB secreted from hiHs were comparable to those obtained from the mice that had been transplanted with hESC-derived hepatocytes [14], demonstrating that hiPSCs also can function in vivo as hEHs do.
Conclusion
In summary, we have generated a relatively homogeneous population of differentiated cells from hiPSCs, which not only demonstrate the phenotype of human hepatocytes with liver-specific functions but also, for the first time, show complete metabolic function comparable to that of freshly isolated hPHs. Moreover, hiHs engrafted, integrated, and proliferated in mouse livers after transplantation, and human liver-specific protein could be detected in mouse liver and serum. This also demonstrated that human ALB produced by fibroblast-derived hepatocytes was detected in an animal model. Thus, these results represent a significant step in developing patient/disease-specific hepatocytes that may be effective for cell therapy and pharmacologic studies in the future.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grant DK075415 (to M.A.Z.), California Institute of Regenerative Medicine Grant RC1-00359 (to M.A.Z.), and a pilot grant from the University of California Davis Genome Center (to Y.D.). We gratefully acknowledge Dr. Stephen Strom and the NIH-supported Liver Tissue Cell Distribution System for providing human primary hepatocytes.
Author Contributions
X.M.: design, provision of study material, execution and collection of data, data analysis and interpretation; Y.D.: conception and design, execution and collection of data, data analysis and interpretation, manuscript writing, partial financial support; B.T.-S. and G.R.: execution and collection of data, data analysis and interpretation; I.S.B., T.P.A., and S.K.: execution and collection of data; V.T.: execution and collection of data, data analysis and interpretation, partial facility support; C.W.: provision of study material, execution and collection of data; J.M.: provision of study material; J.A.N.: conception and design, provision of study material, review of manuscript; H.W.: conception and design, revision of manuscript; M.A.Z.: conception and design, laboratory and full financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Cai J, Zhao Y, Liu Y, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–1239. doi: 10.1002/hep.21582. [DOI] [PubMed] [Google Scholar]
- 2.Hay DC, Zhao D, Fletcher J, et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 2008;26:894–902. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal S, Holton KH, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117–1127. doi: 10.1634/stemcells.2007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Hay DC, Fletcher J, Payne C, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires Activin A and Wnt3a signaling. Proc Natl Acad Sci USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basma H, Soto-Gutiérrez A, Yannam GR, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Y, Ma X, Zou W, et al. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells. 2010;28:674–686. doi: 10.1002/stem.315. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 10.Song Z, Cai J, Liu Y, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan GJ, Hay DC, Park I, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rashid ST, Corbineau S, Hannan N, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan Y, Catana A, Meng Y, et al. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells. 2007;25:3058–3068. doi: 10.1634/stemcells.2007-0291. [DOI] [PubMed] [Google Scholar]
- 15.Guo D, Fu T, Nelson JA, et al. Liver repopulation after cell transplantation in mice treated with retrorsine and carbon tetrachloride. Transplantation. 2002;73:1818–1824. doi: 10.1097/00007890-200206150-00020. [DOI] [PubMed] [Google Scholar]
- 16.Francis RJ, East PB, Larman J. Kinetics and metabolisms of (+)-, (-)- and (+/-)- bufuralol. Eur J Clin Pharmacol. 1982;23:529–533. doi: 10.1007/BF00637501. [DOI] [PubMed] [Google Scholar]
- 17.Narimatsu S, Takemi C, Kuramoto S, et al. Stereoselectivity in the oxidation of bufuralol, a chiral substrate, by human cytochrome P450s. Chirality. 2003;15:333–339. doi: 10.1002/chir.10212. [DOI] [PubMed] [Google Scholar]
- 18.Mensch J, Noppe M, Adriaensen J, et al. Novel generic UPLC/MS/MS method for high throughput analysis applied to permeability assessment in early drug discovery. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:182–187. doi: 10.1016/j.jchromb.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 19.Wen B, Ma L, Nelson SD, et al. High-throughput screening and characterization of re-active metabolites using polarity switching of hybrid triple quadrupole linear ion trap mass spectrometry. Anal Chem. 2008;80:1788–1799. doi: 10.1021/ac702232r. [DOI] [PubMed] [Google Scholar]
- 20.Gouon-Evans V, Boussemart L, Gadue P, et al. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- 21.Hewitt NJ, Lecluyse EL, Ferguson SS. Induction of hepatic cytochrome P450 enzymes: methods, mechanisms, recommendations, and in vitro-in vivo correlations. Xenobiotica. 2007;37:1196–1224. doi: 10.1080/00498250701534893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.