Abstract
Background
Heart morphogenesis involves sequential anatomical changes from a linear tube of a single channel peristaltic pump to a four-chamber structure with two channels controlled by one-way valves. The developing heart undergoes continuous remodeling, including septation.
Results
Pitx2-null mice are characterized by cardiac septational defects of the atria, ventricles, and outflow tract. Pitx2-null mice also exhibited a short outflow tract, including unseptated conus and deformed endocardial cushions. Cushions were characterized with a jelly-like structure, rather than the distinct membrane-looking leaflets, indicating that endothelial mesenchymal transition was impaired in Pitx2−/− embryos. Mesoderm cells from the branchial arches and neural crest cells from the otic region contribute to the development of the endocardial cushions, and both were reduced in number. Members of the Fgf and Bmp families exhibited altered expression levels in the mutants.
Conclusion
We suggest that Pitx2 is involved in the cardiac outflow tract septation by promoting and/or maintaining the number and the remodeling process of the mesoderm progenitor cells. Pitx2 influences the expression of transcription factors and signaling molecules involved in the differentiation of the cushion mesenchyme during heart development.
Keywords: heart, development, cardiac outflow tract, homeobox, Pitx2
INTRODUCTION
Congenital heart defects are the leading non-infectious cause of death in newborns. Approximately half of all cases are associated with septational malformations in the outflow tract (OT) and/or ventricles (Hoffman and Kaplan, 2002). The mammalian heart develops from cells of four embryonic origins: (1) the cardiac crescent, first lineage or first heart field; (2) the branchial arch (BA)-derived mesoderm, second lineage or secondary heart field; (3) the cardiac neural crest (cNC) cells and (4) the epicardium. The first lineage appears shortly after gastrulation as a population of mesodermal cells that differentiate into endocardial and myocardial types and form a tubular structure (Harvey, 2002). The second lineage arises from a population of splanchnic mesodermal cells that contribute to the formation of OT and right ventricle (RV) (Kelly et al., 2001; Buckingham et al., 2005). The OT is a tubular structure consisting of striated cardiac musculature lined with endocardium. The OT follows a dramatic remodeling to form the aorto-pulmonary (AP) septum that separates the initially single OT vessel to form the ascending aorta (Ao) and the pulmonary trunk (Webb et al., 2003). Between E9.5 and E10.5, endocardial cushions start to form across the common OT, the conotruncus, and the atrioventricular canal. The conotruncal endocardial cushions further divide into proximal and distal conotruncal cushions. The distal conotruncal cushions will form the AP septum at E12.5 – E13.5. The linear heart tube consists of the external layer of myocardium and the internal layer of endocardium, which are separated by the myocardium-produced extracellular matrix, the cardiac jelly. As endocardial cushions develop, endocardial cells proliferate and undergo an epithelial-to-mesenchymal transformation (EMT) and infiltrate the cardiac jelly. As the two cushions come closer, the endocardial cell barrier degenerates and the mesenchymal cells form a bridge to stabilize the fusion of the two cushions (Ray and Niswander, 2012). When tissue alignment, fusion or rotation is disrupted, transposition of the great arteries (TGA) or double-outlet right ventricle (DORV) occurs. Complete failure of OT septation results in persistent truncus arteriosus (PTA).
At the early stages, cNC cells penetrate the second lineage, which is adjacent to the pharyngeal ectoderm, for the formation of the OT. The unspecified mesoderm receives extracellular cues that orchestrate its sequential differentiation into cardiogenic mesoderm, myocardium and smooth muscle. These cues are primarily signaling molecules, such as the bone morphogenetic proteins (BMPs) and the fibroblast growth factors (FGFs). BMP signaling is involved in the induction of the cardiac differentiation. Bmp2 and Bmp4 induce Nkx2.5 and Gata4, which regulate differentiation of cardiac mesoderm into first and second heart lineages (Monzen et al., 1999). BMP signaling promotes specification and differentiation of the second lineage to a cardiac fate by inhibiting FGF signaling (Tirosh-Finkel et al., 2006). Bmp4 is expressed in the splachnic mesoderm, BA mesoderm, and OT myocardium, and is required for OT septation and endocardial cushion remodeling (Liu et al., 2004). FGF signaling is involved in cardiac induction, septation, cell proliferation and OT alignment (Kelly et al., 2001; Ilagan et al., 2006; Park et al., 2008).
Sequence-specific transcription factors (SSTFs) are also involved in guiding proper cardiac cellular proliferation, differentiation and migration. The LIM-homeodomain protein Islet-1 (Isl1) is expressed in the pharyngeal mesoderm and is required for the development of the SHF lineage and its derivatives (Cai et al., 2003). Isl1 marks proliferating, undifferentiated pluripotent cardiovascular progenitors of the second lineage (Cai et al., 2003; Buckingham et al., 2005). Isl1-null mice die at E10 with hearts lacking OT septation. The bHLH protein Mef2c marks a subpopulation of the second lineage (Dodou et al., 2004) and, when mutated, leads to similar defects as Isl1, including defective heart looping and malformed OT (Lin et al., 1997; Buckingham et al., 2005). Tbx1 is expressed in the non-cNC-derived mesoderm of the caudal pharyngeal region, which is part of the second lineage and contributes to the formation of OT and RV (Hu et al., 2004; Xu et al., 2004). Fgf8 and Fgf10 act downstream of Tbx1 in the second lineage (Vitelli et al., 2002; Kelly and Papaioannou, 2007).
Pitx2, a paired-like homeobox SSTF, is transiently expressed on the left side of the cardiac crescent and linear heart tube during early development (E8), and later (E9–E14.5) is expressed in the OT and RV. Genomic screens for inherited atrial fibrillation patients have found deficiencies in the Pitx2 locus (Schnabel, 2011). Pitx2-null embryos are characterized by a non-septated atrium, valvular and OT deficiencies, including PTA, DORV and TGA (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999; Kioussi et al., 2002). Pitx2 controls the specification of cardiac cells within the second lineage by repressing Fgf10 and Isl1 (Galli et al., 2008). Pitx2 and Tbx1 are necessary for proper migration and proliferation of secondary lineage cardiac progenitors (Nowotschin et al., 2006). The observed phenotypes of the knockout mouse models indicated that Pitx2 is critical in regulating the OT formation. Here we report that Pitx2 acts in a network kernel during cardiogenesis and controls the state of the cells of the second lineage as they migrate from the BA to OT and enter the remodeling state to form the valves.
RESULTS
Hypocellular OT Endothelial Cushions in Pitx2 Mutants
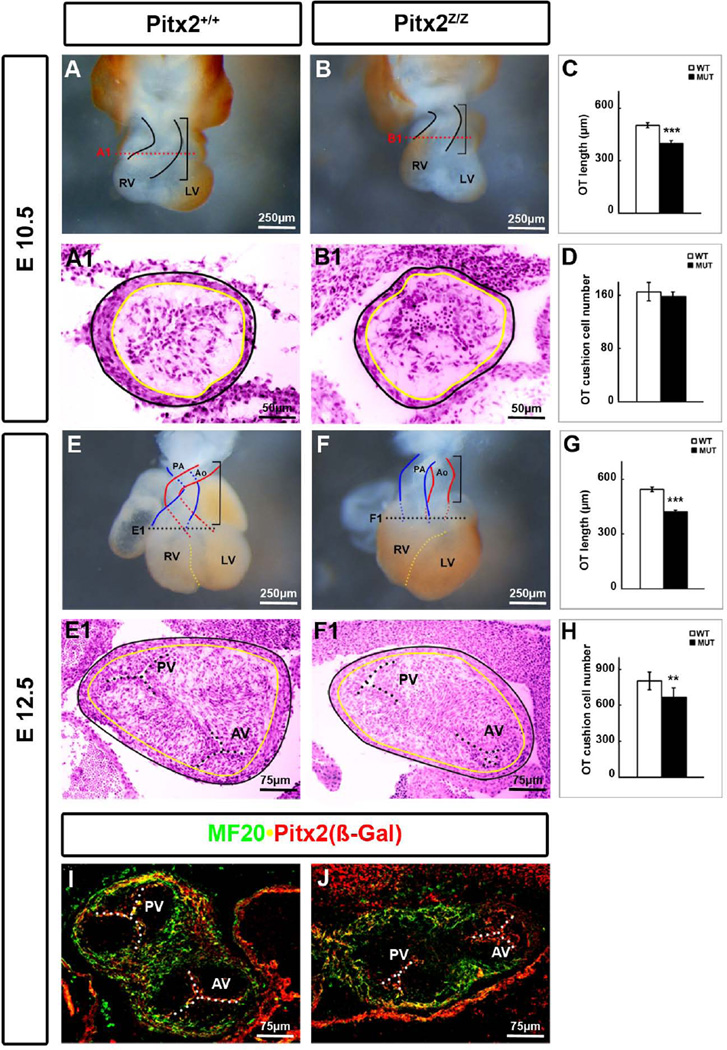
Pitx2-null mice (Pitx2LacZ/LacZ, Pitx2Z/Z) die at E14.5 due to arrest of organ development (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999). The heart is one of the organs that is severely affected, displaying atrial and ventricular septal defects, hypoplastic RV, RA isomerism, DORV, TGA, PTA and abnormal aortic arch remodeling (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999; Campione et al., 2001; Liu et al., 2002). The phenotype of the developing OT was analyzed at E10.5 and E12.5 in mutant and wild type mice (Fig. 1). At E10.5, the heart was already smaller, with delayed looping (Fig. 1A, B). The length of the OT was measured in at least three groups of mice, and a 20% decrease was identified (Fig. 1C), as has been also previously described (Ai et al., 2006). As the heart matures, it undergoes remodeling, and at E12.5 the OT divides and forms two structures, the aorta (Ao) and pulmonary artery (PA) (Fig. 1E). The septation of the OT occurs by (1) the initial division of the aortic sac by the cNC cells, (2) the septation of the distal part of the OT (truncus), and (3) the zipper-like closure of the proximal part (conus) through the fusion of the cushions (Kirby, 2007). By E12.5, septation of the truncus, including the semilunar OT valves, has been completed. The conus closes from distal to proximal towards the ventricles. The ridges beneath the endocardium start to bulge, and when they meet in the middle of the lumen, the endocardium breaks down and a septum is formed (Waldo et al., 1998; Waldo et al., 1999). This conal septum separates the pulmonary and aortic roots. The Pitx2 mutants that exhibited DORV (Fig. 1F) continued to exhibit a shorter OT by 20% (Fig. 1G). Histological analysis on transverse sections at the conus level (Fig. 1E1, F1) indicated a thin and loose epithelium, a non-septated conus with randomly arranged mesenchymal cells within the semilunar valves, in the mutants (Fig. 1F1). Cell-counts of serial transverse sections through the OT of three individual mice for each stage indicated a slow but consistent cell reduction during OT valve formation, starting with no significant reduction at E10.5 (Fig. 1D) followed by a 17.5% reduction at E12.5 (Fig. 1H). Double labeling immunohistochemistry for MF20 and Pitx2(β-Gal) on transverse sections at the level of the pulmonary and aortic roots at E12.5 (Fig. 1I, J) indicated lack of MF20+ cells between the roots in mutants (Fig. 1J). Pitx2 was expressed in the muscularized semilunar pulmonary and aortic valves (Fig. 1I, yellow cells). No MF20+ cells were detected in these cells in mutants (Fig. 1J). These data collectively suggest that Pitx2 is involved in the formation of the OT septum and in the muscularization process of the pulmonary and aortic valves.
Figure 1. Shorter and Hypocellular Cardiac OT in Pitx2 Mutants.
Ventral view of the entire heart at E10.5 (A, B) and E12.5 (E, F) showed a shortened OT with a prominent DORV in Pitx2-mutant mice (F). (C, G) The length of the OT was measured as indicated by brackets. Statistics were based on results from 3 different embryos at each stage. HE staining on 14 µm transverse cryosections at E10.5 (A1, B1) and E 12.5 (E1, F1) mice indicated thinner OT epithelium in the conus. The black and yellow lines correspond to the outer and inner epithelium, respectively. (D, H) Cell counts of a set of 5–8 serial sections along the OT showed reduction of cells in the cushions of mutants. ***: p<0.01, **: p<0.05. (I, J) Double labeling immunohistochemistry on E12.5 mouse transverse sections for MF20 and Pitx2 (β-Gal). No MF20+ cells were detected in the conal septum and semilunar valves in the mutants. Ao, aorta; AV, aortic valve; LV, left ventricle; PA, pulmonary artery; PV, pulmonary valve; RV, right ventricle.
Cell Death and Proliferation Defects of OT Mesenchymal Cells in Pitx2 Mutants
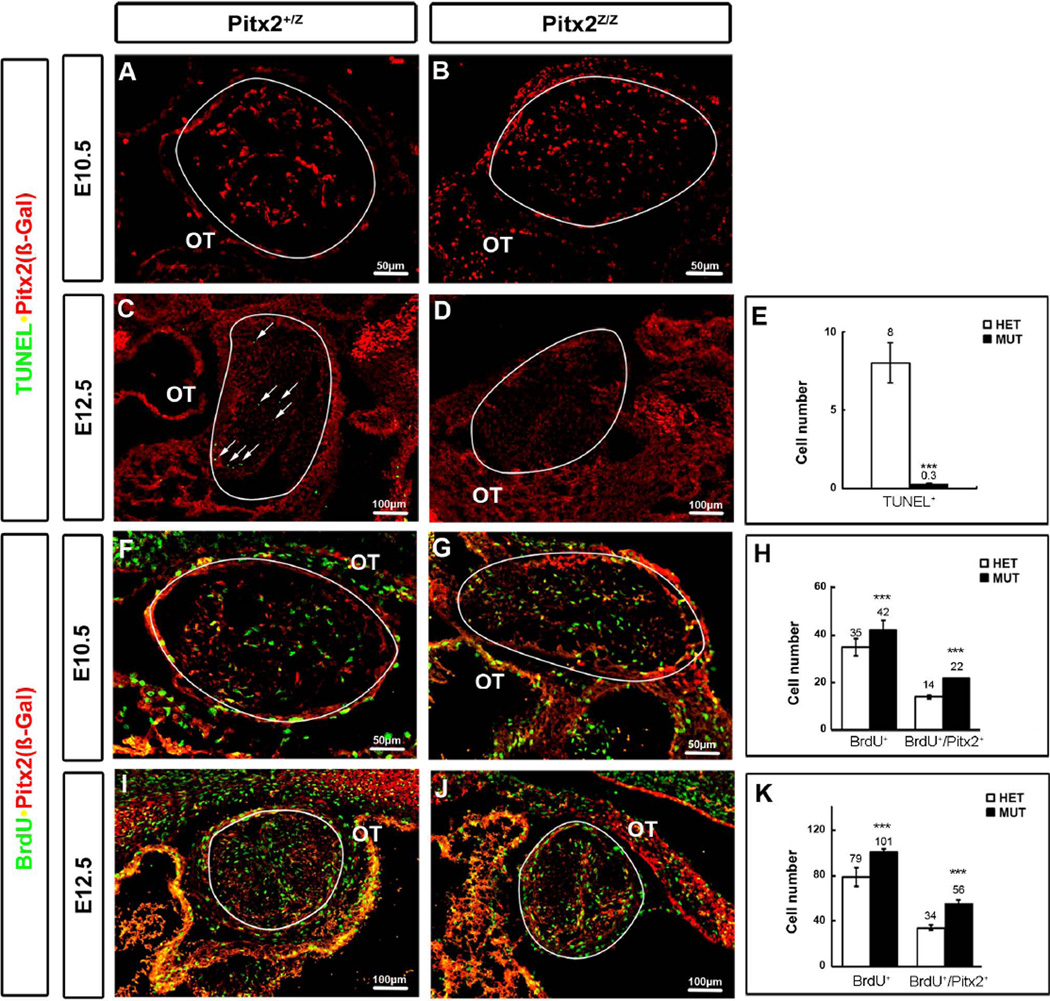
During cardiac remodeling, the OT cushions undergo EMT and become the membranous valves upon activation of cell apoptosis. To determine if Pitx2 is involved in this mechanism, Pitx2-mutant and heterozygote littermates were examined for TUNEL and BrdU incorporation (Fig. 2). No cell death differences were detected at E10.5 (Fig. 2A, B). By E12.5, the OT undergoes remodeling and mesenchymal cells follow programmed cell death (Fig. 2C, arrows). No such cells were detected in the mutants (Fig. 2D). Cell-counts of OT serial sections at E12.5 showed significant decrease of TUNEL+ cells in the mutants (Fig. 2E). To assay for cell proliferation, BrdU+ and BrdU+/Pitx2+(β-Gal+) mesenchymal cells were detected by double labeling immunohistochemistry at E10.5 (Fig. 2F, G) and E12.5 (Fig. 2I, J). Cells were counted in five serial sections from three independent embryos at each stage and genotype. The number of BrdU+ cells was 20% higher in mutants at both stages, while the number of BrdU+/Pitx2+ cells was increased by 57% at E10.5 and 65% at E12.5 (Fig. 2H, K). Collectively these data suggest that Pitx2 maintains the number of mesenchymal cells by inhibiting them from entering the cell cycle and promoting their exit, for further differentiation and/or apoptotic fate.
Figure 2. OT Cushion Mesenchymal Cell Proliferation and Apoptosis Defects in Pitx2 Mutants.
TUNEL assay (A–D) was performed on 14 µm frontal sections to identify the cell apoptosis index during OT remodeling. TUNEL signal was not detected at E10.5 in either OT mesenchyme of the heterozygote or in mutant littermates (A, B). The TUNEL signal was detected in heterozygote OT cushion mesenchyme (C, white arrows) but not in mutant littermate (D) at E12.5. The number of apoptotic cells in the OT cushion was counted based on eight continuous sections for three individual embryos at E12.5 (E). Double labeling of β-gal and Bromodeoxyuridine (BrdU) on 14 µm frontal sections showed Pitx2 effects on cell proliferation (F, G, I, J). The number of proliferative cells was increased in Pitx2 mutants at E10.5 and E12.5, respectively (H, K). The BrdU+ and BrdU+/Pitx2+ cells were counted based on five continuous sections for three individual embryos in each stage. The OT cushion was traced by a white line. Statistics were based on results from 3 different embryos at each stage. ***: p<0.01.
Second Cell Lineage and cNC Cell Defects in Pitx2 Mutants
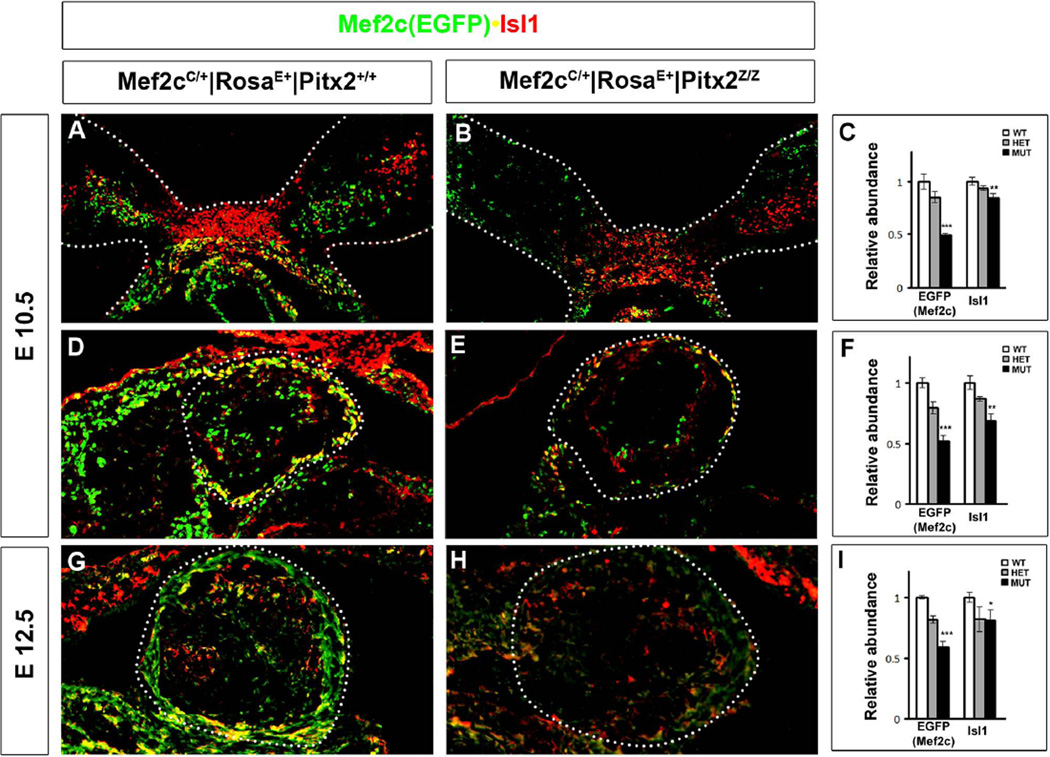
Expression of Pitx2 in the OT cushions is detected as early as E10, and absence of Pitx2 results in a non-septated aorta and pulmonary trunk (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Kioussi et al., 2002). To investigate the cellular events, the Mef2c-lineage tracer mouse was crossed to RosaEGFP to detect the second cell lineage (Verzi et al., 2005). Double labeling immunohistochemistry was used to determine the distribution of EGFP(Mef2c) and Isl1 in BAs (Fig. 3A, B) and OT (Fig. 3D, E) at E10.5. Mef2c is restricted to the second lineage, while Isl1 primarily marks the cardiac progenitor cells (Cai et al., 2003) and, to a lesser extent, the cNC cells (Engleka et al., 2012). EGFP+/Isl1+ cells were detected as they enter the heart tube (Fig. 3A) and were severely reduced in the mutants (Fig. 3B). The EGFP+ cells were also already very much reduced in the BAs (Fig. 3B). The EGFP+/Isl1+ cells located in several layers of the OT epithelium (Fig. 3D) were reduced in mutants (Fig. 3E). The EGFP+ cells that contribute to the formation of the epithelium and leaflets (Fig. 3D) were also reduced in the mutants (Fig. 3E). These cellular defects were more prominent at E12.5 (Fig. 3G, H). Very few EGFP+ and almost no Isl1+ cells were detected in the OT epithelium and leaflets. Quantitative assays indicated almost 50% reduction of EGFP(Mef2c) in BA (Fig. 3C) and the entire heart (Fig. 3F, I) compared to wild type embryos. Isl1 RNA levels were also reduced in the tested biopsies but in less extent (Fig. 3C, F, I). RNA levels for both EGFP and Mef2c were measured and no difference was detected in both BA and OT, suggesting that EGFP levels correspond to the endogenous Mef2c.
Figure 3. Defects of the Second Cardiac Cell Lineage in Pitx2 Mutants.
Double labeling immunohistochemistry of transverse cryosections sections of Mef2cCre/+|RosaEGFP/+|Pitx2+/+ (A, D, G) and Mef2cCre/+|RosaEGFP/+|Pitx2Z/Z (B, E, H) for EGFP (Mef2c) and Isl1 indicated reduction of cell populations in the mutant BA (B) at E10.5 and OT at E10.5 (E) and E12.5 (H). (G, F, I) Quantitative analysis by qPCR for EGFP and Isl1 indicated reduced levels in E10.5 BA (C) and OT (F, I). ***: p<0.01; **: p<0.05; *: p<0.1.
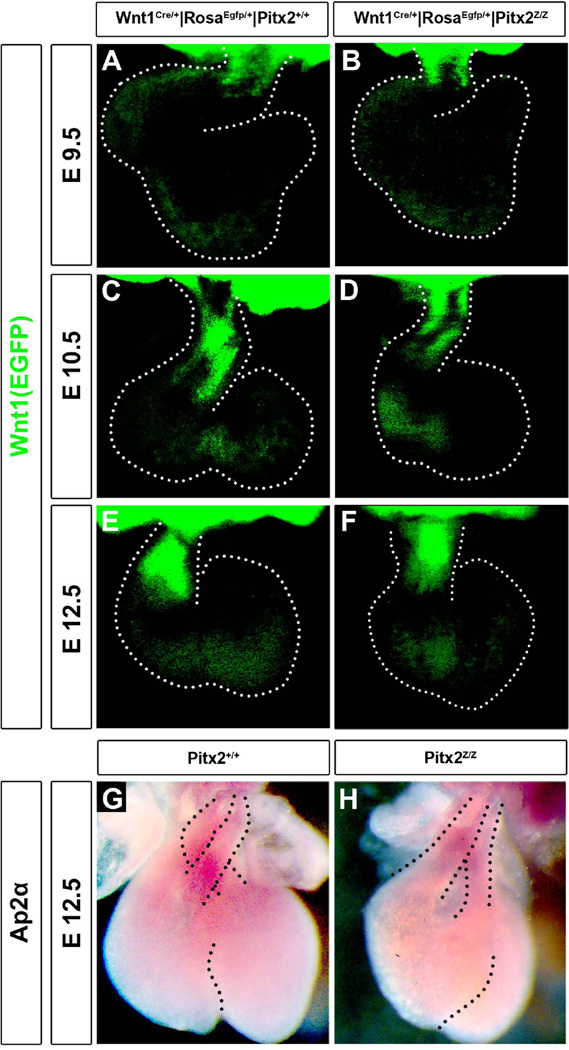
The cNC cells also contribute to the OT endocardial cushions. Postotic NC cells contribute to OT endocardial cushions (Kirby, 2007); preotic NC cells distribute to the contruncus and coronary artery formation (Arima et al., 2012). To test the cNC distribution in the OT of the Pitx2 mutants, the Wnt1Cre|RosaEGFP reporter mouse was crossed to the Pitx2LacZ line. Although the contribution of the Wnt1+ cells to the OT under the Pitx2 influence was previously reported, (Ai et al., 2006), we performed the analysis at earlier developmental stages. At E9.5 the OT is already shorter in the Pitx2 mutants, (Fig. 4B) and the Wnt1+ cells just populated the OT; while in the control wild type mice, they start to fuse and widely populate the area (Fig. 4A). At E10.5, a thick stream of Wnt1+ cells was located in the OT (Fig. 4C); while in the mutants, this population seems restricted to the truncated truncus (Fig. 4D). At E12.5 this delay of the Wnt1+ cells to populate the truncus and conus seems to be recovered in the mutants (Fig. 4E, F). However, the expression levels of another cNC marker, Ap2α, were reduced in the OT in mutants (Fig. 4G, H) at E12.5. These data suggest that Pitx2 influences the distribution of the BA mesoderm derived and cNC cells during OT and endocardial cushion development.
Figure 4. Impaired cNC cells in Pitx2 mutants.
Wnt1Cre/+|RosaEGFP|Pitx2+/+ (A, C, E) and Wnt1Cre/+|RosaEGFP|Pitx2Z/Z (B, D, F) hearts were dissected at E 9.5 (A, B), E 10.5 (C, D) and E 12.5 (E, F). The green fluorescent cNC cells that migrated towards the OT were reduced in mutants, with more prominent phenotype at E9.5 and E10.5. (G, H) Whole mount RNA in situ hybridization at E12.5 hearts for Ap2α expression indicated reduced levels in the great arteries in mutants.
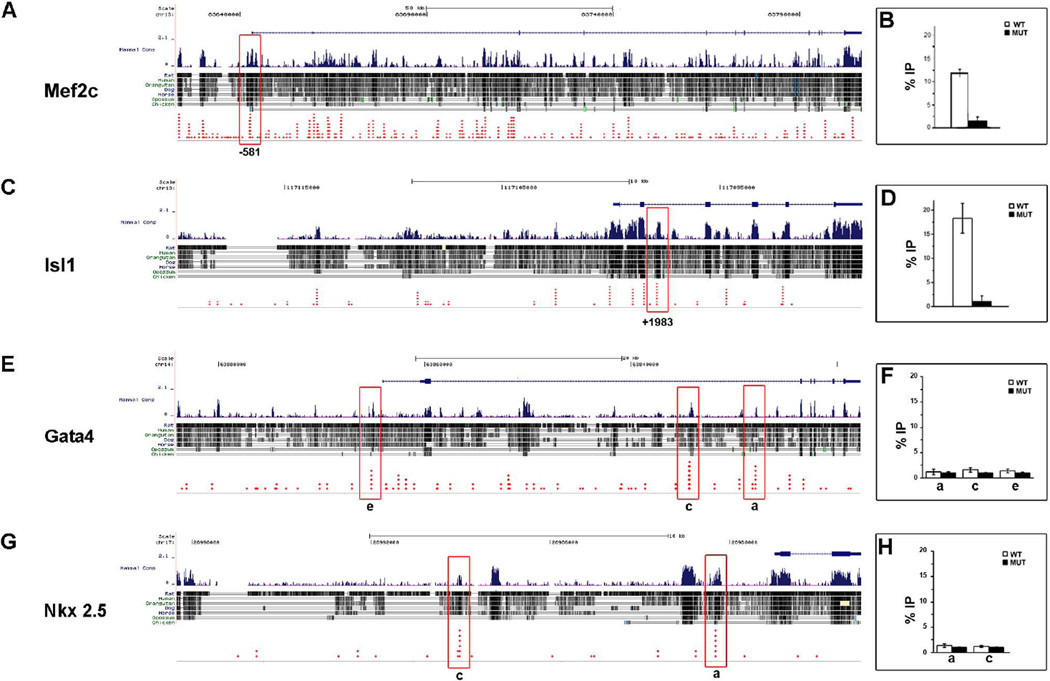
Pitx2 Occupancy at SSTF Loci in BAs and Heart
Our data have shown that Pitx2 regulates the expression of both Mef2c and Isl1 in the BAs and OT, indicating this regulation might be on a transcriptional level. Chromatin-occupancy analyses provided another means to assess the interaction between Pitx2 and the SSTF that were altered in E10.5 Pitx2 mutants. Embryos from approximately 2–3 synchronous litters were rapidly genotyped. Tissue biopsies, including BA (mandibular part of 1st, 2nd and 3rd BA) and heart (OT, atria and ventricles), were dissected from 6–8 embryos of each genotype and pooled for two independent experiments. From each experiment, a pair of wild type and mutant chromatin extract was generated with sufficient material for one immunoprecipitation, using an anti-Pitx2 antibody. Each immunoprecipitate provided enough material for 15 triplicate qPCR analyses. Amplicons within the Mef2c, Isl1, Gat4 and Nkx2.5 loci were identified, as described previously for T-box (Hilton et al., 2010) and Hox (Eng et al., 2012) genes. Core motifs for bicoid class homeodomains (TAATCY) that were embedded in evolutionarily conserved non-coding regions and were, themselves, evolutionarily conserved, were identified (Fig. 5). Each red diamond represents a different species in which the core motif was conserved and expected to be essential for biological function. Core motifs with the most diamonds were selected as candidate cis regulatory modules, and primer pairs were designed to encompass a 70–150 bp context around these sites. The initially selected primer pairs were tested by endpoint PCR on purified genomic DNA. Amplified pairs were selected for SSTF chromatin occupancy analyses by ChIP-qPCR (Table 1). The mutant extract lacks Pitx2 protein and is, therefore, expected to have 0% occupancy. The signal measured in the mutant precipitate, for any given amplicon, is a direct measurement of the background. Pitx2 occupied Mef2c (Fig. 5A, B) and Isl1 (Fig. 5C, D) in the BA biopsies at positions −521 and +1983, respectively. No Pitx2 occupancy was detected on Gata4 (Fig. 5F) at the positions −1065, +29818 and +36199, despite being evolutionarily conserved (Fig. 5E). Pitx2 occupancy was also not detected on Nkx2.5 (Fig. 5H) at the positions −10523 and −1952 (Fig. 5G). Pitx2 occupancy on Tbx1 BA biopsies has previously been reported in E10.5 mice (Shih et al., 2007a). The Pitx2 occupancy on the SSTF Mef2c and Isl1 correlates well with their altered expression profiles in the developing BA. It has been shown that Pitx2 regulates Gata4 expression (Lozano-Velasco et al., 2011), and Nkx2.5 has synergistic activity with Pitx2 (Ganga et al., 2003). However, this might not be due to Pitx2 occupancy at this developmental stage.
Figure 5. Pitx2 Occupancy on SSTF Gene Loci in BA and Heart Chromatin.
Sonically sheared chromatin, isolated from E10.5 mice, was used to detect Pitx2 protein occupancy on Mef2c (A) and Isl1 (B) in BA biopsies and Gata4 (E) and Nkx2.5 (G) in heart biopsies. PCR amplicons of 70–150bp (red boxes) were designed around highly evolutionarily conserved bicoid core motif TAATCY. Each red diamond indicated a single vertebrate species, containing the biocoid core motif. Bar graphs show the average relative amount of signal precipitated from wild type (white bar) and mutant (black bar). Pitx2 was found to occupy the Mef2c and Isl1 gene on the conserved −521 (B) and +1983 (D) sites in E10.5 embryonic BA biopsies, respectively. No significant difference was measured for Pitx2 occupancy in conserved regions of Gata4 (F) and Nkx2.5 (H) in E10.5 heart biopsies.
Table 1.
qPCR primer sets
| Primer | Forward | Reverse |
|---|---|---|
| qPCR | ||
| EGFP | ACGTAAACGGCCACAAGTTC | AAGTCGTGCTGCTTCATGTG |
| Isl1 | ATGATGGTGGTTTACAGGCTAAC | TCGATGCTACTTCACTGCCAG |
| Fgf8 | CCGAGGAGGGATCTAAGGAAC | CTTCCAAAAGTATCGGTCTCCAC |
| Fgf3 | TGCGCTACCAAGTACCACC | CACTTCCACCGCAGTAATCTC |
| Notch2 | ATGTGGACGAGTGTCTGTTGC | GGAAGCATAGGCACAGTCATC |
| Bmp4 | TTCCTGGTAACCGAATGCTGA | CCTGAATCTCGGCGACTTTTT |
| PECAM | ACGCTGGTGCTCTATGCAAG | TCAGTTGCTGCCCATTCATCA |
| NF | ACAGCTCGGCTATGCTCAG | CGGGACAGTTTGTAGTCGCC |
| GAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| ChIP-qPCR | ||
| Isl1 | TTGGGGTGACTCTTCCTTTG | GATGGGCAATTTGATCTGCT |
| Mef2c | CCATGACCATCCAGTTTTGA | GCACACACTTGCTTCATTTCA |
| Nkx2.5a | GGGCGAGGGTCCTGGGAGTC | CGGCCCCCAATATAGCTCCCC |
| Nkx2.5c | ACTGACACACACTGCAGGGGC | GTGGGTGGTCCTCTCTCAGCAGT |
| Gata4a | TGTCCAACAATGGCTGTGGAGTGC | TCCCTAGTTCCTCTGTCCCTTGCC |
| Gata4c | AAGCCCCCATCCCCTGCACTT | ACTGGACAGAACCTTGCCTGCTCA |
| Gata4e | TTCTCTCCCCGGCACCGGTTT | GTCCTCGAACTGCGGGAGCC |
Pitx2 and FGF/BMP Signaling
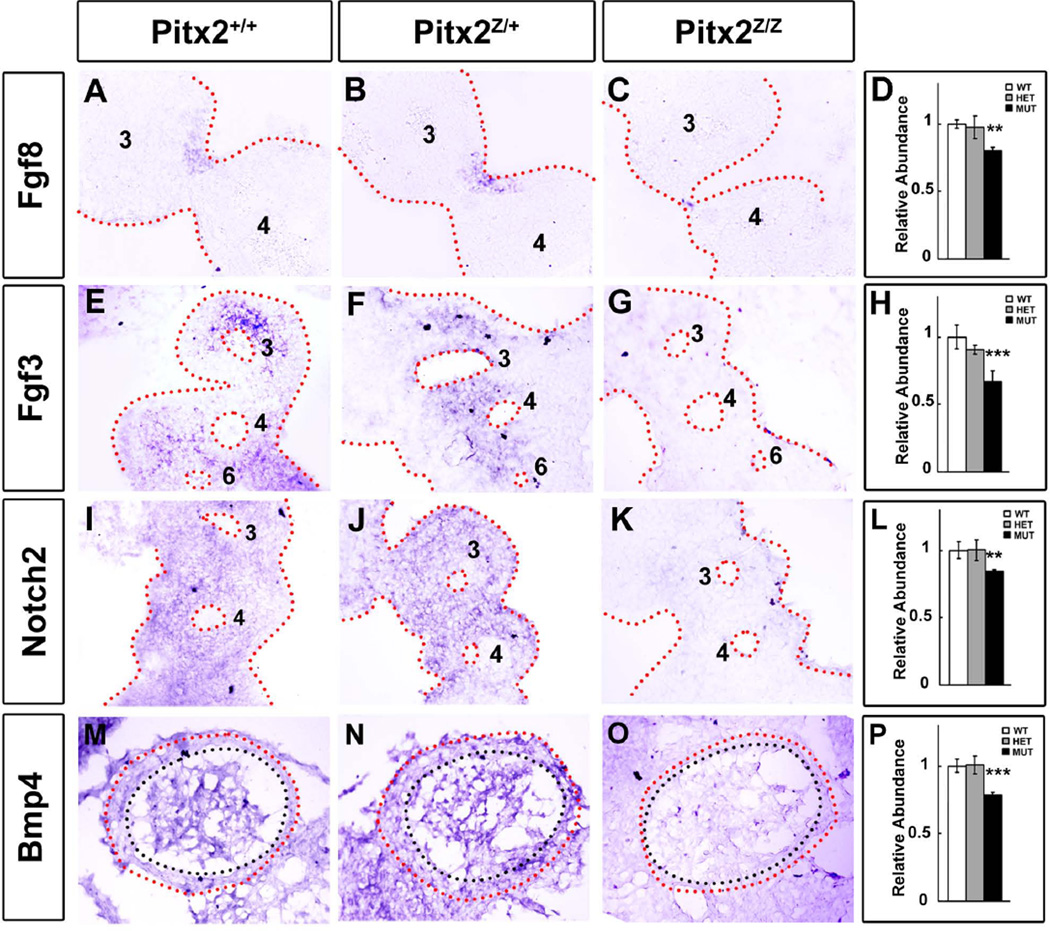
FGF signaling in the second cardiac lineage is essential for OT cushion formation and remodeling. Fgf8 expression in the ectoderm of the 1st and 2nd BA regulates proliferation and differentiation of post-migratory cNC cells. FGF signaling in the OT myocardium controls extracellular matrix formation; while BMP signaling is essential for endothelial cell transformation and invasion of cNC cells (Park et al., 2008). RNA in situ hybridization at E10.5 was used to determine the expression profile of Fgf8, Fgf3, Bmp4 and Notch (Fig. 6). Fgf8 is detected in the ectoderm of the 2nd, 3rd and 4th BA in wild type and heterozygote embryos (Fig. 6A, B) and was barely detectable in the 4th BA in mutants (Fig. 6C), as previously described (Liu et al., 2003). Similarly, the area of Fgf3 expression in the 3rd – 6th BA (Fig. 6E, F) was reduced in mutants (Fig. 6G). The Notch signaling has been implicated in regulating EMT during valve development. Notch2 is expressed in the cNC-derived vascular smooth muscle cells and is critical in mammalian OT development (Niessen and Karsan, 2008). Notch2 was expressed in the 3rd and 4th aortic arch (Fig. 6I, J), with significantly reduced expression levels in the mutants (Fig. 6K). Bmp4 is expressed in the ventral splanchnic mesoderm, BA mesoderm, and OT myocardium. Bmp4 promotes proliferation of cushion mesenchyme and concurrently represses cell proliferation in the OT myocardium (Liu et al., 2004). The Bmp4-distinct area of mesenchymal expression in the OT (Fig. 6M, N) was not detected in mutants (Fig. 6O). The thinner OT epithelium was prominent in all mutants (Fig. 5M, N, O, red and black dotted line). Quantitative PCR analysis further confirmed the lower expression levels of Fgf8 (Fig. 6D), Fgf3 (Fig. 6H), Notch2 (Fig. 6L) and Bmp4 (Fig. 6P) in the mutant BA and heart (only for Bmp4) biopsies at E10.5. No significant difference between wild type and heterozygote was detected. Collectively, these results suggest that the combination of the altered FGF, BMP and Notch signaling results in the hypoproliferative mesenchymal cells in the OT cushions and their delayed EMT.
Figure 6. Alteration of FGF, BMP and Notch Signaling in Pitx2 Mutants.
RNA in situ hybridization on cryostat sections revealed altered Fgf8 (A–C), Fgf3 (E–G), Notch2 (I–K) and Bmp4 (M–O) expression in E10.5 Pitx2 control (wild type and heterozygote) and mutant mice. The Fgf8 expression in 3rd and 4th BA ectoderm was decreased in mutants compared to control embryos (C). Fgf3 expression levels were reduced in the 2nd – 6th BA mutants (G). Notch2 expression levels, detected in the 3rd and 4th BA in wild type (I) and heterozygote (J), were barely detectable in 3rd and 4th BA in mutants (K). No Bmp4 expression was detected in the OT mesenchymal cushions (area inside the black dotted line) in mutants (O) compare to the control (M, N). Thinner OT wall (between red and black dotted line) was consistently observed in Pitx2 mutants (O). Quantitative PCR assay indicated the significantly decreased mRNA expression levels of Fgf8 (D), Fgf3 (H), Notch2 (L) and Bmp4 (P) at E10.5 BA (D, H, L) and heart (P) biopsies, respectively.
Cardiac Vascular and Innervation Defects in Pitx2 Mutants
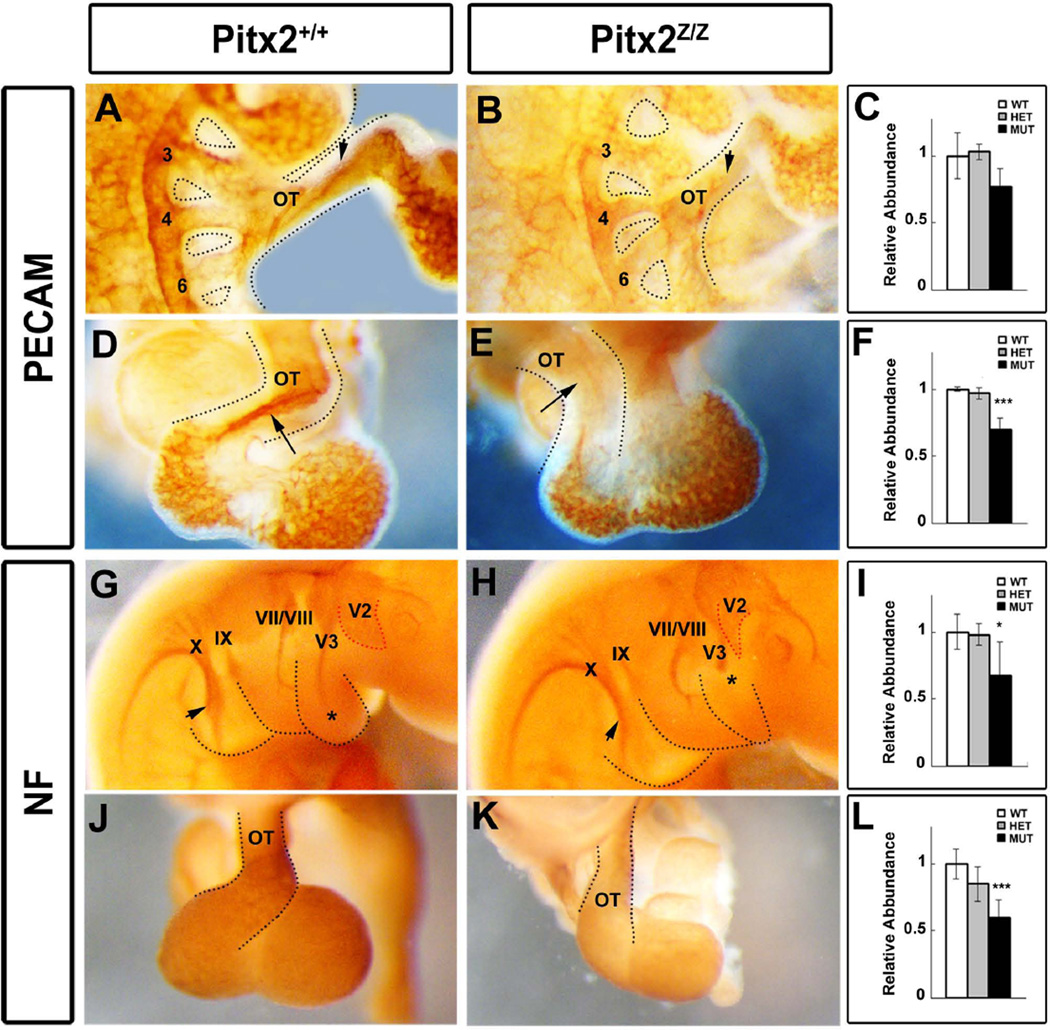
Unilateral ablation of Pitx2 results in asymmetric remodeling of the BA system, which leads to randomized laterality of the aortic arch (Yashiro et al., 2007). We investigated the aortic arch system defects, found in the Pitx2 mutants at the cellular level, by whole mount antibody staining with platelet-endothelial cell adhesion molecule (PECAM) at E10.5 (Fig. 7A, B, D, E). Endothelial cells were detected in the well-developed 3rd and 4th BAs and, to a lesser extent, in the 6th BA (Fig. 7A). An increased number of endothelial cells were extending into the OT and RV in wild type mice (Fig. 7A, D). In contrast, endothelial cells were severely reduced in mutant BAs (Fig. 7B) and OT (Fig. 7E). A similar phenotype was also observed at E11.5 (data not shown). Quantitative PCR further confirmed the reduced PECAM level of expression in mutant BA (Fig. 7C) and heart biopsies (Fig. 7F). The BA innervation process was also affected in Pitx2-null mutant mice as detected by whole mount neurofilament (NF) antibody staining (Fig. 7G, H, J, K). The maxillary (V2) (Fig. 7G, H, red dotted line) and mandibular (V3) (Fig. 7G, H, star) branch of the trigeminal nerve (V) that innervates mastication muscles was not prominent in the mutants. Pitx2 mutants do not form mastication muscles (Shih et al., 2007a); and, thus, V2 and V3 were unable to migrate to their final destinations, as the supportive tissue was missing. The facial nerve (VII) that innervates the facial muscles and receives the sense from the anterior tongue was distorted. It failed to reach the edge of the 2nd BA (Fig. 7G, H). The sensory acoustic nerve (VIII) that migrates parallel to VII had a similar phenotype (Fig. 7G, H). The glossopharyngeal nerve (IX) provides special innervation to the stylopharyngeus. The vagus nerve (X) provides brachiomotor innervation to the majority of laryngeal and pharyngeal muscles and has three nuclei associated with the cardiovascular control the dorsal motor nucleus, the nucleus ambiguous, and the solitary nucleus. The afferent fibers of the autonomic nervous system transmit signals to the medulla by cranial nerves X and IX. Both IX and X nerves, located in the jugular area, were thinner, shorter, and not properly aligned in mutants, possibly as a result of the severe distortion or absence of several facial muscles (Fig. 7G, H). The X nerve innervates the OT to sense the aortic blood pressure and to slow the heart rate (Fig. 7G, arrow). NF expression was detected in the wild type (Fig. 7J) heart but in much lower levels than in mutants (Fig. 7K). This signal reduction may explain the observed arrhythmias and conduction deficiencies in Pitx2-mutant mice and human patients (Schnabel, 2011). Quantitative analyses for NF (Fig. 7I, L) were also performed in BAs (Fig. 7I) and heart (Fig. 7L) biopsies, indicating significant reduction of NF in mutants.
Figure 7. Vascular and Nervous System Defects in Pitx2 Mutants.
Whole mount antibody staining with PECAM (A, B, D, E) and NF (G, H, J, K) was performed on E10.5 Pitx2 wild type and mutant mice. PECAM expression is reduced in BA (3, 4 and 6) and OT (A, B, arrowhead). The dorsal view of the OT (D, E) indicated the septational vasculature (arrow) was disrupted in Pitx2-null mice. (C, F) Quantitative qPCR for PECAM mRNA levels in Pitx2 wild type, heterozygote and mutant in BAs (C) and heart (F) biopsies. The expression levels of PECAM in heart were significantly decreased in E10.5 mutants. Innervation of BAs and OT was detected by NF antibody in both control and mutant embryos (G, H, J, K). V2 (red dotted line) and V3 (stars) and X (arrow head) were shorter and thinner in mutants (H). NF signals were reduced in mutant hearts (K). Quantitative qPCR assay indicated significantly decreased levels in mutants in BA (I) and OT (L) biopsies, with no difference between wild type and heterozygote at this stage. V2: maxillary nerve; V3: Mandibular nerve; VII/VIII: Facial nerve; IX: Glossopharyngeal nerve; X: Vagus nerve.
DISCUSSION
Homeobox genes are key players for cell specification and organ formation as members of network kernels at early developmental stages. Pitx2 specifies tooth, pituitary, (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999; Shih et al., 2007b), facial (ocular and mastication) (Gage et al., 2005; Shih et al., 2007a) and abdominal (Hilton et al., 2010) muscle development, while regulating the developmental process of organs including heart, intestine and lung (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999; Shih et al., 2007b). The human Axenfeld-Rieger syndrome associated with mutations in PITX2 locus 4q25 is characterized by umbilical hernia, glaucoma, myopathies and cardiac arrhythmias (Perveen et al., 2000; Schnabel, 2011). The close correlation of mouse phenotypes to the human syndrome demonstrates the evolutionarily conserved functions of Pitx2. Pitx2 loss of function results in severe cardiovascular defects, including atrial isomerism, DORV, TGA, PTA and abnormal aortic arch remodeling (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999; Campione et al., 2001; Liu et al., 2002). Genetic studies have shown that Pitx2-mediated signaling during cardiogenesis is conducted within BA mesoderm cells (Ai et al., 2006), pharyngeal arch mesenchyme (Franco and Campione, 2003) and cNC cells (Hamblet et al., 2002; Kioussi et al., 2002). Our data show that Pitx2 is a member of a network kernel, including Mef2c, Isl1, Tbx1, Gata4 and Nkx2.5, that synergistically regulates endocardial cushion development and separation of the great arteries. We have demonstrated that Pitx2 occupies cis regulatory elements of Mef2c, Isl1 and Tbx1 (Harel I, 2012) in BA mesoderm.
Conotruncal defects, including TGA, DORV, tetralogy of Fallot, and PTA, result in abnormal OT development, including hypocellular cushions, altered conotruncal rotation, and misalignment of septal components. Endocardial cushion formation starts with a swelling of the OT region at E9.5 and their formation is induced by signals from the myocardium that inhibit expression of chamber-specific genes and the active expression of extracellular matrix genes. BMPs are the major myocardial signals that initiate endocardial cushion formation and remodeling by promoting EMT (Lyons et al., 1990; Ma et al., 2005). Bmp4 is required for endocardial cushion expansion and OT septation (McCulley et al., 2008). Loss of Bmp4 in the second cardiac lineage results in a limited number of cells in the developing OT cushions and a defective remodeling process, a very similar phenotype to the one observed in the Pitx2-mutant mice. BMP signaling in the endocardium and cNC cells is vital to OT septation and formation of the aorta and pulmonary arteries.
TGFβs are among the early signaling molecules implicated in endocardial cushion development. TGFβ ligands and receptors are expressed in the OT during cushion formation and EMT (Brown et al., 1996). TGFβ signaling acts through SMADs to induce expression of the SSTF Slug that, in turn, promotes endocardial cushion formation of the antrioventricular canal (AVC) via EMT mechanisms (Romano and Runyan, 2000). Wnt/β-catenin regulates cardiac valve formation (Hurlstone et al., 2003) and, together with TGFβ, regulates cushion EMT. Notch signaling induces the expression of the pro-migratory SSTF Snail in AVC and OT endocardial cushion endothelial cells undergoing EMT. Snail inhibits VE-cadherin activity, and mesenchymal cells break contact with their neighboring cells. Notch signaling is also required for TGFβ2 and several TGFβ receptors in AVC and OT to further support endocardial EMT.
Fgf10, a target of the Wnt-β-catenin pathway in the cardiac mesoderm, is expressed in the second cardiac lineage (Kelly et al., 2001). However, Fgf10-null mice do not exhibit apparent cardiac defects. Fgf8 is also expressed in the second cardiac lineage and, when mutated, results to DORV and PTA (Abu-Issa et al., 2002; Frank et al., 2002). Fgf8 is essential for mesoderm-derived cell proliferation and survival during OT elongation. Reduced expression of Fgf8 in mesoderm- and ectoderm-derived cells, resulting in apoptotic cNC cell death in the developing pharyngeal arches (Ilagan et al., 2006).
Cardiac NC cells migrate into the OT endocardial cushions and contribute to the formation of aortic and pulmonary valves. Ablation of cNC cells results in OF defects, including shortening in length, delayed rotation and caudal displacement, dextroposed aorta (DORV), PTA, and interruption of the aortic arch. The shorter OT is also characterized by decreased second cardiac lineage cell migration (Waldo et al., 2005). This reciprocal interaction between the two cell lineages is essential for the OT septation.
Pitx2 acts upstream of the Wnt11/TGFβ2 signaling pathway that regulates extracellular matrix composition, cytoskeletal rearrangements and polarized cell movement required for tissue morphogenesis (Zhou et al., 2007). BMP and Notch expression was Pitx2-dependet in the OT of the linear heart tube in areas where cushions will be formed (Fig. 6I–K, M–O). This further supports the involvement of Pitx2 in a multi-signaling network during cushion formation and EMT induction and maintenance. Pitx2-mutant mice also exhibit remodeling malformations of intraventricular septum and ventricular myocardium (Tessari et al., 2008). The formation of AVC and the ventricular septation is another type of cell fusion that requires EMT.
Thus, we conclude that Pitx2 regulates the maintenance and epithelial-mesenchymal transitions of the BA mesoderm cells as they enter the linear heart tube to form the OT endocardial cushions. Pitx2 promotes the healthy interaction of the mesoderm-derived and cNC cells for proper OT septation by acting as a node of a sophisticated network kernel.
EXPERIMENTAL PROCEDURES
Mice
ICR Pitx2+/LacZ (Pitx2+/Z) mice (Lin et al., 1999) were bred and females were checked for the presence of a vaginal plug (E0). Embryos were isolated at different developmental stages and the yolk sacs were used for genotyping. Mef2cCre mice (Verzi et al., 2005) and Wnt1Cre mice (Jackson Lab) (Echelard et al., 1994) were crossed with Rosa26EGFP(RosaEGFP) (Jackson Lab) (Mao et al., 2001) mice to obtain Mef2cCre|RosaEGFP and Wnt1Cre|RosaEGFP double heterozygotes, respectively. Pitx2+/Z mice were crossed with Mef2cCre|RosaEGFP or Wnt1Cre|RosaEGFP to generate green Pitx2 wild type and mutant mice. PCR analysis from tail genomic DNA identified the 380 bp EGFP and 400 bp Cre bands.
Immunohistochemistry, TUNEL and BrdU
Immunohistochemistry on cryosections was performed as described by (Shih, 2007). Sections were photographed on an AxioImager Z1, Zeiss microscope. TUNEL assay was also performed as recommended by the manufacturer (Dead End kit; Promega). Pregnant female Pitx2+/Z mice were injected with 5 mg/ml BrdU 2 hr before dissection. Embryos from injected mice were processed, as previously described (Shih et al., 2007a). Immunohistochemistry of whole-mount embryos was performed according to standard protocol (Joyner and Wall, 2008). Whole embryos were photographed with a discovery V8, Zeiss microscope. Primary antibodies are listed as follows: MF20 (Mouse, 1:50, DHSB), β-galactosidase (Rabbit, 1:1000, Cappel), BrdU (Rat, 1:100, Accurate Chemical Scientific Corporation), EGFP [Rat, 1:1500, (Shih et al., 2007b)], Isl1 (Mouse, 1:30, DHSB), PECAM (Rat, 1:10, BD Biosciences), Neurofilament 200 (Rabbit, 1:100, Sigma).
Quantitative Real-time PCR(qPCR)
cDNA from BA (n=5) and heart (n=5) were prepared by RNeasy Micro Kit (Qiagen). cDNA (25ng) was analyzed by qPCR using SYBR Green I methodology as previously described (Hilton et al., 2010). All samples were analyzed in triplicate and normalized by glyceraldehyde-3-phosphate dehydrogenase. All qPCR primer sets are listed in Table 1.
Pitx2-Binding Site Analysis
An in house Perl script, binding_site_compare.pl was used for identifying the absolute location and evolutionary conservation of potential Pitx2-binding sites TAATCY (Amendt et al., 1998; Eng et al., 2010; Campbell et al., 2012; Eng and Dubovoy, 2012). The alignments for each gene, along with the 20kb region upstream of the gene, were download from the UCSC Genome Browser on Mouse July 2007 (NCBI37/mm9) Assembly, available at http://genome.ucsc.edu/. The alignments were then formatted for our script, which identified the absolute location of potential Pitx2 binding sites and the species conserved for each binding site. Excel was used to map binding site locations and species to each gene.
Chromatin Immuno-Precipitation (ChIP)
Heart and BA biopsies from 6–8 embryos of E10.5 Pitx2 wild type and Pitx2Z/Z mice were harvested per ChIP. Samples were collected and processed as previously described (Hilton et al., 2010). Primers were designed for binding sites identified as conserved sites. Control primers were designed for regions on the genome with no putative binding site within a minimum of a 1kb window on the mouse genome. All primer sets are listed in Table 1.
RNA in situ Hybridization
Whole-mount RNA in situ hybridization was performed according to standard procedures (Oliver et al., 1995). RNA in situ hybridization on sections was performed on 16 µm cryosections, as previously described (Kyrylkova et al., 2012). Digoxigenin-labeled antisense RNA in situ probes were generated by an in vitro transcription kit (Dig RNA labeling kit, Roche Molecular Biochemicals). AP-conjugated anti-DIG antibody (1:500) was used to detect the hybridization signals (Roche Molecular Biochemicals).
Pitx2 controls cellularity of conotruncus.
Pitx2 regulates inductive signaling for endocardial cushion formation.
Pitx2 is a node of the network kernel for OT development.
ACKNOWLEDGMENTS
The authors thank Brian Black, UCSF for providing the Mef2cCre mice and R. Kelly, W. Pu, P. Sepúlveda, and M. Goulding for providing RNA in situ probes. This research was supported by the Oregon State University College of Pharmacy, the American Heart Association, the March of Dimes and the NIH-NIAMS grant awards to CK. We thank Denny Weber for help editing the manuscript.
AHA, 0550179Z, MOD FY05-120, NIH-NIAMS AR054406
REFERENCES
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, Black BL, Brown NA, Martin JF. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J Biol Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- Arima Y, Miyagawa-Tomita S, Maeda K, Asai R, Seya D, Minoux M, Rijli FM, Nishiyama K, Kim KS, Uchijima Y, Ogawa H, Kurihara Y, Kurihara H. Preotic neural crest cells contribute to coronary artery smooth muscle involving endothelin signalling. Nat Commun. 2012;3:1267. doi: 10.1038/ncomms2258. [DOI] [PubMed] [Google Scholar]
- Brown CB, Boyer AS, Runyan RB, Barnett JV. Antibodies to the Type II TGFbeta receptor block cell activation and migration during atrioventricular cushion transformation in the heart. Dev Biol. 1996;174:248–257. doi: 10.1006/dbio.1996.0070. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AL, Eng D, Gross MK, Kioussi C. Prediction of gene network models in limb muscle precursors. Gene. 2012;509:16–23. doi: 10.1016/j.gene.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Developmental biology. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Eng D, Campbell A, Hilton T, Leid M, Gross MK, Kioussi C. Prediction of regulatory networks in mouse abdominal wall. Gene. 2010;469:1–8. doi: 10.1016/j.gene.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng D, Dubovoy A. High Left Ventricular Vent Return After Left and Right Ventricular Assist Device Placement in a Patient With a Mechanical Aortic Valve. J Cardiothorac Vasc Anesth. 2012 doi: 10.1053/j.jvca.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Eng D, Ma HY, Xu J, Shih HP, Gross MK, Kiouss C. Loss of abdominal muscle in Pitx2 mutants associated with altered axial specification of lateral plate mesoderm. PLoS One. 2012;7:e42228. doi: 10.1371/journal.pone.0042228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka KA, Manderfield LJ, Brust RD, Li L, Cohen A, Dymecki SM, Epstein JA. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res. 2012;110:922–926. doi: 10.1161/CIRCRESAHA.112.266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco D, Campione M. The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc Med. 2003;13:157–163. doi: 10.1016/s1050-1738(03)00039-2. [DOI] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Rhoades W, Prucka SK, Hjalt T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest Ophthalmol Vis Sci. 2005;46:4200–4208. doi: 10.1167/iovs.05-0691. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, Buckingham ME. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development. 2008;135:1157–1167. doi: 10.1242/dev.014563. [DOI] [PubMed] [Google Scholar]
- Ganga M, Espinoza HM, Cox CJ, Morton L, Hjalt TA, Lee Y, Amendt BA. PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J Biol Chem. 2003;278:22437–22445. doi: 10.1074/jbc.M210163200. [DOI] [PubMed] [Google Scholar]
- Hamblet NS, Lijam N, Ruiz-Lozano P, Wang J, Yang Y, Luo Z, Mei L, Chien KR, Sussman DJ, Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- Harel I MY, Avraham R, Rinon A, Ma HY, Cross JW, Leviatan N, Hegesh JT, Roy A, Jacob-Hirsch J, Rechavi G, Carvajal J, Tole S, Kioussi C, Quaggin S, Tzahor E. Proceedings of the National Academy of Sciences. PNAS; 2012. Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- Hilton T, Gross MK, Kioussi C. Pitx2-dependent occupancy by histone deacetylases is associated with T-box gene regulation in mammalian abdominal tissue. J Biol Chem. 2010;285:11129–11142. doi: 10.1074/jbc.M109.087429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131:5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, Clevers H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- Ilagan R, Abu-Issa R, Brown D, Yang YP, Jiao K, Schwartz RJ, Klingensmith J, Meyers EN. Fgf8 is required for anterior heart field development. Development. 2006;133:2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- Joyner A, Wall N. Immunohistochemistry of whole-mount mouse embryos. CSH Protoc. 2008 doi: 10.1101/pdb.prot4820. 2008:pdb prot4820. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Papaioannou VE. Visualization of outflow tract development in the absence of Tbx1 using an FgF10 enhancer trap transgene. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:821–828. doi: 10.1002/dvdy.21063. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Kirby M. Cardiac Development. New York: Oxford University Press; 2007. [Google Scholar]
- Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- Kyrylkova K, Kyryachenko S, Kioussi C, Leid M. Determination of gene expression patterns by in situ hybridization in sections. Methods Mol Biol. 2012;887:23–31. doi: 10.1007/978-1-61779-860-3_3. [DOI] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Lu MF, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Wang D, Lu MF, Moses KA, Schwartz RJ, Martin JF. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4489–4494. doi: 10.1073/pnas.0308466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Velasco E, Chinchilla A, Martinez-Fernandez S, Hernandez-Torres F, Navarro F, Lyons GE, Franco D, Aranega AE. Pitx2c modulates cardiac-specific transcription factors networks in differentiating cardiomyocytes from murine embryonic stem cells. Cells Tissues Organs. 2011;194:349–362. doi: 10.1159/000323533. [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Lyons KM, Pelton RW, Hogan BL. Organogenesis and pattern formation in the mouse: RNA distribution patterns suggest a role for bone morphogenetic protein-2A (BMP-2A) Development. 1990;109:833–844. doi: 10.1242/dev.109.4.833. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- McCulley DJ, Kang JO, Martin JF, Black BL. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev Dyn. 2008;237:3200–3209. doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzen K, Shiojima I, Hiroi Y, Kudoh S, Oka T, Takimoto E, Hayashi D, Hosoda T, Habara-Ohkubo A, Nakaoka T, Fujita T, Yazaki Y, Komuro I. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol. 1999;19:7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen K, Karsan A. Notch signaling in cardiac development. Circ Res. 2008;102:1169–1181. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- Oliver G, Wehr R, Jenkins NA, Copeland NG, Cheyette BN, Hartenstein V, Zipursky SL, Gruss P. Homeobox genes and connective tissue patterning. Development. 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perveen R, Lloyd IC, Clayton-Smith J, Churchill A, van Heyningen V, Hanson I, Taylor D, McKeown C, Super M, Kerr B, Winter R, Black GC. Phenotypic variability and asymmetry of Rieger syndrome associated with PITX2 mutations. Invest Ophthalmol Vis Sci. 2000;41:2456–2460. [PubMed] [Google Scholar]
- Ray HJ, Niswander L. Mechanisms of tissue fusion during development. Development. 2012;139:1701–1711. doi: 10.1242/dev.068338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano LA, Runyan RB. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev Biol. 2000;223:91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- Schnabel RB. Comparison of approaches to fine-map first-generation genome-wide association study results at chromosome 9p21. Circ Cardiovasc Genet. 2011;4:577–578. doi: 10.1161/CIRCGENETICS.111.961466. [DOI] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proceedings of the National Academy of Sciences of the United States of America. 2007a;104:5907–5912. doi: 10.1073/pnas.0701122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr Patterns. 2007b;7:441–451. doi: 10.1016/j.modgep.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, Schneider MD, Lembo G, Campione M. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res. 2008;102:813–822. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Elhanany H, Rinon A, Tzahor E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development. 2006;133:1943–1953. doi: 10.1242/dev.02365. [DOI] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Human molecular genetics. 2002;11:915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev Biol. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Hutson MR, Stadt HA, Zdanowicz M, Zdanowicz J, Kirby ML. Cardiac neural crest is necessary for normal addition of the myocardium to the arterial pole from the secondary heart field. Dev Biol. 2005;281:66–77. doi: 10.1016/j.ydbio.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Lo CW, Kirby ML. Connexin 43 expression reflects neural crest patterns during cardiovascular development. Dev Biol. 1999;208:307–323. doi: 10.1006/dbio.1999.9219. [DOI] [PubMed] [Google Scholar]
- Webb S, Qayyum SR, Anderson RH, Lamers WH, Richardson MK. Septation and separation within the outflow tract of the developing heart. J Anat. 2003;202:327–342. doi: 10.1046/j.1469-7580.2003.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Shiratori H, Hamada H. Haemodynamics determined by a genetic programme govern asymmetric development of the aortic arch. Nature. 2007;450:285–288. doi: 10.1038/nature06254. [DOI] [PubMed] [Google Scholar]
- Zhou W, Lin L, Majumdar A, Li X, Zhang X, Liu W, Etheridge L, Shi Y, Martin J, Van de Ven W, Kaartinen V, Wynshaw-Boris A, McMahon AP, Rosenfeld MG, Evans SM. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat Genet. 2007;39:1225–1234. doi: 10.1038/ng2112. [DOI] [PMC free article] [PubMed] [Google Scholar]