Abstract
Despite great advances in our understanding of the driving events involved in malignant transformation, only a small number of oncogenic drivers have been targeted and translated into tangible clinical benefit. Moreover, even when a targeted therapy can be shown to effectively inhibit an oncogenic driver, leading to cancer remission, disease persistence and/or relapse is typically inevitable. Reemergence of the cancer can result from either intrinsic or acquired resistance mechanisms that result in failure to eliminate all cancer cells. Intrinsic mechanisms of resistance include tumor heterogeneity and pathways that can compensate for the inhibition of the oncogenic driver. Acquired resistance mechanisms include mutation of the oncogenic driver to directly prevent drug-mediated inhibition and the activation of compensatory survival pathways. RNA interference (RNAi)-based screening provides a powerful approach for the interrogation of both intrinsic and acquired resistance mechanisms. The availability of short interfering (si)RNA libraries targeting all human and mouse genes has made it possible to perform large-scale unbiased screens to identify pathways that are specifically required in cancer cells of particular genotypes or following particular treatments, facilitating the design of potential new therapeutic strategies that may limit resistance mechanisms. In this review, we will discuss how RNAi screens can be used to uncover critical growth and survival pathways and aid in the identification of novel therapeutic targets for improved treatment of hematological malignancies.
Keywords: Hematological Malignancies, Leukemia, Lymphoma, RNAi, Synthetic Lethal Screens, shRNA, Targeted Cancer Therapy
Introduction
Hematological malignancies comprise a broad spectrum of diseases involving multiple cell types, broadly classified as leukemia, lymphoma or myeloma. More specifically, they are classified by cell lineage, morphology, genotype/karyotype, and immunophenotype, as well as by clinical features including tumor site (for a review on classification of hematological malignancies, see [1–3]). The complexity of these diseases has limited the development of targeted therapies for hematological malignancies. A few small molecule inhibitors and specific monoclonal antibodies have demonstrated substantial clinical benefit, including targeted small molecule inhibitors of BCR-ABL kinase activity for the treatment of BCR-ABL+ leukemia [4–6], all-trans retinoic acid (ATRA) for the treatment of PML-RARα expressing acute promyelocytic leukemia (APL) [7, 8], and antibodies targeting CD20 for B-cell malignancies [9–11]. Still, in most cases, even when the driving oncogenic mutations have been identified, targeted therapies have failed to live up to their anticipated potential. Most hematologic malignancies are currently treated with non-targeted, highly cytotoxic drugs, radiation and/or bone marrow transplantation. These non-targeted therapies are often toxic, elicit incomplete responses, and may have severe long-term negative effects [12, 13]. In this review, we will discuss how RNAi screens can be utilized to identify novel targets for the treatment of hematological malignancies, including the development of strategies that target multiple pathways at once.
Why do most targeted therapies fail?
Current targeted therapies can provide a significant, although usually transient, clinical response. These responses are often insufficient to completely eradicate tumor cells and eventually the malignancy relapses. While the treatment of chronic myeloid leukemia (CML) with BCR-ABL tyrosine kinase inhibitors, like imatinib mesylate, has revolutionized CML therapy and can effectively control the disease in its chronic phase, most patients will require lifetime therapy, as leukemia stem cells persist in most patients [14, 15]. A small subset of patients that achieved sustained complete molecular responses (CMR) with imatinib were able to remain in CMR after discontinuation of treatment, which suggests that for at least the best responders, there is hope that imatinib might be safely discontinued [16]. In most patients that relapsed after imatinib discontinuation, relapse occurred within 3 months, which suggests that there is rapid expansion of a small number of residual leukemic cells. Moreover, advanced BCR-ABL+ leukemias, such as CML in blast crisis phase and BCR-ABL+ acute lymphoblastic leukemias (ALL), exhibit only transient responses to BCR-ABL inhibitors, and disease relapse is likely even if therapy is maintained [17]. Still, the addition of BCR-ABL tyrosine kinase inhibitors to traditional conventional chemotherapeutic regiments, have improved the outcomes for childhood BCR-ABL+ B-cell ALL dramatically [18]. In some ways, chronic phase CML, which can be effectively controlled by imatinib in more than 80% of patients, is the exception in its response to targeted therapy, and this disease is better characterized as a myeloproliferative disorder (driven largely by BCR-ABL without other apparent oncogenic mutations [19]) rather than a true leukemia. In contrast, the more advanced CMLs and BCR-ABL+ ALLs show numerous additional genetic changes beyond the BCR-ABL translocation [19], typical of most hematologic malignancies and other cancers.
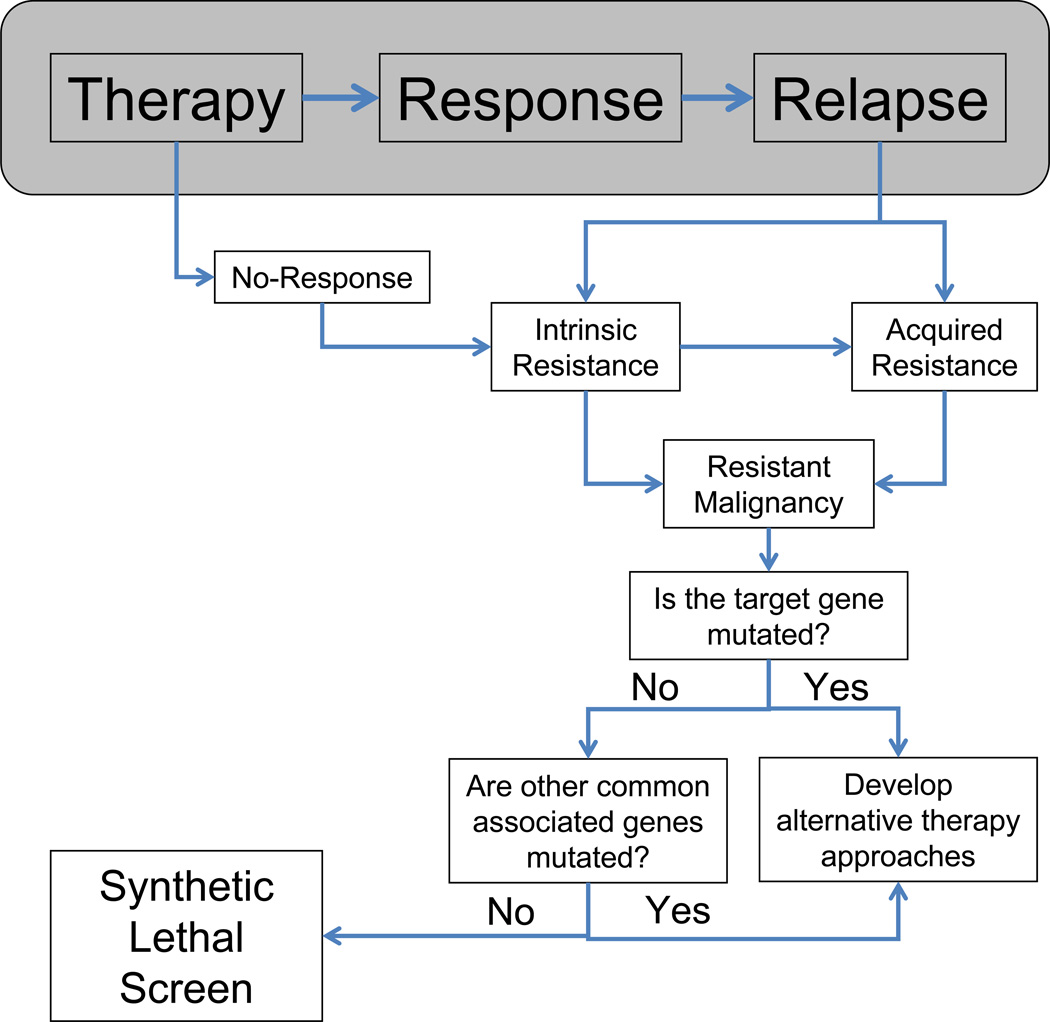
Numerous mechanisms of drug resistance have been identified and classified as either intrinsic or acquired resistance (Figure 1). Intrinsic resistance refers to a cell property innate to a cancer cell and present in the bulk of the cancer population prior to therapy that prevents an optimal response. Acquired resistance refers to a cell property that is gained (or selected for) during therapy and leads to the loss of a clinical response. Several mechanisms of intrinsic resistance in hematologic malignancies have been identified including those that mediate cell quiescence [20–22]; downregulation or increased instability of pro-apoptotic signals including BCL2-associated X protein (BAX) [23]; overexpression of anti-apoptotic signals including BCL2 family members [24–27]; mechanisms that regulate cell adhesion-mediated resistance including upregulation of integrins and chemokines on the cell surface [28–31]; and mechanisms that promote autophagy [32, 33]. In addition, intrinsic resistance can represent a characteristic of the leukemia hierarchy, with the leukemia stem cells exhibiting resistance to therapies like imatinib, due to stem cell-like characteristics of quiescence and high drug-efflux pump activity [14, 34]. Moreover, a subset of cancer cells appear to reside in a reversible, epigenetic drug-tolerant state, providing protection from both conventional and targeted therapies, which can be reversed by manipulating chromatin-modifying enzymes [35]. Although this observation has not yet been extended to hematological malignancies, DNA methyltransferase inhibitors have been successfully utilized for the treatment of myelodysplastic syndromes for which they received FDA approval and are currently in clinical trials for the treatment of several types of leukemia, lymphoma and other solid malignancies [36–40].
Figure 1. Using Functional Genomics to discover mechanisms to overcome intrinsic and acquired resistance.
When the mechanisms of resistance are unknown and not mediated by mutations in the target gene or other associated genes, synthetic lethal screens can be used to identify the causes.
Acquired resistance mechanisms have been associated with amplification or mutations of the gene encoding the targeted protein, amplification of other genes leading to activation of compensatory pathways, increased expression of cellular efflux pumps, and activation of redundant pathways or “oncogenic bypass” [41–46]. In addition, acquired resistance can be mediated by expression of binding partners that sequester the drug or upregulation of enzymes that accelerate the metabolism of the drug in vivo [47–49]. A common cause of acquired resistance to BCR-ABL inhibitors involves mutations in the tyrosine kinase domain (TKD) of BCR-ABL that prevent drug binding [50–54] or expansion of clones that pre-existed therapy harboring mutations in the kinase domain that become dominant upon relapse [55–59]. Soverini et al. [59] found that the majority of the BCR-ABL mutations detected at diagnosis are not associated with resistance or relapse upon BCR-ABL inhibitor therapy, which suggests that most of these mutations do not provide a survival advantage and are most likely eliminated during therapy. Importantly, they found that detection of known BCR-ABL resistance mutations at diagnosis usually, but not always, preclude a primary response. Similar mechanisms of acquired resistance involving TKD mutations have been identified in fms-like tyrosine kinase receptor-3 (FLT3) in acute myeloid leukemia (AML) treated with FLT3 inhibitors [60, 61]. Mutations in transcriptional co-activators, transcription factors and histone genes that are present at diagnosis or acquired at relapse have been shown to mediate acquired resistance in ALL [62]. When the cause underlying the acquired resistance is unknown and not caused by mutation/amplification of the target gene or other commonly implicated partners, functional genomics may be utilized to elucidate the culprit (Figure 1).
The failure of targeted therapies to lead to stable remissions is of course not limited to hematologic malignancies. While targeted inhibition of the epidermal growth factor receptor (EGFR) has shown benefit in patients with non-small cell lung cancer (NSCLC) bearing activating EGFR mutations, these therapies rarely result in long-term control of cancer burden. At least two factors may contribute to the inability of EGFR inhibitors to better control lung cancers [63, 64], and analogous factors likely contribute to targeted therapy failures for hematological malignancies. First, these cancer cells may not be sufficiently dependent on EGFR-mediated signaling for their survival and/or proliferation. While this is certainly the case for lung cancers without activation or gene amplification of EGFR, even cancers with these activating events appear to possess intrinsic compensatory survival pathways that can mediate partial cancer maintenance, preventing complete responses and leading to progression of disease. Second, while many cancers develop mutations in the kinase domain of EGFR that confer resistance to EGFR small molecule inhibitors [65, 66], acquired activation of alternative signaling pathways is frequent [64, 67]. For example, amplification of MET is a common cause of resistance to the EGFR inhibitors erlotinib and gefitinib in lung cancer [68–70]. Evidence from multiple biopsies along the course of targeted therapies in lung cancer suggests dynamic phenotypic and genotypic changes that evolve under the selective pressures of targeted therapy [66], and this is a likely consequence of intratumoral heterogeneity that exists prior to therapy (discussed in more detail below).
As a similar example, while BRAF kinase inhibitors lead to dramatic remissions for patients with melanomas bearing activating BRAF mutations (V600E), disease relapse is inevitable [71]. Melanomas escape inhibition of BRAF via upregulation of alternative pathways including upregulation of PDGFRβ receptor tyrosine kinase, N-RAS mediated reactivation of the MAPK pathway or activation mutations in MEK1 downstream of BRAF [72, 73]. Other mechanisms of acquired resistance have been described that are specific to epithelial cancers, including epithelial-to-mesenchymal transition, but these are beyond the scope of this review [66, 74]. Intrinsic signaling complexity, microenvironmental factors, and the mutational activation of multiple survival/proliferation pathways in advanced cancers likely reduces dependence on a single activated kinase, at least in a significant enough fraction of the tumor to prevent durable responses. The failure of kinase inhibitor therapies to better control advanced cancers upfront likely contributes to selection for drug tolerant cells by providing a larger pool of cancer cells from which to select for resistance. As many cancers are typically diagnosed at advanced phases and appear to possess inherent or acquired survival mechanisms that can protect the cells from inhibition of an oncogenic driver, the discovery of pathways that mediate these compensatory survival mechanisms may reveal novel therapeutic targets that could render oncogene inhibition more effective (Figure 1).
Tumor heterogeneity as a barrier to cancer therapy
It has long been appreciated that cancers are genetically complex diseases, however, only recently has there been a true appreciation of intratumoral genetic heterogeneity [75–77]. A recent study of renal cell carcinoma has revealed that only one third of mutations are uniformly detected throughout various regions of the same tumor [77]. A further example of intratumoral genetic heterogeneity is provided by glioblastomas, which display evidence of amplification of up to three different receptor tyrosine kinases in different tumor cell subclones in a mutually exclusive fashion [78]. Such genetic heterogeneity is likely present in almost all tumor types, and could represent a major obstacle for targeted therapeutic strategies [79]. Tumors are well known to be associated with frequent chromosomal rearrangements and a substantial mutational load, with the majority of these mutations thought to be “passengers” as opposed to oncogenic drivers. As the mechanisms of resistance to targeted therapies have become clearer, it is now even more apparent that individual tumors likely contain multiple genetically distinct clones that rely on differing oncogenic driver mutations for their survival and progression, and this heterogeneity may preclude the long-term efficacy of targeted therapies directed toward individual oncogenic drivers. An example of this phenomenon is provided by patients with activating mutations in EGFR who show a significant clinical response to EGFR-targeted therapies (i.e. the EGFR kinase inhibitors gefitinib and erlotinib), but they rapidly relapse with tumors displaying amplification of MET [68]. While initial hypotheses may have considered that such mutational events are newly acquired during the course of therapy, another interpretation is that MET-amplified EGFR-independent clones existed prior to EGFR-targeted therapy and facilitated relapse. Indeed, Turke et al. identified subpopulations of tumor cells with MET amplification in EGFR mutant NSCLC patient samples prior to EGFR therapy and provide evidence for clonal selection of these cells upon therapy relapse [70]. Although extensive studies examining target-independent mechanisms of targeted therapy resistance are not yet prevalent, intratumoral genetic heterogeneity is likely to be a common determinant of therapy resistance in most, if not almost all, tumor types. For example, amplification of c-KIT was recently observed as a mechanism of resistance in ALK-rearranged lung cancers treated with the ALK kinase inhibitor crizotinib [80]. Furthermore, Doebele et al. have shown that resistance to crizotinib therapy in ALK+ NSCLC can be associated with mutations in either KRAS or EGFR without evidence of persistent ALK rearrangement, suggesting that such mutations existed, independently of ALK mutation, prior to and were selected for during crizotinib therapy [81]. The implications of such tumor heterogeneity for targeted therapy are important: successful therapeutic strategies may need to rely on combination therapies that target multiple oncogenes. In other words, differing clones within an individual tumor may have differing oncogenic “addictions” and targeting all of these might be necessary to effectively eradicate most or all tumor cells and lead to truly durable remissions.
Given the spatial complexity of solid tumors, with differing microenvironmental influences and selective forces, it is perhaps not surprising that these distinct microenvironments would engender selection for distinct genotypes, leading to substantial genetic diversity within the cancer. It was perhaps wishful thinking that promoted the hypothesis that such genetic diversity would be limited to solid tumors and would not apply to hematological malignancies. This ideal was likely further fueled by the outstanding success of BCR-ABL inhibitors in treating CML that, as previously discussed, is a rather genetically simple leukemia, in which BCR-ABL is both the initiating mutation and the principle driver, at least in its early phases [5, 82]. However, recent studies of both BCR-ABL+ and ETV6-RUNX1+ ALLs reveal that individual leukemic tumors show evidence of branching evolution leading to complex genetic variegation [76, 83], with differing genetic components within unique tumor cell clones likely contributing to disease maintenance. As with solid tumors, these observations would seem to have severe implications for the success of targeted therapies. Unless therapies are directed toward initiating, as opposed to secondary mutations within subclones, combination therapies directed toward multiple genetic targets will likely be necessary. In addition, tumor heterogeneity may also be a barrier to the efficacy of standard genotoxic chemotherapies for leukemia. Indeed, whole-genome sequencing of AMLs has provided evidence for the existence of unique tumor cell subclones that survive initial genotoxic therapy and, upon additional mutation, become dominant at relapse [84].
Synthetic Lethal Screens
Synthetic lethal screens often take advantage of RNAi technology to inhibit sets of genes, either as genome-wide or smaller more targeted panels. RNAi technology comprises multiple different modalities including small interfering RNA (siRNA), enzymatically prepared siRNA (esiRNA) and short hairpin RNA (shRNA) (for a review on RNAi technologies, see [85, 86]) that mimic endogenous expressed microRNAs (miRNA) used by eukaryotic cells to silence genes at transcriptional and post-transcriptional levels. siRNAs are chemically synthesized RNA duplexes. esiRNA are produced from in vitro transcribed long dsRNA [87]. These latter two modalities provide a transient “knockdown” of gene expression, which limits their applications. The main difference between siRNAs and hairpin-based shRNAs are the mode of delivery and the duration of gene silencing. shRNAs are ~65 nt RNAs containing complementary sequences. Upon transcription, shRNAs fold to form a short hairpin that can be recognized by the Dicer complex, which cleaves them into siRNA within the target cell. Vector-mediated expression of shRNAs provides for stable incorporation into cells and can confer robust phenotypes. Both shRNAs and siRNAs are commercially available individually, as libraries targeting the whole human and mouse genomes, or as focused subsets directed toward, for example, solely phosphatases or kinases [88–90].
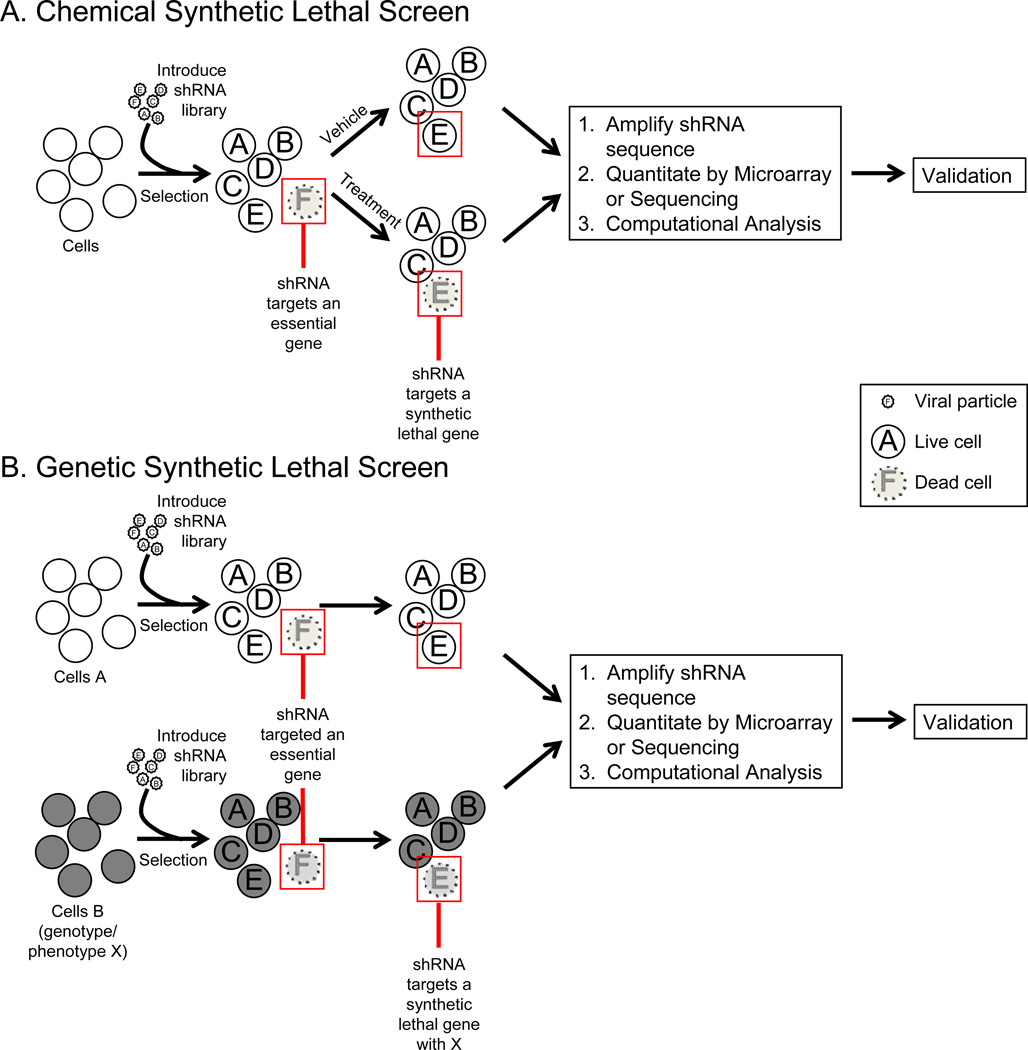
Synthetic lethal (SL) screens provide an unbiased approach to identify novel drug targets and elucidate functional relationships between genes in tumor cells. Moreover, loss of function mediated by RNAi-based knockdown can, at least partially, mimic how drug inhibitors work, allowing for the identification of potential therapeutic targets [91]. There are two major types of SL screens: genetic and chemical (Figure 2). Both types of screens utilize RNAi technology to knockdown a target gene and identify a specific phenotype. In the case of genetic SL screens, knocking down a particular gene confers sensitivity to a genotype already present in the cell. As an example, this strategy was successfully employed to identify genes that when knocked down resulted in cell death only in the presence of oncogenic KRAS [92–94], and to identify genes that mediate the survival of activated B-cell like diffuse large B-cell lymphoma (DLBCL) but not germinal center DLBCL [95].
Figure 2. Synthetic Lethal Screen Modalities.
A. In a pooled chemical SL screens, a population of cells (cell line, primary cells) is transduced with an shRNA library, and after selection of transduced cells, divided into vehicle or treatment. After treatment, the shRNAs are recovered, amplified and quantitated by DNA microarray or sequencing. Differential shRNA representation is compared between the vehicle and treatment groups. B. In a pooled genetic SL screens, two populations of cells, differing in genotype, are transduced with an shRNA library. The shRNAs are recovered, amplified and quantitated. Differential shRNA representation between the cell populations is determined.
Chemical SL screens are used to identify genes than when knocked down confer sensitivity to a drug, targeted or otherwise. For examples, chemical SL screens have been used to identify gene targets whose inhibition sensitizes lung cancer cells to the spindle breakdown inhibitor paclitaxel [96], sensitizes osteosarcoma cells to the HDAC inhibitor suberoylanilide hydroxamic acid, and sensitizes CML cells to the BCR-ABL tyrosine kinase inhibitor imatinib [97, 98]. Chemical SL screens can also be used to identify targets that sensitize tumor cells to non-specific therapy, such as genotoxic chemotherapy or radiation therapy, to identify genetic factors that augment the effect of such therapy, or to identify novel gene targets of a drug [99–102]. Thus, SL screens can be used to identify second hits that can better eliminate cancer cells either in combination with inhibition of a known oncogene (like BCR-ABL) or when a particular oncogenic pathway is activated (as for KRAS).
A potential weakness of RNAi screening for therapeutic targets in cancer, at least as it is currently being applied, is the inability to take into account tumor heterogeneity. RNAi screens have typically relied on the employment of tumor-derived cell lines that, at least presumably, are reasonably genetically clonal. That is not to discount the potential of these RNAi screens in revealing targets that can improve treatment efficacy in genetically complex cancers. A focus on initiating mutations may help to ensure the ultimate success of identifying adjuvant targets in such screens. For example, a recent RNAi screen was able to identify targets that are synthetic lethal with BCR-ABL inhibitor therapy (imatinib and dasatinib) in both CML and Ph+ ALLs, with evidence of nuclear factor of activated T-cells (NFAT) being an effective sensitizing target [98]. Given that the BCR-ABL mutation is strongly implicated as an initiating mutation in these diseases, NFAT-targeted adjuvant therapy will hopefully effectively sensitize most tumor cell clones to BCR-ABL inhibition. In AML, so-called “type II” mutations, exemplified by chromosomal translocations involving MLL that block hematopoietic differentiation through altered epigenetic regulation, are considered to be early events in leukemogenesis [103]. An RNAi screen of chromatin modifiers revealed bromodomain-containing protein 4 (BRD4) as a potential therapeutic target that seems to play a critical role in maintenance of MLL-AF9+ AML [104]. Finally, screens to identify pathways whose inhibition cooperates with the standard-of-care therapy for a cancer could accelerate the translation of discovered combination therapies.
Future screens may have the potential to identify targets that will further sensitize AML cells to therapies targeting pathways dependent on type II initiating mutations such as MLL and IDH1 and 2 [105]. Still, screening for targets that sensitize cells to inhibition of later mutational events in hematological tumors, such as “type I” FLT3 mutations in AML, should not be discounted. Activating mutations in FLT3 are detected in about one-third of AMLs and are associated with aggressive disease and a poor outcome [103]. Kinase inhibitor therapies targeting FLT3 have already demonstrated impressive efficacy in treatment of FLT3+ AML, although relapse is currently inevitable with FLT3 inhibitors as monotherapy [106]. RNAi screening could reveal therapeutic targets that will further sensitize FLT3+ AML cells to FLT3 inhibition and could be incorporated into therapeutic regimens that may also include standard genotoxic chemotherapy to eradicate less aggressive FLT3-independent AML clones. Similarly, multiple RNAi screens have discovered targets that sensitize lung cancer cells to therapies targeting EGFR [107–109], whose mutation is also a later event in lung tumorigenesis. Such targets could potentially be integrated into combination targeted therapy regimens that may more effectively control disease. Thus, while tumor heterogeneity presents a clear challenge for identifying therapeutic targets using SL screens, an appreciation of this heterogeneity should facilitate the design of more complex therapeutic approaches to target advanced malignancies.
Important considerations when designing a screen
The main advantage of genome-wide screening is the ability to discover previously uncharacterized or unsuspected genes with minimal a priori predictions of what should represent a good target. The downside is that genome-wide screens are more technically challenging to ensure appropriate library representation. They require high-throughput data acquisition platforms and complex computational analyses. In contrast, smaller pathway focused screens use a select sub-genomic set of constructs to target specific pathways (apoptosis panels, kinase, phosphatases, tumor suppressors, T-cell activation, B-cell activation panels, etc.). SL screens can be performed either in arrayed (multiwell format, in which a single shRNA or siRNA is added to each well) or pooled format, or a hybrid of the two. Arrayed screens depend on a quantifiable phenotype, such as a reporter assay or measured parameter, to identify targets. In addition, they require high-throughput equipment, complex statistical analysis and are typically more costly than pooled screens. An important consideration for pooled screens is the maintenance of proper representation of a diverse pool of shRNAs in the cell population, as the goal is for changes in shRNA representation to reflect the experimental condition (such as drug treatment) rather than stochastic changes. Thus, targeted screens using smaller pools of shRNAs can reduce the chances for stochastic loss of shRNAs, which could obscure shRNA losses that truly result from treatment. Still, genome-wide shRNA screens can reliably produce valuable information if stochastic changes in shRNA representation are minimized by maintaining large cell population sizes, avoiding population bottle-necks (such as during cytotoxic treatment), the use of multiple replicates and cell lines, and by the application of appropriate statistical and bioinformatic analyses [110].
Many factors can affect library representation and successful gene silencing, including optimal bacterial culture, lentiviral packaging, transfection and transduction efficiency, and cell culture conditions of the target cell line [111]. Library representation becomes especially important for screens with negative selection, because a large percentage of cells will be eliminated from the population by the treatment, and a number of shRNAs will be lost by chance alone. Cells that express shRNAs against essential and non-redundant genes will be eliminated from the population (Figure 2). After selection, cells are divided into experimental groups. For chemical screens, cells are treated with the drug of interest.
In the case of arrayed screens, data acquisition is performed by quantification of a particular phenotype (cell survival, apoptosis, migration, invasion, senescence, etc) [91]. For pooled screens, genomic DNA or mRNA is isolated from the different groups, and following amplification of the shRNA sequences, shRNA representation in the different experimental groups is quantified using DNA microarrays or high-throughput “deep” sequencing. Commercially available DNA hybridization microarrays utilize a sequence-specific probe and measure the relative abundance of each shRNA hybridized to the array. Alternatively, each shRNA and adaptor sequence can be sequenced using deep sequencing. Deep sequencing allows the identification and quantitation of millions of single DNA reads per run. Multiple analysis pipelines have been developed to analyze genome-wide sequencing data from RNAi-based screens, including the BiNGS! (Bioinformatics for Next Generation Sequencing) [110], GARP (Gene Activity Ranking Profile) [112], RIGER (RNAi Gene Enrichment Ranking) [113] and RSA (Redundant siRNA Activity) [114] methods. The major advantages of deep sequencing, compared with microarray-based detection, are improved data coverage, quantitation, and signal to noise ratio. Comparison between experimental groups allows for the reliable identification of shRNA sequences lost or enriched upon drug treatment or other manipulations. Results can be further categorized into functional pathways utilizing KEGG pathway analysis [115] or other pathways analyses. Analysis of SL targets into functional pathways can point to other druggable targets downstream or upstream of the synthetic lethal hits. Staudt and colleagues identified several components of B-cell receptor signaling pathway in a screen in diffuse large B-cell lymphoma [95]. Even though the Bruton’s tyrosine kinase (BTK) was not a hit, several downstream components including CARD11 were identified as part of the screen. Based on this results, inhibitors of BTK are currently in clinical trials for the treatment of relapsed/refractory activated B-cell (ABC) and germinal-cell B-cell (GCB) Diffuse Large B-cell Lymphoma (DLBCL).
Prioritization of targets for validation should include top ranked hits, targets in pathways with multiple hits, and those with druggable potential. Target validation can be performed by knocking down the target gene utilizing a unique set of shRNA constructs distinct from those used in the screen, pharmacological inhibition, or, in some cases, antagonistic antibodies. Each approach is complementary to the others since knockdown of a gene can have very distinct effects compared to pharmacologic inhibition of the target; and vice-versa. Moreover, pharmacologic inhibition may have off-target effects not recapitulated by the genetic inhibitory approaches. This is exemplified by a genome-wide shRNA screen by Scholl et al. [92] in which they identified STK33 as a synthetic lethal target in the presence of oncogenic KRAS. Simultaneously, two other groups performed similar screen and identified different targets other than STK33 that are crucial to mutated KRAS survival [93, 94]. At the time of these initial screens, there were no pharmacologic inhibitors of STK33. Two other groups developed specific STK33 inhibitors and demonstrated that they were not effective at killing KRAS mutated cells [116, 117]. One of the groups was able to recapitulate that the shRNA knockdown of STK33 selectively eliminated KRAS mutant cells (the other group was not). This example illustrates a case in which pharmacologic inhibition does not equate to genetic ablation, which may be important when a protein plays a structural or scaffolding role in addition to a catalytic function. Furthermore, the paucity of overlap in terms of the SL interactions with KRAS discovered by multiple groups indicates that these screens are either far from saturating and/or that false positive rates may be high. Genetic SL screens allow the identification of a relationship between decreased level of a target gene, but they do not establish a relationship involving the function of the encoding protein [116]. The STK33 example illustrates the importance of extensive target validation as a key step in screen design.
In addition to pharmacologic and knockdown inhibitory approaches, targets should be further validated by activating the gene target directly or indirectly and observing the opposite effect. Also, identification of previously implicated genes that influence the observed phenotype or interact with other identified targets will increase confidence in the results. Finally, high throughput validation can be accomplished by screening a curated candidate gene list in a sub-library format against a panel of cell lines or in patient samples [102]. The low complexity of a smaller library allows for pooling of multiple samples into a single lane of sequencing, greatly reducing the cost and expediting the validation process.
shRNA Screens in Hematological Malignancies
As described above, while major advances have been made over the last few decades in cancer research, the diagnosis of cancer in most cases still constitutes a death sentence, even if therapy can delay this outcome. Loss of function shRNA screens have the potential to identify novel therapeutic targets, the inhibition of which could improve therapeutic outcomes with currently used drugs. These screens can also be used to identify pathways important for maintenance of the cancer phenotype, as well as to reveal unique sensitivities of cancer cells (such as “non-oncogene addictions” [118]). Panel screens have been utilized to identify genes required for the proliferation and survival of diffuse B-cell lymphomas [95], potential mechanisms of resistance to IKKβ inhibitors in activated B-cell like diffuse large B cell lymphoma [119], chromatin regulators in AML [104], functional profiles of primary leukemia samples [120], and functional characterization of chemotherapeutic targets [121]. For a comprehensive list of RNAi screens in hematologic malignancies, see Table 1.
Table 1. siRNA based screens in hematologic malignancies.
For the Screen Strategy, the source of siRNAs and basic strategy are listed in 1., and the method of determining relevant siRNA or shRNA is described in 2.
| Cancer Type (Cell Line) |
Screen strategy | Results | Clinical relevance | Reference |
|---|---|---|---|---|
| Chemical Screens | ||||
| APL (NB4 and NB4-R2) |
1. Large-scale SL with ATRAa,c 2. Colony Forming Assay and Sequencing |
Identify mediators of ATRA-induced differentiation and growth arrest of APL including UBE2D3 | ATRA is FDA approved for treatment of hematologic malignancies | [137] |
| Murine Eµ-Myc p19ARF−/− Burkitt's lymphoma | 1. Cancer 1000 SL with doxorubicin and camptothecinb,c,d,e 2. DNA microarray and deep sequencing |
Identified genes that mediate response to doxorubicin and camptothecin | Doxorubin is FDA approved for treatment of hematologic malignancies | [99] |
| Activated B-cell like DLBCL (OCI-Ly3) |
1. Kinome SL screen with IKKβ inhibitorsb,c 2. DNA microarray |
Identified that targeting both IKKα and IKKβ is more potent on the classical NF-κB pathway than IKKβ alone | [119] | |
| AML (HL-60) |
1. Kinome SL screen to identify off-target effects of gefitinibb,d 2. GE-HTS |
Identify SYK as an off-target effect of gefitinib responsible for anti-AML effect | Syk inhibitor fostamatinib disodium successfully completed Phase I/II (NCT00446095) for relapsed/refractory B cell malignancies [138] | [100] |
| CML (K562) |
1. Genome wide SL with imatinibb,c 2. DNA microarray |
Non-canonical Wnt/Ca+2/NFAT pathway mediates cell intrinsic resistance to imatinib. | Resulting combination therapy is in Phase I (NCT01456988) | [98] |
| Murine Eµ-Myc p19ARF−/− Burkitt's lymphoma | 1. Small scale SL screen with multiple genotoxic drugsb,d,f 2. Flow cytometry |
Identified drug signatures based on a subset of knockdown effects | Developed a small screen to functional characterize new genotoxic drugs | [121] |
| MM (KMS11) |
1. Human Druggable Genome SL screen with bortezomiba,d 2. Cell viability |
CDK5 sensitizes multiple myeloma cells to bortezomib | Non-specific CDK inhibitors are in Phase I for solid tumors in combination with nucleoside analogs (NCT00999401) | [123] |
| Murine Eµ-Myc p19ARF−/− Burkitt's lymphoma and p185 BCR-ABL+ p19ARF−/− B-ALL | 1. In vivo targeted screen with doxorubicinb,c,g 2. Luminex bead-based |
Identified tissue-specific modifier of doxorubicin response | Doxorubicin is FDA approved for treatment of hematologic malignancies | [101] |
| AML (Molm13 and MV4-11) |
1. Genome wide SL with cytarabineb,c 2. Deep sequencing |
WEE1 is a critical mediator of AML survival after cytarabine | WEE1 inhibitor MK-1775 is currently in Phase II clinical trials for ovarian cancer (e.g. NCT01164995) | [102] |
| AML (TF-1 and THP-1) |
1. Kinome SL with cytarabinea,d 2. Cell viability |
WEE1 is a critical mediator of AML survival after cytarabine | WEE1 inhibitor MK-1775 is currently in Phase II clinical trials for ovarian cancer | [124] |
| Genetic Screens | ||||
| Activated B-cell like DLBCL and germinal center DLBCL (OCI-Ly3, OCI-Ly10 and OCI-Ly7, OCI-Ly19) |
1. Small scale SL screenb,c 2. DNA microarray |
Identified CARD11 as a key upstream signaling component responsible for the constitutive IKKβ kinase activity in activated B-cell like DLBCL | Bruton's Tyrosine Kinase (Btk) Inhibitor is currently in Phase II clinical trials (NCT01325701) |
[95] |
| Primary leukemia samplesh | 1. Kinome SL screena,d 2. Cell viability |
Identified targets including activating mutations in JAK2, K-RAS and novel insertion in MPL1186InsGG critical to the survival of the malignancy | Patient specific oncogenic target identification | [120] |
| Murine Eµ-Myc p19ARF−/− Burkitt's lymphoma | 1. In vivo/in vitro Cancer 1000 SL screenb,c,e 2. Deep sequencing |
Identified genes that mediate in vivo survival compared to in vitro survival | [125] | |
| Murine Eμ-Myc p19Arf−/− HSPC | 1. In vivo Cancer 1000 SL screenb,c,e 2. Deep sequencing |
Identified several tumor suppressor genes and components of DNA damage response | MEK1/2 inhibitor in Phase II in cancers with BRAF mutations (NCT00888134) and Phase I in combination with PI3K/mTOR inhibitors (NCT01337765, NCT01378377) | [126] |
| AML (MLL-AF9/NrasG12D leukemia) |
1. Targeted SL screenb,c,i 2. Deep Sequencing |
Brd4 is required for disease maintenance of AML | Small molecule inhibitor of Brd4, JQ1, effective in mouse model | [104] |
| AML (HL-60) |
1. Kinome SL screenb,d 2. GE-HTS |
Serine-threonine kinase GSK3 plays a role in AML differentiation | GSK3 inhibitor completed Phase I/II (NCT00948259, NCT01049399 NCT01350362) for Alzheimer’s disease and Progressive Supranuclear Palsy |
[122] |
siRNA,
shRNA,
Pooled,
Arrayed,
Cancer 1000 set of genes containing putative cancer-related genes in breast cancer [136],
Library targeting BCL2 and p53 family members,
Library targeting BCL-2 family member,
Samples included ALL, CMML, AML, CNL,
Library targeting chromatin regulators.
Abbreviations: APL, acute promyelocytic leukemia; ATRA, all-trans retinoic acid; DLBCL, diffuse large B-cell lymphoma; AML, acute myeloid leukemia; GE-HTS, Gene Expression-based High-throughput Screening; CML, chronic myelogenous leukemia; MM, multiple myeloma; B-ALL, B-cell acute lymphoblastic leukemia; ALL, acute lymphoblastic leukemia; CMML, chronic myelomonocytic leukemia; CNL, Chronic neutrophilic leukemia; HSPC, hematopoietic stem and progenitor cells
Several groups have taken distinct approaches for the discovery of potential therapeutic targets utilizing functional genomics, in some cases in combination with conventional approaches, such as gene expression profiling. Jiang et al. [121] utilized a panel of shRNAs targeting BCL2 family members and p53 related proteins, including its activating kinases, to screen several chemotherapies in Burkitt’s lymphoma and Bcr-Abl+ ALL, assigning these targets into functional categories based on their shRNA profile. This approach could be used to assign novel compounds or derivatives of existing compounds into functional categories and determine if they share the same specificity as the parent drug. More importantly, the authors were able to define a genotoxic signature utilizing as few as 8 shRNAs to predict genotoxic drugs that could be utilized to rapidly screen libraries of compounds. shRNA screens can also identify specific dependencies of malignancies. Banerji et al. [122] utilized a combination of shRNA SL screen and small molecule kinase inhibitor screen to identify a GSK-3α pathways involved in AML differentiation. As the inhibition of GSK-3α potentiates AML differentiation, this study reveals a novel differentiation pathway for this leukemia. Similarly, Zuber et al. [104] utilized a custom shRNA library targeting all known chromatin regulators to identify novel drug targets in AML. As noted, they were able to identify a novel target, BRD4, which specifically mediates survival of AML but not normal cells. Importantly, while their screen utilized a mouse AML model, their follow up experiments demonstrated similar roles for BRD4 in human AMLs, including primary patient samples. Their work highlights one of the main advantages of functional genomics: to identify previously uncharacterized genetic dependencies for specific malignancies.
Other groups have utilized functional genomics to identify mechanisms of intrinsic drug resistance. Lam et al. [119] utilized an shRNA panel library targeting all kinases to identify putative mechanism of drug resistance to IKKβ inhibitors in diffuse large B-cell lymphoma. In addition, functional genomic screens can be utilized to identify pathways whose inhibition will synergize with a current therapy. We performed a large-scale shRNA screen to identify pathways that sensitize CML cells to imatinib, revealing components of the non-canonical WNT/calcineurin/NFAT pathway as synthetic lethal upon imatinib treatment [98]. Most importantly, inhibition of calcineurin using cyclosporine A (CsA) enhanced the efficacy of BCR-ABL inhibitor therapy for mice with BCR-ABL+ leukemia [98], leading to a clinical trial testing this combination therapy in humans (Table 1). Importantly, as components of the WNT/Ca+2/NFAT pathway (and transcriptional targets) do not exhibit obvious deregulation or mutational activation in CML, these results highlight the ability of unbiased screens to reveal cancer cell dependencies that would be missed by analyses of gene expression or mutational changes alone.
Similarly, Zhu et al. [123] utilized the Human Druggable Genome siRNA library to identify targets that sensitize multiple myeloma cells to bortezomib, revealing the importance of CDK5 in multiple myeloma survival. Chemical SL screens have also been utilized to identify off-target effects of targeted therapies. This approach was successfully employed to identify SYK as the off-target effect of gefitinib (EGFR tyrosine kinase inhibitor) responsible for its anti-AML effects [100]. Screens to identify additional functionally relevant pathways are not limited to targeted therapies. Porter el al. [102] and Tibes et al. [124] performed genome-wide and kinome screens, respectively, to identify genes that when inhibited potentiate AML cell killing with cytarabine, a cytosine analog utilized to treat certain types of leukemia including AML, ALL, and CML. These studies revealed a critical role for the WEE1 dependent cell cycle checkpoint in allowing AML cell survival during cytarabine induced damage in S-phase. Genetic or pharmacological inhibition of Wee1 prevented S-phase stalling, leading to increased AML cell death, thereby suggesting a therapeutic strategy to increase cytarabine effectiveness in AML treatment. This example illustrates the use of SL libraries to develop combination therapies to enhance the efficacy of non-targeted therapies.
In vivo shRNA screens have been used to identify genes involved in tumor cell homeostasis and metastasis in mouse models of cancer. Such in vivo studies have the advantage of identifying functionally relevant genes under more physiological conditions, which could reveal pathways important for tumor cell interactions with their microenvironment and the immune system. In vivo shRNA screens are limited by the number of transplanted RNAi library-transduced cells that are required to ensure maintenance of sufficient library representation, but can still provide valuable information. Meacham et al. [125] utilized a B cell lymphoma model (Eµ-Myc mouse lymphoma) to identify shRNAs that are depleted or enriched in vivo during tumorigenesis. After tumor formation, they were able to retrieve 26–40% of their library in vivo compared to 71% in vitro. As expected, the in vivo samples were depleted of shRNAs targeting cell motility and cell adhesion genes, dependencies that might have been missed in vitro. Similarly, Bric et al. [126] transduced Eµ-Myc hematopoietic stem and progenitor cells (HSPC) with small pools of shRNAs targeting the Cancer 1000 shRNA set and transplanted these cells into mice. Their screen revealed that the knockdown of multiple tumor suppressor genes and DNA damage response associated genes accelerated lymphomagenesis, thus identifying novel tumor suppressive pathways for Myc driven lymphomas. In vivo shRNA screens have also been utilized to identify targets that mediate survival to topoisomerase inhibitors [99]. A screen in hepatocellular carcinoma (HCC) is worth noting given that an analogous approach could be extended to hematologic malignancies. Zender et al. [127] also used an in vivo mouse model of HCC and a series of small pooled shRNA subsets targeting the mouse orthologs of genes recurrently deleted in human HCC to functionally query potential tumor suppressors. Since many genes deleted in human cancers will be passengers (their deletion does not clearly contribute to the cancer phenotype), this study provides a blueprint for distinguishing driver and passenger mutations in cancers, such as those identified via The Cancer Genome Atlas. As another prototypical screen performed in a non-hematopoietic malignancy, Possemato et al. [128] utilized a subset library targeting metabolic genes in a human breast cancer xenograft model, together with comparisons to regions of genomic copy number gains in breast cancers, to reveal an important role for the serine synthesis pathway in these cancers. Thus, by either determining which shRNAs are enriched [127] or lost [128] in the cancers grown in mice, these studies have revealed unanticipated tumor suppressive and oncogenic pathways, respectively.
Finally, RNAi screens have been utilized to construct functional profiles of primary leukemia samples. Tyner et al. [120] utilized an arrayed siRNA screen targeting all members of the tyrosine kinase family to identify sensitivities and driving mutations in AML patient samples, a strategy that they dubbed RNAi assisted protein target identification (RAPID). They were able to identify common mutations present in AMLs and other novel mutations including the novel insertion in MPL1186InsGG. Importantly, this screen allowed the identification of tyrosine kinases driving these malignancies beyond those containing genetic abnormalities.
As illustrated with the examples above, multiple distinct synthetic lethal approaches can be used to discover potential therapeutic targets.
Applications to other immunologic processes
The use of RNAi screening is not confined to the field of cancer biology. Several groups have used both genome-wide and targeted screens to identify regulatory pathways involved in lymphocyte signaling [129], host factors involved in influenza pathogenesis [130], host genes required for viral entry and early stages of HIV infection [131, 132], and mechanisms that regulate tissue specific MHC Class II expression, peptide loading and transport in dendritic cells [133]. Astier et al. used a sub-genomic arrayed siRNA library focused on kinases and phosphatases in primary human T cells to identify novel genes that increase/decrease levels of IL-10, IL-13 and/or IFNγ. Using this approach, they identified FLT3 and FLT3 ligand as negative regulators of IL-10 in activated T cells [129]. Two independent groups performed genome-wide arrayed siRNA screen to identify host factors required for HIV entry and early stages of replication. In addition to host factors previously implicated in HIV pathogenesis, Brass et al. [132] identified nucleocytoplasmic transporter activity genes, helicases, transcription factors and retrograde golgi transport proteins as playing important roles in viral entry and early replication. Konig et al. [134] performed parallel screens utilizing a single-cycle HIV-1 reporter virus, MuLV retroviral vector and a AAV vector to identify factors specific to HIV (and not just to retroviruses and viruses in general). Their analysis incorporated gene-based scored (Redundant siRNA Analysis), gene expression signatures, gene ontology data, cellular protein-protein interaction data and virus-host interaction data. Interestingly, the screens from the two groups have limited overlap which might be attributed to the different models they utilized as well as their reporter assays and their analysis. In addition to the examples described above, synthetic lethal screens should also be useful for to the identification of novel pathways involved in lymphocyte differentiation, activation and migration.
Conclusions
Loss-of -function genetic screens, until recently, were limited to model organisms such as yeast and worms. The discovery that RNAi can effectively be employed to perform functional genetic screening in mammalian cells has already had a major impact in interrogating the genetic factors that influence human disease. RNAi-based screening has paved the way for the efficient and effective discovery of genetic targets that can selectively elicit elimination of tumor cells, through the targeting of particular oncogene-mediated dependencies alone, or through combination targeting of multiple tumor survival pathways. Thus RNAi-mediated screening has great potential for identification of therapeutic targets that can sensitize any tumor/disease cell to therapy, targeted or otherwise. Tumor cells seem to employ multiple genetic/epigenetic mechanisms to resist therapy, and these changes, either intrinsic or acquired, can be effectively exposed through RNAi screens. Furthermore, the continued development and improved efficacy of RNAi within large libraries, minimizing “off-target” and maximizing “on-target” effects, should dramatically enhance our current ability to screen for potential therapeutically-effective gene targets in many cancers, including hematological malignancies, and improve treatment of these diseases. From a therapeutic standpoint, a major advantage of genome-scale screening is that it can reveal multiple targets within the same genetic pathway, some of which may be inhibited by drugs already available in our current therapeutic arsenal. The discovery of already “drugged” targets, such as with CsA for inhibiting NFAT in CML treatment, should accelerate our ability to move potential treatments to the clinic. With further development, the potential applications of RNAi screening technology for exploration of other diseases, in addition to cancer, seems unlimited.
Genome-wide shRNA screens have the potential to identify thousands of target genes including both oncogenes and non-oncogenes. Multiple non-oncogene addictions are observed in cancer cells, including those mediating DNA damage/replication stress, proteotoxic stress, mitotic stress, metabolic stress and oxidative stress [118, 135]. These non-oncogenes have the potential to be ubiquitous targets available for multiple different cancer types. Genome-wide screens offer the advantage of being able to identify non-oncogene co-dependencies whose inhibition is selectively lethal in particular cancer types or during drug treatment.
Future prospects: personalized screens?
As targeted therapies become the standard of care for cancer treatment, diagnostic tests that include molecular and genetic identification of the driving oncogenic processes will become even more important. With falling costs of deep sequencing, better functional understanding of the genome, and advanced computational approaches, it is possible that one day we will be able to screen a newly diagnosed patient sample with shRNA libraries to identify a priori which therapeutic combinations that a patient will likely benefit from. By understanding the basic biology driving the particular cancer type, we will be able to more effectively personalize cancer therapy, hopefully decreasing therapy-related toxicity and improving relapse-free survival.
Acknowledgments
These studies were supported by grants from the National Institutes of Health (R01-CA157850 to J.D, K01-CA133182 to M.A.G and F31-CA157166 to F.A.C) and the Leukemia Lymphoma Society. We would like to thank Matias Casas, Courtney Fleenor, Andriy Marusyk, Jennifer Salstrom, Aik-Choon Tan, and Christopher Porter for their comments and suggestions.
Footnotes
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- 1.Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg OK, et al. Clinical characterization of acute myeloid leukemia with myelodysplasia-related changes as defined by the 2008 WHO classification system. Blood. 2009;113(9):1906–1908. doi: 10.1182/blood-2008-10-182782. [DOI] [PubMed] [Google Scholar]
- 3.Campo E, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117(19):5019–5032. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. The New England journal of medicine. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112(13):4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 6.Hughes TP, et al. Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS) Blood. 2010;116(19):3758–3765. doi: 10.1182/blood-2010-03-273979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang ME, et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72(2):567–572. [Google Scholar]
- 8.Sanz MA, et al. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113(9):1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- 9.Alas S, Bonavida B. Rituximab inactivates signal transducer and activation of transcription 3 (STAT3) activity in B-non-Hodgkin's lymphoma through inhibition of the interleukin 10 autocrine/paracrine loop and results in down-regulation of Bcl-2 and sensitization to cytotoxic drugs. Cancer research. 2001;61(13):5137–5144. [PubMed] [Google Scholar]
- 10.Vega MI, et al. Rituximab (chimeric anti-CD20) sensitizes B-NHL cell lines to Fas-induced apoptosis. Oncogene. 2005;24(55):8114–8127. doi: 10.1038/sj.onc.1208954. [DOI] [PubMed] [Google Scholar]
- 11.Vega MI, et al. Rituximab-induced inhibition of YY1 and Bcl-xL expression in Ramos non-Hodgkin's lymphoma cell line via inhibition of NF-kappa B activity: role of YY1 and Bcl-xL in Fas resistance and chemoresistance, respectively. Journal of immunology. 2005;175(4):2174–2183. doi: 10.4049/jimmunol.175.4.2174. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong GT, et al. Late mortality among 5-year survivors of childhood cancer: a summary from the Childhood Cancer Survivor Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(14):2328–2338. doi: 10.1200/JCO.2008.21.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reulen RC, et al. Long-term risks of subsequent primary neoplasms among survivors of childhood cancer. JAMA : the journal of the American Medical Association. 2011;305(22):2311–2319. doi: 10.1001/jama.2011.747. [DOI] [PubMed] [Google Scholar]
- 14.Corbin AS, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. The Journal of clinical investigation. 2011;121(1):396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurtz C, et al. BCL6-mediated repression of p53 is critical for leukemia stem cell survival in chronic myeloid leukemia. The Journal of experimental medicine. 2011;208(11):2163–2174. doi: 10.1084/jem.20110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahon F-X, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. The lancet oncology. 2010;11(11):1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 17.Foà R, et al. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;118(25):6521–6528. doi: 10.1182/blood-2011-05-351403. [DOI] [PubMed] [Google Scholar]
- 18.Schultz KR, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children's oncology group study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(31):5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 20.Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 21.Lemoli RM, et al. Molecular and functional analysis of the stem cell compartment of chronic myelogenous leukemia reveals the presence of a CD34-cell population with intrinsic resistance to imatinib. Blood. 2009;114(25):5191–5200. doi: 10.1182/blood-2008-08-176016. [DOI] [PubMed] [Google Scholar]
- 22.Kumari A, et al. Low BCR-ABL expression levels in hematopoietic precursor cells enable persistence of chronic myeloid leukemia under imatinib. Blood. 2012;119(2):530–539. doi: 10.1182/blood-2010-08-303495. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal SG, et al. Increased proteasomal degradation of Bax is a common feature of poor prognosis chronic lymphocytic leukemia. Blood. 2008;111(5):2790–2796. doi: 10.1182/blood-2007-10-110460. [DOI] [PubMed] [Google Scholar]
- 24.Konopleva M, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer cell. 2006;10(5):375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Paoluzzi L, et al. The BH3-only mimetic ABT-737 synergizes the antineoplastic activity of proteasome inhibitors in lymphoid malignancies. Blood. 2008;112(7):2906–2916. doi: 10.1182/blood-2007-12-130781. [DOI] [PubMed] [Google Scholar]
- 26.Paoluzzi L, et al. Targeting Bcl-2 family members with the BH3 mimetic AT-101 markedly enhances the therapeutic effects of chemotherapeutic agents in in vitro and in vivo models of B-cell lymphoma. Blood. 2008;111(11):5350–5358. doi: 10.1182/blood-2007-12-129833. [DOI] [PubMed] [Google Scholar]
- 27.Balakrishnan K, et al. AT-101 induces apoptosis in CLL B cells and overcomes stromal cell-mediated Mcl-1 induction and drug resistance. Blood. 2009;113(1):149–153. doi: 10.1182/blood-2008-02-138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bélanger SD, St-Pierre Y. Role of selectins in the triggering, growth, and dissemination of T-lymphoma cells: implication of L-selectin in the growth of thymic lymphoma. Blood. 2005;105(12):4800–4806. doi: 10.1182/blood-2004-04-1406. [DOI] [PubMed] [Google Scholar]
- 29.Buchner M, et al. Spleen tyrosine kinase inhibition prevents chemokine- and integrin-mediated stromal protective effects in chronic lymphocytic leukemia. Blood. 2010;115(22):4497–4506. doi: 10.1182/blood-2009-07-233692. [DOI] [PubMed] [Google Scholar]
- 30.De Toni-Costes F, et al. A New alpha5beta1 integrin-dependent survival pathway through GSK3beta activation in leukemic cells. PloS one. 2010;5(3):e9807. doi: 10.1371/journal.pone.0009807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenouille N, et al. Persistent activation of the Fyn/ERK kinase signaling axis mediates imatinib resistance in chronic myelogenous leukemia cells through upregulation of intracellular SPARC. Cancer research. 2010;70(23):9659–9670. doi: 10.1158/0008-5472.CAN-10-2034. [DOI] [PubMed] [Google Scholar]
- 32.Bellodi C, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. The Journal of clinical investigation. 2009;119(5):1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amrein L, et al. p53 and autophagy contribute to dasatinib resistance in primary CLL lymphocytes. Leukemia research. 2011;35(1):99–102. doi: 10.1016/j.leukres.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 34.Quintas-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113(8):1619–1630. doi: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun T, et al. Molecular predictors of response to decitabine in advanced chronic myelomonocytic leukemia: a phase 2 trial. Blood. 2011;118(14):3824–3831. doi: 10.1182/blood-2011-05-352039. [DOI] [PubMed] [Google Scholar]
- 37.Lubbert M, et al. Low-dose decitabine versus best supportive care in elderly patients with intermediate- or high-risk myelodysplastic syndrome (MDS) ineligible for intensive chemotherapy: final results of the randomized phase III study of the European Organisation for Research and Treatment of Cancer Leukemia Group and the German MDS Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(15):1987–1996. doi: 10.1200/JCO.2010.30.9245. [DOI] [PubMed] [Google Scholar]
- 38.Stathis A, et al. Phase I study of decitabine in combination with vorinostat in patients with advanced solid tumors and non-Hodgkin's lymphomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(6):1582–1590. doi: 10.1158/1078-0432.CCR-10-1893. [DOI] [PubMed] [Google Scholar]
- 39.Cashen AF, et al. Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(4):556–561. doi: 10.1200/JCO.2009.23.9178. [DOI] [PubMed] [Google Scholar]
- 40.Issa JP, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103(5):1635–1640. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 41.Beck WT, Mueller TJ, Tanzer LR. Altered surface membrane glycoproteins in Vinca alkaloid-resistant human leukemic lymphoblasts. Cancer research. 1979;39(6 Pt 1):2070–2076. [PubMed] [Google Scholar]
- 42.Cole SP, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258(5088):1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 43.le Coutre P, et al. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95(5):1758–1766. [PubMed] [Google Scholar]
- 44.Thomas J, et al. Active transport of imatinib into and out of cells: implications for drug resistance. Blood. 2004;104(12):3739–3745. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 45.White DL, et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood. 2007;110(12):4064–4072. doi: 10.1182/blood-2007-06-093617. [DOI] [PubMed] [Google Scholar]
- 46.Aceves-Luquero CI, et al. ERK2, but not ERK1, mediates acquired and "de novo" resistance to imatinib mesylate: implication for CML therapy. PloS one. 2009;4(7):e6124. doi: 10.1371/journal.pone.0006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delva L, et al. Resistance to all-trans retinoic acid (ATRA) therapy in relapsing acute promyelocytic leukemia: study of in vitro ATRA sensitivity and cellular retinoic acid binding protein levels in leukemic cells. Blood. 1993;82(7):2175–2181. [PubMed] [Google Scholar]
- 48.Cornic M, et al. In vitro all-trans retinoic acid (ATRA) sensitivity and cellular retinoic acid binding protein (CRABP) levels in relapse leukemic cells after remission induction by ATRA in acute promyelocytic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 1994;8(6):914–917. [PubMed] [Google Scholar]
- 49.Ozpolat B, Mehta K, Lopez-Berestein G. Regulation of a highly specific retinoic acid-4-hydroxylase (CYP26A1) enzyme and all-trans-retinoic acid metabolism in human intestinal, liver, endothelial, and acute promyelocytic leukemia cells. Leukemia & lymphoma. 2005;46(10):1497–1506. doi: 10.1080/10428190500174737. [DOI] [PubMed] [Google Scholar]
- 50.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 51.Shah NP, Sawyers CL. Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene. 2003;22(47):7389–7395. doi: 10.1038/sj.onc.1206942. [DOI] [PubMed] [Google Scholar]
- 52.Al-Ali HK, et al. High incidence of BCR-ABL kinase domain mutations and absence of mutations of the PDGFR and KIT activation loops in CML patients with secondary resistance to imatinib. The hematology journal : the official journal of the European Haematology Association / EHA. 2004;5(1):55–60. doi: 10.1038/sj.thj.6200319. [DOI] [PubMed] [Google Scholar]
- 53.Muller MC, et al. Dasatinib treatment of chronic-phase chronic myeloid leukemia: analysis of responses according to preexisting BCR-ABL mutations. Blood. 2009;114(24):4944–4953. doi: 10.1182/blood-2009-04-214221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherbenou DW, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116(17):3278–3285. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roche-Lestienne C, et al. A Mutation Conferring Resistance to Imatinib at the Time of Diagnosis of Chronic Myelogenous Leukemia. New England Journal of Medicine. 2003;348(22):2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- 56.Hofmann W-K, et al. Presence of the BCR-ABL mutation Glu255Lys prior to STI571 (imatinib) treatment in patients with Ph+ acute lymphoblastic leukemia. Blood. 2003;102(2):659–661. doi: 10.1182/blood-2002-06-1756. [DOI] [PubMed] [Google Scholar]
- 57.Willis SG, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106(6):2128–2137. doi: 10.1182/blood-2005-03-1036. [DOI] [PubMed] [Google Scholar]
- 58.Soverini S, et al. Philadelphia-positive patients who already harbor imatinib-resistant Bcr-Abl kinase domain mutations have a higher likelihood of developing additional mutations associated with resistance to second- or third-line tyrosine kinase inhibitors. Blood. 2009;114(10):2168–2171. doi: 10.1182/blood-2009-01-197186. [DOI] [PubMed] [Google Scholar]
- 59.Soverini S, et al. Philadelphia-positive acute lymphoblastic leukemia patients already harbor BCR-ABL kinase domain mutations at low levels at the time of diagnosis. Haematologica. 2011;96(4):552–557. doi: 10.3324/haematol.2010.034173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagrintseva K, et al. Mutations in the tyrosine kinase domain of FLT3 define a new molecular mechanism of acquired drug resistance to PTK inhibitors in FLT3-ITD-transformed hematopoietic cells. Blood. 2004;103(6):2266–2275. doi: 10.1182/blood-2003-05-1653. [DOI] [PubMed] [Google Scholar]
- 61.Moore AS, et al. Selective FLT3 inhibition of FLT3-ITD(+) acute myeloid leukaemia resulting in secondary D835Y mutation: a model for emerging clinical resistance patterns. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2012 doi: 10.1038/leu.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunn PA., Jr Can acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors be overcome by different small-molecule tyrosine kinase inhibitors? Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25(18):2504–2505. doi: 10.1200/JCO.2007.11.3258. [DOI] [PubMed] [Google Scholar]
- 64.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nature reviews. Cancer. 2010;10(11):760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 66.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herbst RS, Bunn PA., Jr Targeting the epidermal growth factor receptor in non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(16 Pt 1):5813–5824. [PubMed] [Google Scholar]
- 68.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 69.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer cell. 2010;17(1):77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(22):3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomson S, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer research. 2005;65(20):9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 75.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et biophysica acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 77.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 79.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. British journal of cancer. 2010;103(8):1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Science translational medicine. 2012;4(120):120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doebele RC, et al. Mechanisms of Resistance to Crizotinib in Patients with ALK Gene Rearranged Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(5):1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 83.Notta F, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469(7330):362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 84.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews. Genetics. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 86.Czech MP, Aouadi M, Tesz GJ. RNAi-based therapeutic strategies for metabolic disease. Nature reviews. Endocrinology. 2011;7(8):473–484. doi: 10.1038/nrendo.2011.57. [DOI] [PubMed] [Google Scholar]
- 87.Kittler R, Buchholz F. Functional genomic analysis of cell division by endoribonuclease-prepared siRNAs. Cell Cycle. 2005;4(4):564–567. [PubMed] [Google Scholar]
- 88.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature genetics. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 89.Root DE, et al. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nature methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 90.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 91.Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28(50):4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- 92.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137(5):821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 93.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441(7089):106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 96.Whitehurst AW, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446(7137):815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 97.Fotheringham S, et al. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer cell. 2009;15(1):57–66. doi: 10.1016/j.ccr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 98.Gregory MA, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer cell. 2010;18(1):74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burgess DJ, et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105(26):9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hahn CK, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer cell. 2009;16(4):281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pritchard JR, et al. Bcl-2 family genetic profiling reveals microenvironment-specific determinants of chemotherapeutic response. Cancer research. 2011;71(17):5850–5858. doi: 10.1158/0008-5472.CAN-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Porter CC, et al. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2012 doi: 10.1038/leu.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology / the Education Program of the American Society of Hematology. American Society of Hematology. Education Program. 2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 104.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fathi AT, Chabner BA. FLT3 inhibition as therapy in acute myeloid leukemia: a record of trials and tribulations. The oncologist. 2011;16(8):1162–1174. doi: 10.1634/theoncologist.2011-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Astsaturov I, et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Science signaling. 2010;3(140):ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bivona TG, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471(7339):523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Casas-Selves M, et al. Genome wide shRNA screens reveal that tankyrase and the canonical Wnt pathway protects lung cancer cells from EGFR inhibition. doi: 10.1158/0008-5472.CAN-11-2848. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim J, Tan AC. BiNGS!SL-seq: a bioinformatics pipeline for the analysis and interpretation of deep sequencing genome-wide synthetic lethal screen. Methods Mol Biol. 2012;802:389–398. doi: 10.1007/978-1-61779-400-1_26. [DOI] [PubMed] [Google Scholar]
- 111.Sims D, et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome biology. 2011;12(10):R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marcotte R, et al. Essential Gene Profiles in Breast, Pancreatic, and Ovarian Cancer Cells. Cancer discovery. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(51):20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nature methods. 2007;4(10):847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 115.Kanehisa M, et al. The KEGG databases at GenomeNet. Nucleic acids research. 2002;30(1):42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Luo T, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc Natl Acad Sci USA. 2012;109(8):2860–2865. doi: 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Babij C, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer research. 2011;71(17):5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- 118.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lam LT, et al. Compensatory IKKalpha activation of classical NF-kappaB signaling during IKKbeta inhibition identified by an RNA interference sensitization screen. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(52):20798–20803. doi: 10.1073/pnas.0806491106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tyner JW, et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(21):8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang H, et al. A mammalian functional-genetic approach to characterizing cancer therapeutics. Nat Chem Biol. 2011;7(2):92–100. doi: 10.1038/nchembio.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Banerji V, et al. The intersection of genetic and chemical genomic screens identifies GSK-3α as a target in human acute myeloid leukemia. The Journal of clinical investigation. 2012;122(3):935–947. doi: 10.1172/JCI46465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu YX, et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117(14):3847–3857. doi: 10.1182/blood-2010-08-304022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tibes R, et al. RNAi screening of the kinome with cytarabine in leukemias. Blood. 2012 doi: 10.1182/blood-2011-07-367557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meacham CE, et al. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nature genetics. 2009;41(10):1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer cell. 2009;16(4):324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Astier AL, et al. RNA interference screen in primary human T cells reveals FLT3 as a modulator of IL-10 levels. Journal of immunology. 2010;184(2):685–693. doi: 10.4049/jimmunol.0902443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463(7282):818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 131.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 133.Paul P, et al. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell. 2011;145(2):268–283. doi: 10.1016/j.cell.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 134.König R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130(6):986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 136.Witt AE, et al. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res. 2006;5(3):599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hattori H, et al. RNAi screen identifies UBE2D3 as a mediator of all-trans retinoic acid-induced cell growth arrest in human acute promyelocytic NB4 cells. Blood. 2007;110(2):640–650. doi: 10.1182/blood-2006-11-059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Friedberg JW, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115(13):2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]