Abstract
Lung cancer is the leading cause of death worldwide. Adenocarcinomas, the most common histological subtype of non-small cell lung cancer (NSCLC), are frequently associated with activating mutations in the epidermal growth factor receptor (EGFR) gene. Although these patients often respond clinically to the EGFR tyrosine kinase inhibitors erlotinib and gefitinib, relapse inevitably occurs, suggesting the development of escape mechanisms that promote cell survival. Using a loss-of-function, whole genome shRNA screen, we identified that the canonical Wnt pathway contributes to the maintenance of NSCLC cells during EGFR inhibition, particularly the poly-ADP-ribosylating enzymes tankyrase 1 and 2 that positively regulate canonical Wnt signaling. Inhibition of tankyrase and various other components of the Wnt pathway with shRNAs or small molecules significantly increased the efficacy of EGFR inhibitors both in vitro and in vivo. Our findings therefore reveal a critical role for tankyrase and the canonical Wnt pathway in maintaining lung cancer cells during EGFR inhibition. Targeting the Wnt-tankyrase-β-catenin pathway together with EGFR inhibition may improve clinical outcome in patients with NSCLC.
Keywords: tankyrase, β-catenin, EGFR, casein kinase, CK1, CK2, synthetic lethal
Introduction
Lung cancer is the leading cause of cancer death in the United States (1). NSCLC account for ~82% of lung cancers (2). Because most NSCLC are diagnosed at late stages, prognosis is quite poor. Even when combining newer agents, such as bevacizumab with traditional chemotherapy, the median overall survival of patients with metastatic NSCLC remains less than 1 year, and only 3.5% of NSCLC patients survive 5 years after diagnosis (3, 4).
Adenocarcinomas, the most common histological type of NSCLC, are frequently (40–80%) associated with mutations and/or amplifications in the EGFR gene (5). For patients with activating mutations in EGFR, the EGFR tyrosine kinase inhibitors (TKI) erlotinib and gefitinib provide better treatment than conventional chemotherapy, with impressive response rates (>70%) and prolonged progression-free survival (2, 6–8). However the duration of responses are short-lived, and all patients eventually relapse.
The dysregulation of the Wnt/β-catenin pathway has been observed in various forms of cancer. Approximately 90% of sporadic colon cancers exhibit aberrant Wnt signaling activity, usually as a result of Adenomatous Polyposis Coli (APC) gene mutations, but also due to mutations in genes encoding β-catenin or Axin (9). Importantly, it has been recently shown that altered β-catenin expression, or Wnt1, Wnt3a and Wnt5a overexpression, are associated with poor prognosis in NSCLC patients (10). Thus, the Wnt/β-catenin pathway may play an important role in NSCLC pathogenesis and resistance to therapy.
Secreted Wnt family proteins bind to specific Frizzled (Fzd) receptor complexes on cells and activate intracellular pathways (11, 12). The canonical Wnt pathway regulates the activity of β-catenin and its ability to drive activation of specific gene targets. In the absence of a Wnt signal, β-catenin is targeted for degradation. APC and Axin compose a scaffold that permits β-catenin phosphorylation by Casein Kinase 1α (CK1α) and Glycogen Synthase Kinase 3β (GSK3β), resulting in β-catenin ubiquitinylation and proteosomal degradation. Conversely, upon binding of a Wnt ligand to the Fzd receptor, Axin is shuttled to the cell membrane, preventing β-catenin degradation. Accumulating β-catenin can then enter the nucleus, and upon interaction with members of the Tcf/Lef transcriptional activators, facilitates the expression of various gene targets. Interestingly, the poly-ADP-ribosylating enzymes tankyrases 1 and 2, which play various roles in the cell including in telomere maintenance (13), have recently been shown to positively regulate canonical Wnt signaling (14).
While targeted therapies, such as using EGFR TKI, have shown clinical promise, these therapies do not produce durable remissions for NSCLC. While in some cases secondary mutations in the kinase may prevent inhibitor binding, additional escape mechanisms are mostly poorly defined. As lung cancers are typically diagnosed at advanced phases and appear to possess inherent or acquired survival mechanisms that can protect cells from EGFR inhibition, the discovery of pathways that mediate compensatory survival mechanisms could reveal novel therapeutic targets that render pharmacological EGFR inhibitors more effective for the treatment of lung cancer. For this reason, we carried out a genome-wide shRNA screen to identify gene products whose inhibition synergizes with the EGFR inhibitor gefitinib to eliminate NSCLC cells. Our screen identified a number of druggable gene products within the Wnt/tankyrase/β-catenin pathway as mediators of NSCLC maintenance during EGFR inhibition.
Materials and Methods
Genome-wide functional genetic screening
1×107 cells from each cell line were transduced with the SBI shRNA library (multiplicity of infection of ~0.3) and 72h later subjected to puromycin selection (1 µg/mL) for 10 days. Cells (1×107) were treated in triplicate with gefitinib at IC70 (0.6 µM for H322C; 0.05 µM for HCC4006) or vehicle (DMSO) for 48h followed by culture for 96h without drugs. Total RNA was isolated from each group using Trizol (Invitrogen) and reverse transcribed (using vector-specific primer) with M-MLV reverse transcriptase (Epicenter). The cDNA was amplified by nested PCR, with addition of Illumina-specific adapter sequences. After sequencing on an Illumina GenomeAnalyzer (Illumina, San Diego, CA), shRNAs were identified and the number of clusters for each shRNA sequence quantified.
Bioinformatics analysis
We developed BiNGS! (Bioinformatics for Next Generation Sequencing) for analyzing and interpreting synthetic lethal screen data (15). A preprocessing step filtered out erroneous and low quality reads. Filtered reads were mapped against the shRNA reference library using Bowtie (16). Output from this step is a P×N matrix, where P and N represent shRNA counts and samples, respectively. A secondary filtering step removes shRNA reads mapping to sequences without gene annotations. We also filtered out shRNAs where the median raw count in the control group is greater than the maximum raw count in the treatment group if the shRNA is enriched in the control group, and vice versa. We then employed Negative Binomial to model the count distribution in the sequencing data using edgeR (17). We computed the q-value of False Discovery Rate for multiple comparisons for these shRNAs, and performed meta-analysis by combining adjusted p-values for all shRNAs representing the same gene using weighted Z-transformation (18). We used the associated p-value [P(wZ)] to sort lists of genes with differentially represented shRNAs.
Cell lines
Calu-3 cells were obtained from ATCC (Rockville, MD) in 2002. Drs. John Minna and Adi Gazdar (University of Texas Southwestern Medical School, Dallas, TX) provided HCC4006 cells in 2006. H322C were obtained from Dr. Al Moustada (Biotechnology Research Institute, Montreal, Quebec, Canada) in 2003. H3255 and H3122 were obtained from Dr. Bruce Johnson and Dr. Pasi Janne (Dana-Farber Cancer Institute, Boston, MA) in 2004 and 2008, respectively. Calu-3, HCC4006, H322C, H3255 and H3122 were last authenticated by STR DNA profiling in February 2012 by our Molecular Biology Core. The STR profiles for these lines matched the profiles on file at the ATCC (Calu-3) and profiling data provided by Drs. Minna and Gazdar (HCC4006, H322C, H3255 and H3122).
Nude mouse xenograft tumor model
Athymic nude mice (4–6 week-old females) were obtained from the National Cancer Institute (Bethesda, MD). Cultured cells (2×106) were injected into the flanks of mice at day 0. Vehicle or gefitinib was injected i.p. daily (5/7 days). Tumor volumes were evaluated by caliper measurement and calculated by the formula: π × (short diameter)2 × (long diameter)/6. H&E and Ki67 staining were performed using standard protocols.
Graphing and statistical analyses
Graphing and statistical analysis were carried out using PRISM-5 software. Unless otherwise indicated, 2-tailed Student’s t-test was used.
See Supplemental Methods for descriptions of methods for lentivirus preparation, cell culture, colony forming assays, cell viability/cell cycle/apoptosis/senescence assays, western blotting, RT-PCR and chemicals used.
Results
In order to identify gene targets whose inhibition cooperates with gefitinib to more effectively eliminate NSCLC cells, we designed a genome-wide RNAi-based loss-of-function screen (Figure 1A). We generated EGFR TKI sensitive NSCLC cell lines H322C and HCC4006 expressing lentiviral-encoded shRNAs targeting all known human genes, with most cells expressing a single shRNA. H322C, a bronchioalveolar carcinoma with wild-type EGFR, exhibits intermediate sensitivity to gefinitib (IC50 0.25µM). HCC4006, an adenocarcinoma which possesses an amplified EGFR gene with exon 19 deletion (an activating mutation), exhibits high sensitivity to gefitinib (IC50 0.02µM) (19).
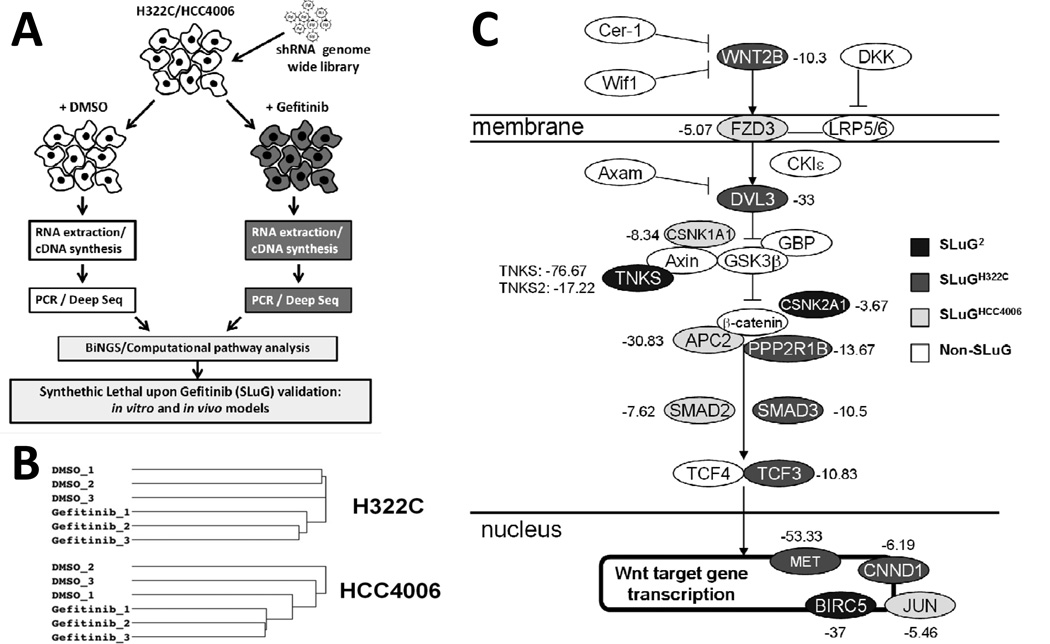
FIGURE 1. Genome-wide RNAi-based screen for synthetic lethal interactions with EGFR inhibition.
A) Overview of screen and data analyses. B) Unsupervised hierarchical clustering of mapped sequences from individual samples. C) Mapping of SLuGs onto the Wnt/Tankyrase/β-catenin pathway. The number listed to the side of each component is the average fold-change of the most differentially represented shRNA targeting the indicated gene from gefitinib-treated NSCLC cells relative to untreated.
Each cell line was divided into 6 populations: 3 were treated with DMSO and 3 were treated with gefitinib for 48 hr at doses that inhibit expansion by ~70%, followed by 96 hours of culture without drug. shRNA sequences were amplified, sequenced, identified and statistically analyzed as described in Materials and Methods. Over 5 million shRNAs were read per sample, representing more than 60,000 unique shRNAs. Unsupervised hierarchical clustering of mapped shRNA sequences from the individual samples revealed that DMSO samples clustered together and gefitinib treated samples clustered together (Figures 1B and S1). Thus, gefitinib treatment is reproducibly affecting the representation of shRNAs; therefore, this screen should reveal shRNA targeted gene products whose knockdown either increases or decreases gefitinib mediated inhibition of NSCLC cells.
For synthetic lethality, we considered genes targeted by shRNAs that were depleted in the treatment group with p-values [P(wZ)] of <0.05 as synthetic lethal hits (1237 and 748 genes each for H322C and HCC4006; Table S1). Importantly, we identified 104 genes with significant underrepresentation in both NSCLC cell lines (Table S1). As the inhibition of shRNA targeted genes sensitizes NSCLC cells to gefitinib, we have dubbed these genes as “SLuG”s, for Synthetic Lethal upon Gefitinib. The 104 synthetic lethal genes found in both cell lines are denoted as SLuG2, and those identified in a single cell line will be labeled SLuGH322C and SLuGHCC4006. Note that SLuG2s are enriched in cancer-related genes (see legend of Table S1).
Among identified SLuGs were multiple genes with known roles in protecting NSCLC or other cancer cells from EGFR inhibition, which provides confidence in the screening results. Examples include: i) MET (c-Met; SLuGH332C): amplification of which has been shown to contribute to pre-existing and acquired clinical resistance to EGFR inhibitors (20–22). ii) PIK3CA (SLuGH332C and phosphoinositide-3-kinase α subunit): pharmacological inhibition of the PI3K/Akt/mTOR pathway restores sensitivity in gefitinib resistant tumor cell lines, forming the basis of a current Phase I trial combining gefitinib with everolimus (PI3K/Akt/mTOR pathway inhibitor) (23). iii) Fibroblast growth factor 2 (FGF2) and the FGF receptor FGFR2 (both SLuGH332C): FGFR signaling has been shown to confer intrinsic resistance to EGFR inhibition (24–26) . iv) BIRC5 (SLuG2): overexpression of BIRC5/survivin has been shown to protect lung cancer cells from gefitinib-induced apoptosis, and has been implicated in conferring resistance to gefitinib (27) . v) PTPN11 (SHP2; SLuG2): Inhibition of SHP2 has been shown to sensitize to gefitinib (28), and vi) SOS2 (SLUGHCC4006): SOS2 was recently discovered in a synthetic lethal siRNA screen with EGFR inhibition in a cervical adenocarcinoma cell line (29).
Importantly, multiple SLuG gene products mapped throughout the Wnt/tankyrase/β-catenin pathway (Figure 1C). In particular, TNKS1 was present as a SLuG2 and TNKS2 was identified as a SLuGHCC4006. These poly-ADP-ribosylating enzymes have recently been shown to be key positive regulators of β-catenin dependent transcription by promoting the degradation of Axin (14). The SLuG2 CSNK2A1 (casein kinase 2α; CK2α) has been shown to promote canonical Wnt signaling by phosphorylation of β-catenin, leading to decreased degradation and increased transcriptional activity (30). Moreover, high expression of CSNK2A1 is predictive of poor prognosis in NSCLC patients (31). Additional SLuGs in this Wnt pathway include SLuGH322C genes TCF3, DVL3, SMAD3, CCND1, WNT2B, PP2A and DVL3, as well as SLuGHCC4006 genes LRP8, FZD3, SMAD2, JUN, CSNK1A1 (CK1α), and APC2. Note that pathway genes include several β-catenin/TCF target genes, including the SLuG2 BIRC5. Thus, the inhibition of numerous components of the canonical Wnt signaling pathway appears to potentiate gefitinib mediated elimination of NSCLC cells.
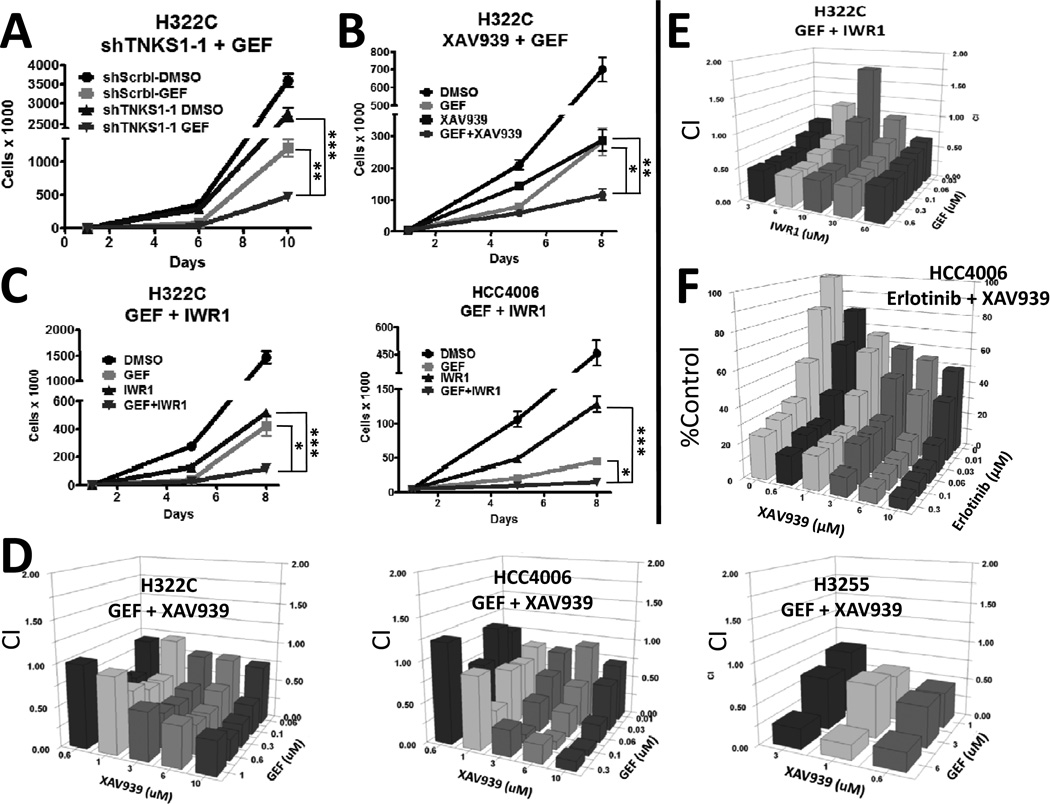
To validate screening results, we generated H322C sublines expressing either negative control shRNAs or shRNAs targeting TNKS1. Vectors from the TRC collection were used for validation of TNKS1 and other SLuGs (in each case, pools of retroviral integrants), with shRNA sequences distinct from those used for the screen. As expected, knockdown of TNKS1 decreased mRNA levels of AXIN-2 (Figure S2B), a well characterized target of Wnt/β-catenin-dependent transcription (32). While TNKS1 knockdown had a mild effect on cell expansion by itself, when coupled with gefitinib-mediated EGFR inhibition, NSCLC cell expansion was greatly inhibited (Figure 2A and S2A). We next asked whether pharmacological inhibition of tankyrase activity, using the small molecule inhibitor XAV939 (14), could also sensitize NSCLC cells to EGFR inhibition. As shown in Figure 2B and S2C, while exposure of H322C and HCC4006 cells to XAV939 or gefitinib alone moderately inhibited proliferation, combination of the two drugs led to more potent inhibition of cell expansion. Additionally, pharmacological stabilization of Axin using endo-IWR1, which also reduces β-catenin levels by inhibiting tankyrase (14, 33), also potentiated gefitinib mediated elimination of H322C and HCC4006 cells (Figure 2C). We validated the synergistic interaction of gefitinib and XAV939 or endo-IWR1 using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays in multiple NSCLC lines with varying sensitivities to gefitinib (Figure 2D, 2E, S2D and S2E), including H3255 (with activating EGFR mutation L858R) and Calu3 (with increased wild-type EGFR gene copy number) (19). Note that synergism is strongest in the EGFR mutant lines. Importantly, we further demonstrated that treatment with the FDA approved EGFR inhibitor erlotinib also synergized with XAV939 to reduce NSCLC cell expansion (Figure 2F and S2E). Interestingly, cell lines that are not driven by EGFR mutation or amplification, such as H3122 (ALK fusion) and human foreskin fibroblasts (HFF) are not affected by the combinatorial treatment (Figure S2F and S2G) and tankyrase inhibition does not synergize with the first line NSCLC chemotherapeutic paclitaxel (Figure S2H), thus suggesting a unique reliance on Wnt signaling in EGFR-dependent cancers following EGFR inhibition. Finally, we showed that combined treatment of H322C or HCC4006 cells with gefitinib and either XAV939 or endo-IWR1 resulted in enhanced elimination of colony forming ability (Figure 3A).
FIGURE 2. Combined inhibition of EGFR and tankyrase leads to synergistic inhibition of NSCLC cells.
A) H322C cells expressing either negative control shRNA (shScrbl) or shRNAs targeting TNKS1 (TNKS1-1) were treated in triplicate with vehicle (DMSO) or 500 nM gefitinib (GEF) for 4 days, followed by replating without drug for 4 more days. Viable cells were counted by flow cytometry using PI-exclusion. B) H322C cells were treated in triplicate with DMSO or 500 nM gefitinib and/or the TNKS inhibitor XAV939 (5 µM) for 4 days, and then replated without drugs for 3 additional days. Viable cells were counted. C) H322C and HCC4006 cells were treated in triplicate with DMSO or gefitinib (500 nM for H322C or 20 nM for HCC4006) and/or the TNKS inhibitor endo-IWR1 (IWR1; 20 µM) for 4 days, and then replated without drugs for 3 days. Viable cells were counted. For A–C, * indicates p<0.05, ** p<0.01, and *** p<0.001. D) The indicated NSCLC cell lines were treated in triplicate with DMSO, gefitinib and/or XAV939 at the indicated concentrations for 5 days, and cell viability assessed by MTT assays. Graphs were plotted for Combination Indices (CI) using to determine additivity (CI=1), synergism (CI<1) and antagonism (CI>1). E) As in D, H322C cells were treated in triplicate with DMSO, gefitinib and/or endo-IWR1. F) HCC4006 cells were treated in triplicate with DMSO, erlotinib and/or XAV939 as in D. Graph was plotted in comparison to untreated cells.
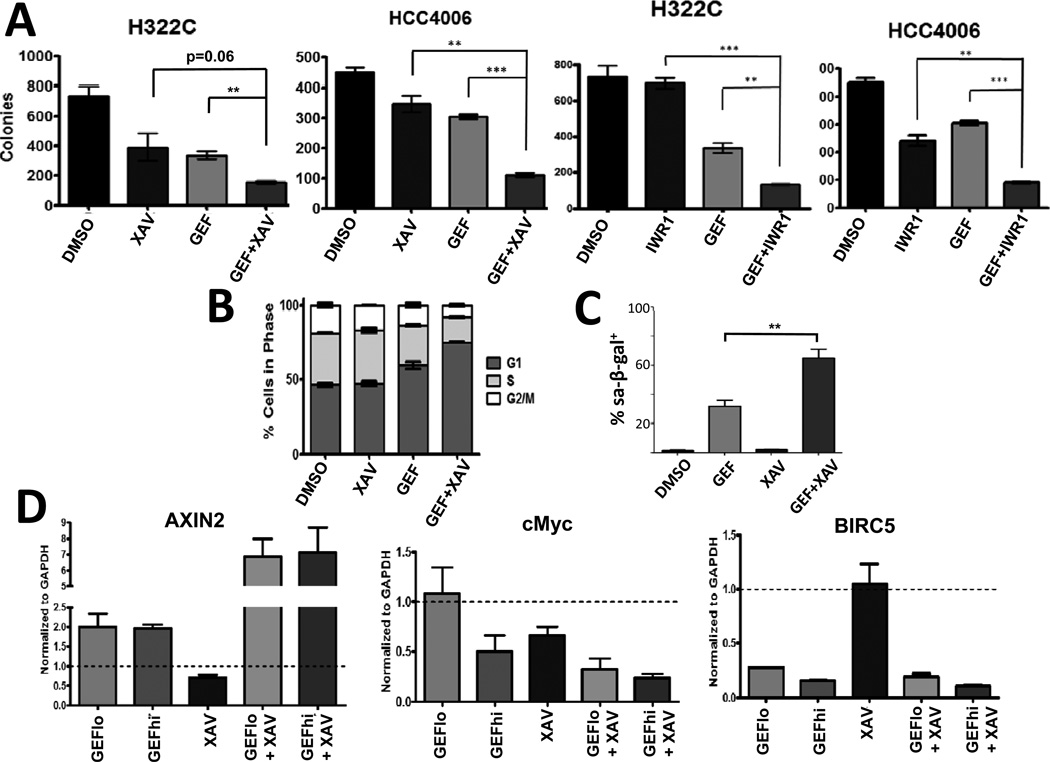
FIGURE 3. Combined inhibition of EGFR and tankyrase permanently impairs clonogenic outgrowth of NSCLC cells.
A) H322C and HCC4006 cells were treated in triplicate with DMSO or gefitinib (500 nM for H322C; 20 nM for HCC4006) and/or the TNKS inhibitors XAV939 (5 µM) or endo-IWR1 (20 µM) for 4 days, and then replated without drugs for colony forming assays. B) H322C cells were treated as in Figure 2B for 72h and cell cycle profiles analyzed by saponin/PI staining. C) H322C cells were treated with DMSO or gefitinib (500 nM) and/or XAV939 (5 µM) for 5 days, and then stained for senescence-associated β-galactosidase (sa-β-gal) activity. For A and C, * indicates p<0.05, ** p<0.01, and *** p<0.001. D) HCC4006 cells were treated for 24 hours with DMSO or gefitinib (30 nM/GEFlo or 60 nM/GEFhi) and/or XAV939 (3 µM) (n=3/condition), and AXIN-2, BIRC5 and MYC mRNA levels determined by RT-PCR. Dotted line at y=1 represents levels in DMSO-treated samples.
While XAV939 alone did not affect cell cycle profiles, H322C cells treated with gefitinib exhibited an increase of cells in G1 and a decrease in S phase (Figure 3B), as expected. Notably, cells treated with both gefitinib and XAV939 exhibited an even greater number of cells in G1 phase with a corresponding decrease in S and G2/M phases relative to gefitinib alone. While gefitinib treatment alone does increase the proportion of H322C cells undergoing apoptosis (Figure S3A), combined treatment with XAV939 only modestly increased the presence of apoptotic cells detected by Annexin-V staining. But notably, the combination treatment resulted in more cells with senescence-associated β-galactosidase staining than gefitinib alone (Figure 3C and S3B), indicating that the combinatorial treatment causes increased senescence to H332C cells. Moreover, known Wnt/β-catenin targets (AXIN-2, Myc and BIRC5) are differentially modulated by the inhibition of EGFR and TNKS (Figure 3D). As expected, AXIN-2 mRNA levels are decreased by XAV939 treatment, but surprisingly increased two-fold by gefitinib. Interestingly, the combination of gefitinib and XAV939 induce a synergistic surge of AXIN-2 mRNA levels. While Myc mRNA levels are reduced by either gefitinib or XAV939 alone, the combinatorial treatment results in further reductions in Myc message levels. Lastly, BIRC5 mRNA levels are reduced by gefitinib treatment, but appear unaffected by XAV939 alone. In summary, our results indicate that inhibiting both EGFR and TNKS synergistically impairs the proliferation of NSCLC cells, correlating with a marked increase in senescence, and altered mRNA levels of various Wnt target genes (model in Figure S3C).
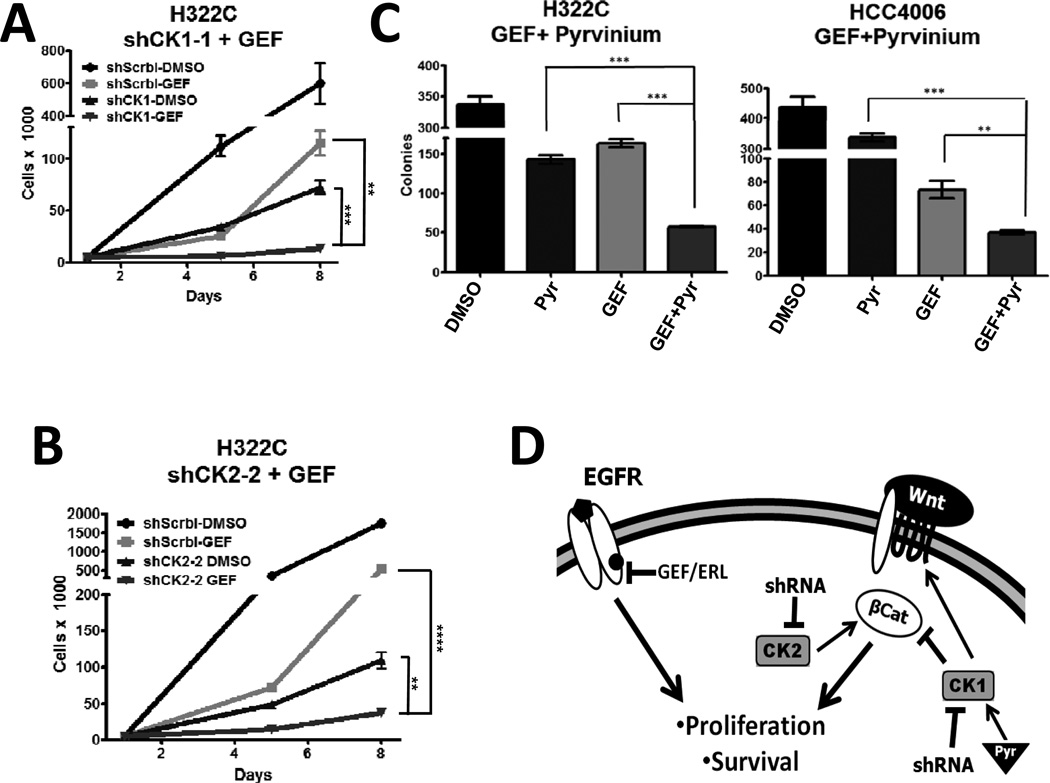
We next sought to validate other SLuGs that also mapped to the canonical Wnt pathway. Wnt signaling is tightly controlled with multiple levels of feedback. For example, whereas CK1α can phosphorylate β-catenin and thus target it for destruction (34), it also can destabilize the destruction complex, thus promoting canonical Wnt signaling (35). As shown in Figure 4A–4B and S4A–S4E, knockdown of either CSNK1A1 (CK1α) or CSNK2A1 (CK2α) impaired the proliferation and colony forming ability of H322C cells, and sensitized these cells to EGFR inhibition with gefitinib. Interestingly, pyrvinium, a small-molecule CK1α activator recently demonstrated to inhibit the Wnt/β-catenin pathway (36) (EC50~10 nM), synergistically increases gefitinib-mediated inhibition of H322C and HCC4006 cells (Figure 4C, S4F–S4G). Pyrvinium has been an FDA approved an anti-helminth drug for >50 years, and is a potent, specific and direct activator of CK1α (36). While it may seem counterintuitive that both activation (with pyrvinium) and inhibition (with shRNAs) of CK1α would sensitize NSCLC cells to gefitinib, as mentioned above, previous studies have also suggested dual roles for CK1α in both positive and negative modulation of Wnt signaling (37). Notably, pyrvinium does not synergize with paclitaxel (Figure S4H) in NSCLC cells or with gefitinib in HFF (Figure S4I).These data reinforce the importance of appropriate regulation of Wnt signaling for NSCLC maintenance during EGFR inhibition (Figure 4D).
FIGURE 4. Simultaneous disruption of EGFR and either CK1 or CK2 signaling act synergistically to inhibits NSCLC cell expansion.
A) H322C cells expressing a negative control shRNA (shScrbl) or shRNAs targeting CSNK1A1 (CK1α) were treated with DMSO or 500 nM gefitinib for 4 days, followed by replating without drug for 3 days. Viable cells were counted. B) As in A, except H322C cells expressed either shScrbl or shRNAs targeting CSNK2A1 (CK2α). C) H322C cells and HCC4006 were treated in triplicate with DMSO or gefitinib (500 nM for H322C; 20 nM for HCC4006) and/or pyrvinium (Pyr; 10 nM for H322C; 2 nM for HCC4006) for 4 days and then replated without drugs for colony forming assays. For A–C, ** indicates p<0.01, and *** p<0.001. D) Proposed model.
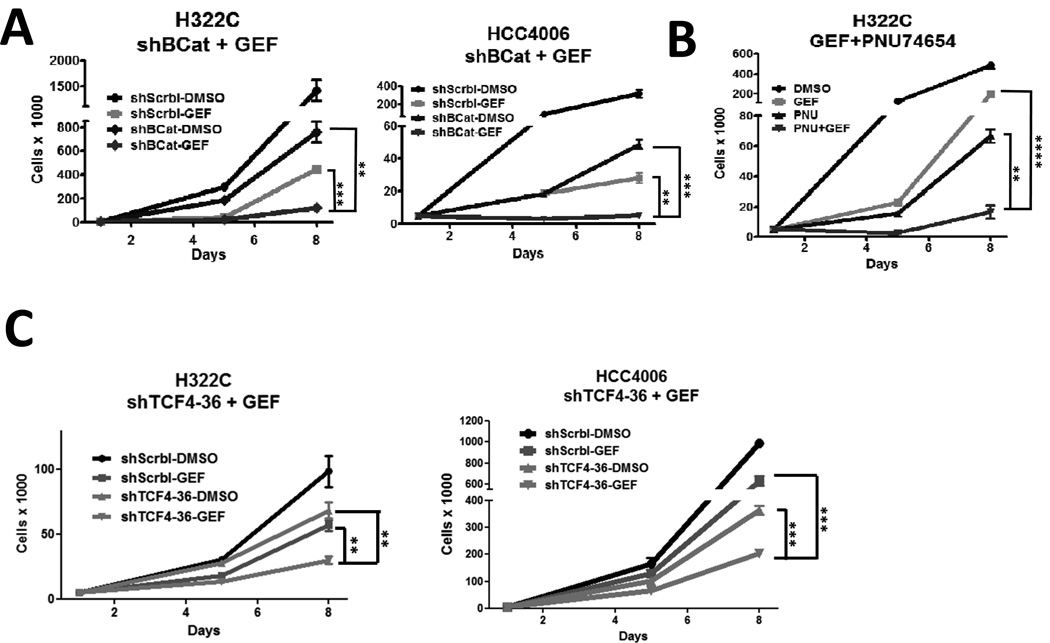
Given the apparent importance of Wnt signaling in residual cell maintenance during gefitinib treatment, we directly assessed the contribution of β-catenin, even though it was not identified as a SLuG. Even modest knockdown of β-catenin had an inhibitory effect on cell expansion by itself, but when coupled with gefitinib, cell expansion was greatly inhibited (Figure 5A, S5A and S5B). Combined treatment with gefitinib and a recently described small molecule, PNU74654, that can disrupt the interaction between β-catenin and TCF4 (38), similarly led to enhanced inhibition of NSCLC expansion (Figure 5B). Moreover, knockdown of TCF4 also potentiated gefitinib efficacy in both H322C and HCC4006 lines (Figure 5C and S5C). In summary, our results indicate that blocking both EGFR and Wnt/β-Catenin pathways leads to enhanced inhibition of NSCLC cells.
FIGURE 5. Combined inhibition of EGFR and β-catenin suppresses NSCLC cell expansion.
A) H322C and HCC4006 cells expressing either shScrbl or shRNAs targeting β-catenin (shBCat-4) were treated with DMSO or gefitinib (500 nM for H322C; 20 nM for HCC4006) for 4 days, followed by replating without drug for 3 more days. B) H322C cells were treated in triplicate with DMSO or gefitinib (500 nM) and/or PNU74654 (PNU; 20 µM) for 4 days and then replated without drugs for 3 days. Viable cells were counted. C) H322C and HCC4006 cells expressing either shScrbl or shRNAs targeting TCF4 (shTCF4-36) were treated with DMSO or gefitinib as in Figure 5A. ** indicates p<0.01, and *** p<0.001.
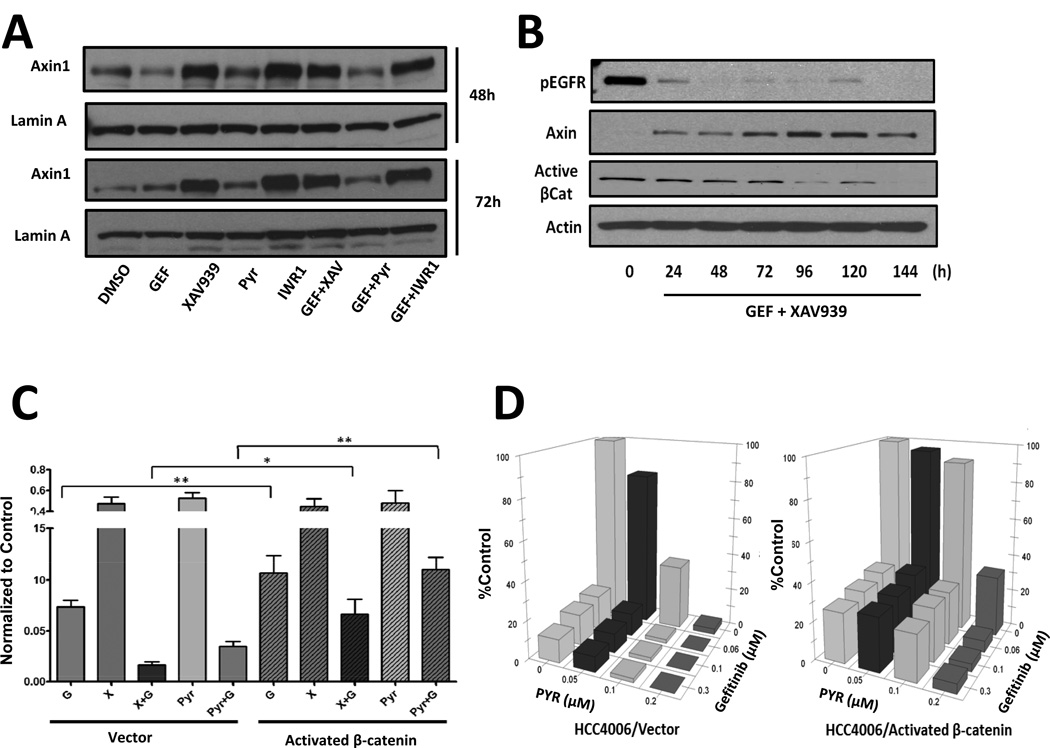
We next asked whether pharmacological inhibition of the Wnt/tankyrase/β-catenin pathway could cooperate with gefitinib by further inhibiting EGFR phosphorylation. As shown in Figure S6A, the inhibition of activating phosphorylation of EGFR and ERK, which is downstream of EGFR, following gefitinib treatment in HCC4006 and H322C cells was not affected by the addition of XAV939, pyrvinium or PNU74654. Thus, the ability of Wnt/tankyrase/β-catenin pathway inhibitors to sensitize NSCLC cells to gefitinib does not appear to be due to augmented EGFR pathway inhibition. Moreover, XAV939 and endo-IWR1 treatments resulted in the expected increase in AXIN-1 protein levels (Figure 6A and 6B). Finally, all small molecules used to disrupt Wnt signaling (XAV939, pyrvinium and PNU74654) decrease AXIN-2 mRNA levels (Figure S6B).
FIGURE 6. Activated β-catenin protects from combinatorial therapies.
A) HCC4006 cells were treated as in Figure 3A and 4C with the indicated drugs for 48 or 72 hours, and then analyzed by western blotting for AXIN-1 or Lamin-A. B) HCC4006 cells were treated with gefinitib plus XAV939 as in A for the indicated times, and blotted for AXIN-1, activated β-catenin or Actin. C) HCC4006 cells expressing vector or activated β-catenin were treated as in A with the indicated drugs for 4 days and replated for 3 days without drugs. Viable cell counts were graphed, normalized to DMSO treatment. D) HCC4006 cells expressing vector or activated β-catenin were treated in sextuplicate with DMSO, gefitinib and/or pyrvinium as indicated for 5 days, and cell viability assessed by MTT assays. * indicates p<0.05, ** p<0.01, and *** p<0.001.
While pharmacological manipulation of the Wnt/tankyrase/β-catenin pathway has clear relevance for potential clinical applications, a caveat with the use of small-molecule inhibitors is their inevitable off-target effects. Moreover, the inhibition of enzymes like tankyrase and CK1α will impact cellular processes beyond Wnt signaling (37, 39). In order to determine the extent to which the efficacy of these inhibitors in cooperating with gefitinib to impair NSCLC cell proliferation was through inhibition of the canonical Wnt pathway, we generated HCC4006 cells that stably express an activated form of β-catenin (S33A/S37A/T41A/S45A) (40) (Figure S6C). As shown in Figure 6C, expression of activated β-catenin not only partially protects NSCLC cells from gefitinib treatment, but provides substantial protection from combined treatments with either XAV939+gefitinib or pyrvinium+gefitinib. This β-catenin rescue is evident over a range of concentrations for gefitinib, XAV939 and pyrvinium (Figure 6D and S6D). Thus, the expression of activated β-catenin obviated the effects of pyrvinium, and to a partial extent XAV939, in terms of increasing HCC4006 inhibition following gefitinib treatment. Nonetheless, it is notable that activation of β-catenin cannot fully rescue growth inhibition by high dose pyrvinium or XAV939 in the absence of gefitinib (Figure 6C and 6D), which could represent off-target effects of these drugs or impacts of tankyrase inhibition beyond the β-catenin pathway. Still, the ability of activated β-catenin expression to largely bypass effects of the tankyrase inhibitor XAV939 and the CK1α activator pyrvinium on enhancing gefitinib-mediated NSCLC inhibition indicates that these small molecules, at least in good measure, act by inhibiting the Wnt/β-catenin pathway.
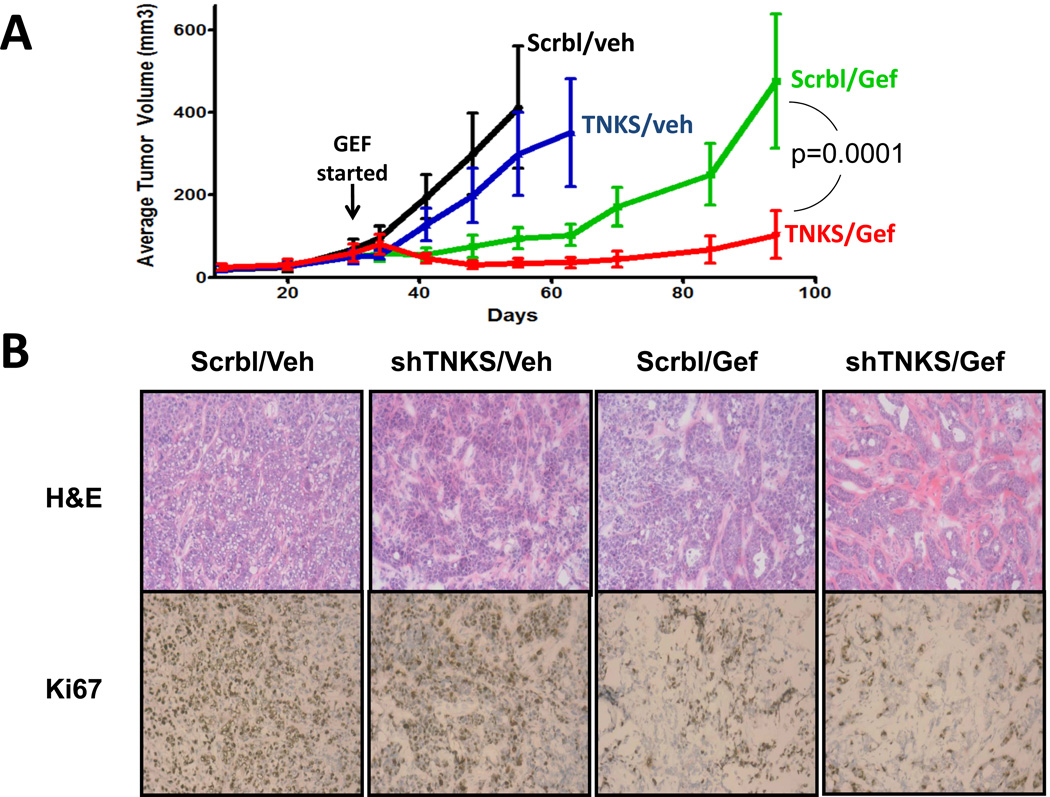
Finally, we asked whether the inhibition of tankyrase could sensitize NSCLC cells to EGFR inhibition in vivo. We injected nude mice subcutaneously with H322C cells expressing either negative control shRNAs (Scrbl) or shRNAs targeting TNKS1. As shown in Figure 7A and S7, gefitinib treatment delayed, but did not prevent, the outgrowth of the Scrbl shRNA expressing tumor cells. In contrast, tumors expressing shRNAs targeting TNKS1 showed a much more substantial inhibition of tumor growth with gefitinib. H&E and Ki67 staining revealed that tumors from gefitinib treated mice exhibited reduced cell density and proliferation, with greater reductions in cell density for TNKS1 shRNA expressing tumors (Figure 7B).
FIGURE 7. Knockdown of Tankyrase-1 sensitizes NSCLC tumors to EGFR inhibition in mice.
A) Nude mice with H322C xenografts expressing shRNAs targeting TNKS or control shRNAs were treated with vehicle (veh) and/or gefitinib (25 mg/kg/d), and tumors measured at least weekly (n=8/group).B) H&E (top) and Ki67 (bottom) staining of tumors from the indicated treatment group. Anova statistical comparison of curves is shown.
Discussion
Our results reveal that the Wnt/β-catenin pathway is critical for NSCLC cell maintenance upon EGFR inhibition. Given that key enzymatic regulators of the Wnt/β-catenin pathway, including tankyrase, can be inhibited by small molecules, these studies reveal potential mechanisms to increase the efficacy of clinically-used EGFR inhibitors like gefitinib and erlotinib for the treatment of NSCLC. It is important to emphasize that our screen was not designed to discover acquired resistance mechanisms, but to reveal pathways that maintain some EGFR-dependent NSCLC cells despite EGFR inhibition. Clinical studies thus far have shown efficacy for small molecule EGFR inhibitors only for patients with NSCLC bearing activating EGFR mutations (~15% of cases) (2, 6), and even in these cases relapse is inevitable. Moreover, our experiments indicate that targeting tankyrase and the Wnt/β-catenin pathway may increase the efficacy of EGFR inhibitors for NSCLC that overexpress wild-type EGFR, which could expand the number of patients that benefit from anti-EGFR therapies.
RNAi screens provide powerful tools to identify novel compensatory pathways upon TKI treatment (29, 41–43). Our results highlight the strength of unbiased, genome-wide analyses, which provides unanticipated insight into the pathways that maintain NSCLC cells. Notably, we did not find significant correlations between our SLuG2 list with genes that are regulated following gefitinib treatment or genes known to be mutated in lung cancers, and we found only very weak correlations with canonical Wnt target genes and gefitinib resistance of NSCLC lines (Figure S5D and legend). These results emphasize the importance of functional screens, as a pathway does not need to be mutated or deregulated to contribute to therapy resistance or maintenance of the cancer phenotype (44). Importantly, tankyrases have been identified by many pharmaceutical companies as attractive targets for drug development (45), and our studies reveal a potentially valuable application of tankyrase inhibitors for the treatment of lung cancers.
Our results suggest that inhibition of EGFR and tankyrases could each impact β-catenin-dependent transcription through divergent mechanisms. While XAV939 (but not gefitinib) treated cells exhibited the expected increase in AXIN-1 protein levels, gefitinib treatment resulted in increases in AXIN-2 mRNA levels, with further increases when combined with XAV939, coinciding with decreased expression of Wnt/β-catenin target genes. Together with previous demonstrations of crosstalk between EGFR and canonical Wnt pathways (46, 47), these data suggest a more complicated impact of EGFR and tankyrase inhibitors on Wnt/β-catenin-dependent transcription than originally anticipated.
Clinical experience thus far has indicated that single-agent, molecularly-targeted chemotherapy will not be curative for NSCLC, or indeed for any advanced cancer. Given the molecular complexity of these cancers, compensatory survival pathways can mediate partial cancer cell maintenance and lead to drug resistance. As shown in a recent report (48), there is considerable heterogeneity in acquired resistance mechanisms in lung cancers which become refractory to EGFR inhibitors, and one might envision that each patient’s cancer will require an individualized therapy. Indeed, activation of the Wnt/tankyrase/β-catenin pathway should be examined as one potential mechanism of acquired resistance to EGFR targeted therapies. Moreover, we would suggest that there are also pathways, such as the Wnt/tankyrase/β-catenin pathway, which may mediate intrinsic resistance to targeted therapies, such as with EGFR inhibitors, in a wider range of NSCLC. Non-oncogene addiction of cancers (44), particularly when specific to a context (such as EGFR inhibition) that the cancer would presumably not have evolved to avoid prior to treatment, may be especially attractive for therapeutic targeting together with the mutationally-activated primary pathway of oncogene addiction.
In conclusion, our studies reveal the effectiveness of genome-wide shRNA screens to discover resistance signaling networks and to help exploit druggable pathways whose inhibition can potentiate current therapeutics for cancer. In particular, our results strongly implicate the Wnt/tankyrase/β-catenin pathway as a promising therapeutic target for NSCLC when combined with EGFR inhibition.
Supplementary Material
Acknowledgements
We thank Mark Gregory, Courtney Fleenor, Joaquin Espinosa, Kelly Sullivan and Lynn Heasley for their suggestions and critical review of the manuscript, and Cancer Center Functional Genomics, Flow Cytometry, and Biostatistics/Bioinformatics Shared Resources. We also thank AstraZeneca for the gift of gefitinib.
Grant support: Studies were supported by RO1-CA157850, pilot funding from NCI SPORE P50-CA058187 and by Uniting Against Lung Cancer. Cancer Center Shared Resources are supported by NIH grant 2-P30-CA46934.
Footnotes
Conflict of interest: The authors have no conflicts of interest relevant to these studies.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapp E, Pater JL, Willan A, Cormier Y, Murray N, Evans WK, et al. Chemotherapy can prolong survival in patients with advanced non-small-cell lung cancer--report of a Canadian multicenter randomized trial. J Clin Oncol. 1988;6:633–641. doi: 10.1200/JCO.1988.6.4.633. [DOI] [PubMed] [Google Scholar]
- 4.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 5.Fujino S, Enokibori T, Tezuka N, Asada Y, Inoue S, Kato H, et al. A comparison of epidermal growth factor receptor levels and other prognostic parameters in non-small cell lung cancer. Eur J Cancer. 1996;32A:2070–2074. doi: 10.1016/s0959-8049(96)00243-2. [DOI] [PubMed] [Google Scholar]
- 6.Bunn PA., Jr Can acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors be overcome by different small-molecule tyrosine kinase inhibitors? J Clin Oncol. 2007;25:2504–2505. doi: 10.1200/JCO.2007.11.3258. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117:2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 10.Nakashima T, Liu D, Nakano J, Ishikawa S, Yokomise H, Ueno M, et al. Wnt1 overexpression associated with tumor proliferation and a poor prognosis in non-small cell lung cancer patients. Oncol Rep. 2008;19:203–209. [PubMed] [Google Scholar]
- 11.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 12.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 13.Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Tan AC. BiNGS!SL-seq: A Bioinformatics Pipeline for the Analysis and Interpretation of Deep Sequecing Genome-wide Syntehtic Lethality Screen. In: Wang ACCT Junbai, Tian Tianhai., editors. Methods in Molecular Biology: Next Generation Microarray Bioinformatics : Methods and Protocols. Springer; 2012. pp. 389–398. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 19.Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, Bemis L, et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- 20.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 22.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milton DT, Riely GJ, Azzoli CG, Gomez JE, Heelan RT, Kris MG, et al. Phase 1 trial of everolimus and gefitinib in patients with advanced nonsmall-cell lung cancer. Cancer. 2007;110:599–605. doi: 10.1002/cncr.22816. [DOI] [PubMed] [Google Scholar]
- 24.Kono SA, Marshall ME, Ware KE, Heasley LE. The fibroblast growth factor receptor signaling pathway as a mediator of intrinsic resistance to EGFR-specific tyrosine kinase inhibitors in non-small cell lung cancer. Drug Resist Updat. 2009;12:95–102. doi: 10.1016/j.drup.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ware KE, Marshall ME, Heasley LR, Marek L, Hinz TK, Hercule P, et al. Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One. 2010;5:e14117. doi: 10.1371/journal.pone.0014117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–2803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- 28.Lazzara MJ, Lane K, Chan R, Jasper PJ, Yaffe MB, Sorger PK, et al. Impaired SHP2-mediated extracellular signal-regulated kinase activation contributes to gefitinib sensitivity of lung cancer cells with epidermal growth factor receptor-activating mutations. Cancer Res. 2010;70:3843–3850. doi: 10.1158/0008-5472.CAN-09-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, et al. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal. 2010;3:ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song DH, Dominguez I, Mizuno J, Kaut M, Mohr SC, Seldin DC. CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J Biol Chem. 2003;278:24018–24025. doi: 10.1074/jbc.M212260200. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Liu H, Liu B, Ma W, Xue X, Chen J, et al. Gene expression levels of CSNK1A1 and AAC-11, but not NME1, in tumor tissues as prognostic factors in NSCLC patients. Med Sci Monit. 2010;16:CR357–CR364. [PubMed] [Google Scholar]
- 32.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, et al. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao ZH, Seeling JM, Hill V, Yochum A, Virshup DM. Casein kinase I phosphorylates and destabilizes the beta-catenin degradation complex. Proc Natl Acad Sci U S A. 2002;99:1182–1187. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol. 2010;6:829–836. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knippschild U, Wolff S, Giamas G, Brockschmidt C, Wittau M, Wurl PU, et al. The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development. Onkologie. 2005;28:508–514. doi: 10.1159/000087137. [DOI] [PubMed] [Google Scholar]
- 38.Trosset JY, Dalvit C, Knapp S, Fasolini M, Veronesi M, Mantegani S, et al. Inhibition of protein-protein interactions: the discovery of druglike beta-catenin inhibitors by combining virtual and biophysical screening. Proteins. 2006;64:60–67. doi: 10.1002/prot.20955. [DOI] [PubMed] [Google Scholar]
- 39.Seimiya H. The telomeric PARP, tankyrases, as targets for cancer therapy. Br J Cancer. 2006;94:341–345. doi: 10.1038/sj.bjc.6602951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory MA, Phang TL, Neviani P, Alvarez-Calderon F, Eide CA, O'Hare T, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell. 2010;18:74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bivona TG, Hieronymus H, Parker J, Chang K, Taron M, Rosell R, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471:523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I, et al. Breast Cancer Special Feature: Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papeo G, Forte B, Orsini P, Perrera C, Posteri H, Scolaro A, et al. Poly(ADP-ribose) polymerase inhibition in cancer therapy: are we close to maturity? Expert Opin Ther Pat. 2009;19:1377–1400. doi: 10.1517/13543770903215883. [DOI] [PubMed] [Google Scholar]
- 46.Civenni G, Holbro T, Hynes NE. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO reports. 2003;4:166–171. doi: 10.1038/sj.embor.embor735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krejci P, Aklian A, Kaucka M, Sevcikova E, Prochazkova J, Masek JK, et al. Receptor Tyrosine Kinases Activate Canonical WNT/beta-Catenin Signaling via MAP Kinase/LRP6 Pathway and Direct beta-Catenin Phosphorylation. PLoS One. 2012;7:e35826. doi: 10.1371/journal.pone.0035826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.