Abstract
Background
The intestinal mucus layer plays a key role in the maintenance of host-microbiota homeostasis. To document the crosstalk between the host and microbiota, we used gnotobiotic models to study the influence of two major commensal bacteria, Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii, on this intestinal mucus layer. B. thetaiotaomicron is known to use polysaccharides from mucus, but its effect on goblet cells has not been addressed so far. F. prausnitzii is of particular physiological importance because it can be considered as a sensor and a marker of human health. We determined whether B. thetaiotaomicron affected goblet cell differentiation, mucin synthesis and glycosylation in the colonic epithelium. We then investigated how F. prausnitzii influenced the colonic epithelial responses to B. thetaiotaomicron.
Results
B. thetaiotaomicron, an acetate producer, increased goblet cell differentiation, expression of mucus-related genes and the ratio of sialylated to sulfated mucins in mono-associated rats. B. thetaiotaomicron, therefore, stimulates the secretory lineage, favoring mucus production. When B. thetaiotaomicron was associated with F. prausnitzii, an acetate consumer and a butyrate producer, the effects on goblet cells and mucin glycosylation were diminished. F. prausnitzii, by attenuating the effects of B. thetaiotaomicron on mucus, may help the epithelium to maintain appropriate proportions of different cell types of the secretory lineage. Using a mucus-producing cell line, we showed that acetate up-regulated KLF4, a transcription factor involved in goblet cell differentiation.
Conclusions
B. thetaiotaomicron and F. prausnitzii, which are metabolically complementary, modulate, in vivo, the intestinal mucus barrier by modifying goblet cells and mucin glycosylation. Our study reveals the importance of the balance between two main commensal bacteria in maintaining colonic epithelial homeostasis via their respective effects on mucus.
Keywords: Germ-free rats, Goblet cells, Mucin O-glycosylation, KLF4, Short chain fatty acid
Background
Understanding the crosstalk between the microbiota and the intestinal epithelium is essential because of the involvement of the intestinal microbiota in human health [1]. About 90% of the bacterial community that composes the intestinal microbiota and colonizes the gastrointestinal tract (GIT) are members of the phyla Bacteroidetes or Firmicutes [2-4]. B. thetaiotaomicron is a Gram negative bacterium, belonging to the Bacteroides genus from the Bacteroidetes phylum. Faecalibacterium prausnitzii is a Gram positive bacterium, in the Clostridium leptum group from the Firmicutes phylum. These two species are metabolically complementary: B. thetaiotaomicron is an acetate producer whereas F. praunistzii is an acetate consumer and a butyrate producer [5,6]. Both are major components of the intestinal microbiota and, therefore, contribute to the interactions between the microbiota and the GIT. To establish a relevant simplified model of microbiota, we studied these two commensal bacteria as functional members of the microbiome influencing host metabolism [7].
The intestinal mucus layer is located at the interface between the intestinal epithelium and the microbiota; it is, therefore, a key factor in the crosstalk between the intestinal epithelium and the microbiota and, hence, in the maintenance of intestinal homeostasis. Disruption of this protective layer may lead to inflammation [8]. Mucus is continuously secreted into the GIT lumen by specialized epithelial cells, the goblet cells, and is mainly composed of heavily O-glycosylated proteins called mucins [9]. The number and function of goblet cells are modulated by the microbiota [10-12]. However, neither the species nor the mechanisms involved in this process have been described. The microbiota expresses a wide range of carbohydrate-degrading enzymes which process otherwise indigestible dietary compounds and mucus polysaccharides [13,14]. The host genome does not encode homologs of many of these enzymes, evidence of the complementarity between host and microbiota and illustrating the adaptation of the intestinal microbiota to the host digestive environment [15,16]. B. thetaiotaomicron possesses a large repertoire of genes involved in sensing and hydrolyzing numerous diet- and host-derived polysaccharides [17,18]. According to nutrient availability, B. thetaiotaomicron can redirect its metabolism from dietary polysaccharides to host-derived polysaccharides, including mucus, and vice versa, and further refine its niche specificity [19,20]. This ability of B. thetaiotaomicron to grow on mucus contributes to its colonization and persistence in the GIT [18]. Although the ability of B. thetaiotaomicron to use polysaccharides from mucus has been described, little is known about its effect on goblet cells.
F. prausnitzii has been only poorly described in vivo; it is extremely oxygen sensitive and thus difficult to cultivate. Nevertheless, various observations link F. prausnitzii with health status. F. prausnitzii is of particular physiological importance because it can be considered as a sensor and a marker of human health [21]. Intestinal disorders, such as inflammatory bowel diseases [22-25], irritable bowel syndrome [26], colorectal cancer [27,28] and obesity [29] have all been reported to be associated with a diminished prevalence and abundance of F. prausnitzii. Moreover, several diseases have been shown to be associated with abnormal Bacteroidetes to Firmicutes ratios [1]. Therefore, analysis of the species B. thetaiotaomicron and F. prausnitzii may be informative about the crosstalk between the host and microbiota. The first objective of our work was to determine whether B. thetaiotaomicron affects goblet cell differentiation, mucin synthesis and glycosylation in the colonic epithelium. We then investigated how F. prausnitzii influences the colonic epithelial responses to B. thetaiotaomicron. For this study, we developed a new model based on B. thetaiotaomicron and F. prausnitzii di-associated gnotobiotic rodents to show metabolic cooperation between these species. This work illustrates how two bacteria, representative of the two main phyla, can modulate the mucus profile in the colon.
Results
Establishment of a model of Bacteroides thetaiotaomicron mono-associated rats stable as early as 2 days and up to 30 days after inoculation
B. thetaiotaomicron colonized the GIT of recipient germfree (GF) rats quickly and stably: the maximum count, 1010 CFU/g of content, was attained with one day of inoculation and was maintained for at least 30 days (Figure 1A). Scanning electron microscopy of cecal contents of Bt-2d (2 days after inoculation) and Bt-30d (30 days after inoculation) rats revealed B. thetaiotaomicron as short bacilli with rounded ends (Figure 1B); the bacteria were associated with luminal contents, food-residue particles and shed mucus, consistent with previous descriptions in the distal gut intestine [20]. The cell surface of B. thetaiotaomicron differed between Bt-2d rats and Bt-30d rats. Most of the bacteria in Bt-2d rats but not those in Bt-30d rats exhibited granules on the cell surface (Figure 1B). The presence of B. thetaiotaomicron was associated with the cecal-content pH being much lower than that in controls (Figure 1C). Short-chain fatty acid (SCFA) production by B. thetaiotaomicron was investigated: acetate and propionate was produced within two days of implantation but no butyrate was produced (Figure 1D). Thus, two days after inoculation, B. thetaiotaomicron was stably and abundantly implanted and was metabolically active.
Figure 1.
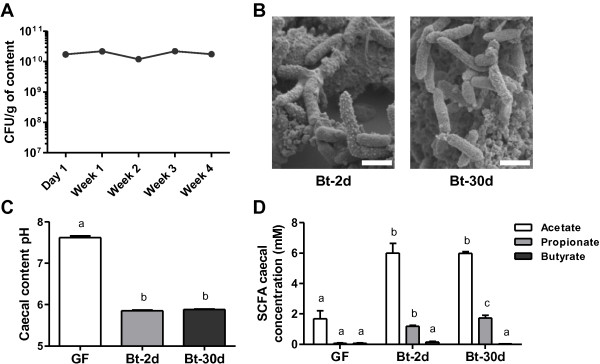
Time course analysis of GIT in B. thetaiotaomicron mono-associated rats. Germfree (GF) rats were inoculated with a culture of B. thetaiotaomicron (107 CFU) and euthanized 2 days (Bt-2d) or 30 days (Bt-30d) after inoculation. (A) Establishment of B. thetaiotaomicron in the gastrointestinal tract (GIT) of Bt-30d rats (n = 13) was monitored weekly by enumeration of the bacterial counts in the feces. (B) Scanning electron microscopy images of B. thetaiotaomicron in the cecum of Bt-2d and Bt-30d rats; scale bars, 1 μm. (C) Measurement of cecal pH in GF (n = 12), Bt-2d (n = 13) and Bt-30d rats (n = 19). (D) Cecal concentration of short-chain fatty acids (SCFA) in GF (n = 16), Bt-2d (n = 13) and Bt-30d rats (n = 19); only results for acetate, propionate and butyrate are shown, other SCFA were not detected; results are expressed in mM. Means with different letters are significantly different (P-value <0.05).
B. thetaiotaomicron modulates the cell signaling pathway in the colon by favoring goblet cell differentiation
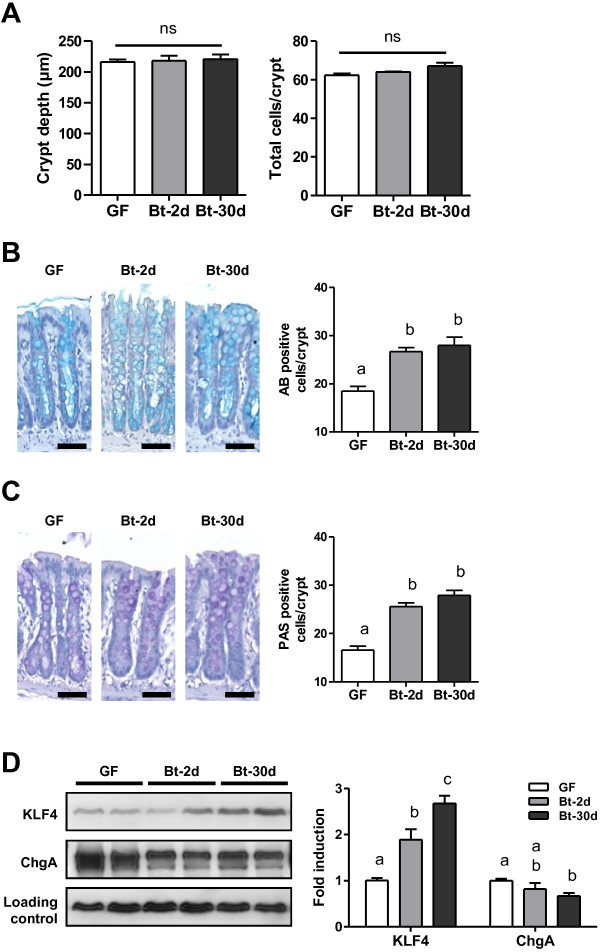
Histological staining of the colonic epithelium from Bt-2d and Bt-30d mono-associated rats and from GF rats showed that there were no significant differences in general morphology, crypt depth or total numbers of cells per crypt (see Additional file 1: Figure S1A and Figure 2A). B. thetaiotaomicron is specialized in the degradation of polysaccharides, notably those of mucus. We, therefore, analyzed goblet cell counts and differentiation. The numbers of goblet cells stained either by Alcian blue (AB) or periodic-acid Schiff (PAS), specific for acidic and neutral mucopolysaccharides, respectively, were significantly higher in Bt-2d and Bt-30d rats than GF rats (Figure 2B, C). Similarly, the amount of KLF4 protein, a transcription factor involved in goblet cell terminal differentiation [30], was higher in Bt-2d and Bt-30d than GF rats (Figure 2D). Also, muc2, muc4, c1galt1 and b4galt4 mRNAs, encoding proteins involved in mucin synthesis and glycosylation, were significantly more abundant in Bt-2d and Bt-30d rats than in GF rats (Table 1). The klf4 mRNA level was higher in Bt-2d, but not in Bt-30d, than in controls (Table 1). The amount of chromogranin A protein, a marker of enteroendocrine cells, another population of differentiated cells of the secretory lineage, was lower in Bt-30d rats than in control rats (Figure 2D). Thus, B. thetaiotaomicron appears to stimulate the goblet cell differentiation pathway of the secretory lineage.
Figure 2.
Characterization of the colonic epithelial response in B. thetaiotaomicron mono-associated rats. (A) Colonic crypt depth and total number of cells per crypt were determined on sections from GF (n = 3), Bt-2d (n = 5) and Bt-30d rats (n = 7) stained with Haematoxylin-Eosin-Safran (HES). (B, C) Representative pictures and graphs showing counts of goblet cells staining with (B) alcian blue (indicated as AB) and (C) periodic acid Schiff (indicated as PAS) in GF (n = 3), Bt-2d (n = 6) and Bt-30d (n = 7) samples. Scale bars, 50 μm. (D) Representative Western blots and densitometric analyses of proteins involved in the differentiation pathway of the secretory lineage, KLF4 and Chromogranin A (ChgA), in GF (n = 7), Bt-2d (n = 6) and Bt-30d (n = 7) samples. Means with different letters are significantly different (P-value <0.05).
Table 1.
Analysis of host gene expression in the colonic epithelium
| GF | Bt-2d | Bt-30d | Bt + Fp-30d | |
|---|---|---|---|---|
|
muc2 |
1.04 ± 0.05 a |
1.52 ± 0.10 a,b |
1.71 ± 0.10 b |
1.79 ± 0.22 b |
|
muc4 |
1.00 ± 0.05 a |
3.51 ± 0.33 b |
2.69 ± 0.29 a,b |
2.83 ± 0.43 b |
|
klf4 |
1.01 ± 0.05 a |
1.30 ± 0.10 b |
1.09 ± 0.04 a,b |
1.08 ± 0.09 a,b |
|
c1galt1 |
1.00 ± 0.03 a |
2.28 ± 0.26 b |
1.23 ± 0.16 a |
1.54 ± 0.24 a,b |
|
b4galt4 |
1.02 ± 0.11 a |
3.89 ± 0.42 b |
2.13 ± 0.20 a,c |
3.00 ± 0.39 b,c |
|
st3gal4 |
1.05 ± 0.14 a |
2.98 ± 0.37 b |
1.39 ± 0.16 a |
1.67 ± 0.22 a |
|
st6galnac3 |
1.07 ± 0.27 a |
0.80 ± 0.29 a |
1,11 ± 0.55 a |
0.41 ± 0.11 a |
|
gal3st3 |
n.d. |
n.d. |
n.d. |
n.d. |
| chst7 | 1.17 ± 0.36 a | 1.84 ± 0.55 a | 1.02 ± 0.23 a | 0.77 ± 0.20 a |
Relative abundance of muc2, muc4, klf4, c1galt1, b4galt4, st3gal4, st6galnac3, gal3st3 and chst7 mRNAs in germfree (GF) (n = 4 to 10), Bt-2d (n = 4), Bt-30d (n = 6) and Bt + Fp-30d (n = 6). Means with different letters (a, b, c) are significantly different (P-value <0.05); n.d., not detected.
Acetate, a bacterial metabolite produced by B. thetaiotaomicron, modulates goblet cells in vitro
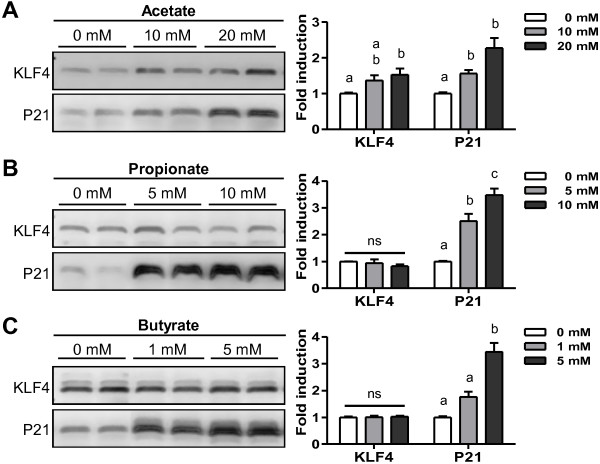
The main metabolites produced by B. thetaiotaomicron in the GIT were acetate (up to 6 mM) and propionate (up to 2 mM) (Figure 1D). We evaluated the effects of these bacterial metabolites on the amount of KLF4 protein produced by the mucus-producing cell line HT29-MTX [31]. The amount of KLF4 protein was higher in the presence than in the absence of acetate (10 and 20 mM) (Figure 3A) whereas propionate (5 and 10 mM) and butyrate (1 and 5 mM) had no effect (Figure 3B, C). The amount of butyrate-responsive protein, P21 [32], was increased by incubation of HT29-MTX cells with butyrate and there were similar trends with propionate and acetate (Figure 3). Thus, consistent with the findings in vivo in the presence of B. thetaiotaomicron (Figure 2), KLF4 protein production in vitro was up-regulated by acetate, the major metabolite produced by B. thetaiotaomicron. However, the stimulation was lower in vitro than in vivo.
Figure 3.
Effects of bacterial metabolites on KLF4 protein in HT29-MTX cells. Representative Western blot and densitometric analyses of KLF4 and P21 proteins in HT29-MTX cells incubated with (A) 0, 10, 20 mM acetate, (B) 0, 5, 10 mM propionate, and (C) 0, 1, 5 mM butyrate. Each graph reports means of three independent experiments with three internal repeats per experiment. Means with different letters are significantly different (P-value <0.05).
Metabolic cooperation between B. thetaiotaomicron and Faecalibacterium prausnitzii in vitro and in vivo
Bacterial metabolites are essential for epithelium homeostasis [33,34]. We investigated whether the effects of B. thetaiotaomicron are modulated in the presence of F. prausnitzii which is a major butyrate-producer and acetate-consumer [5].
We first checked in vitro that B. thetaiotaomicron produced acetate and propionate and that F. prausnitzii consumed acetate and produced butyrate (Figure 4A). When both bacteria were grown together, butyrate was produced rather than acetate. The butyrate production by F. prausnitzii was higher in the presence than absence of B. thetaiotaomicron (10.9 ± 0.3 mM versus 7.8 ± 0.5 mM) suggesting that F. prausnitzii synthesized butyrate from the acetate produced by B. thetaiotaomicron. We thus developed an experimental model of associated B. thetaiotaomicron and F. prausnitzii in rats (Figure 4B). Two single consecutive inoculations of the two bacteria did not lead to a stable di-associated model. Inoculation with F. prausnitzii was thus repeated weekly (arrows on Figure 4B) but it became implanted only after week 4. From week 4 to week 8, a stable balance was maintained between the two bacteria, with B. thetaiotaomicron counts being 100-fold higher. In these Bt + Fp-30d rats, there was less acetate and more butyrate than in Bt-30d rats: 3.6 ± 0.2 mM acetate, 1.0 ± 0.1 mM propionate and 1.3 ± 0.1 mM butyrate in the cecal contents of Bt + Fp-30d rats, versus 6.0 ± 0.2 mM acetate, 1.8 ± 0.2 mM propionate and no butyrate in Bt-30d rats (Figure 4C). F. prausnitzii is an extremely oxygen-sensitive bacterium. We, therefore, monitored the Eh value, reflecting the oxido-reduction potential, in the cecum. The Eh value progressively decreased in the presence of B. thetaiotaomicron (141 ± 9 mV to 46 ± 10 mV for Bt-2d and Bt-30d, respectively) from the value found for GF rats (178 ± 13 mV) and decreased further in the presence of F. prausnitzii, that is, in Bt + Fp-30d rats (-275 ± 13 mV) (Figure 4D). Moreover, the pH of the cecal content in Bt + Fp-30d rats was significantly higher (6.00 ± 0.04) than that in Bt-30d rats (5.88 ± 0.01).
Figure 4.
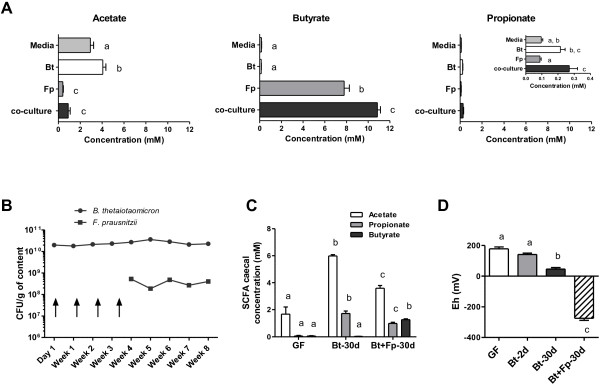
Behavior of B. thetaiotaomicron and F. prausnitzii in combination in vitro and in vivo. (A) Acetate, butyrate and propionate concentrations from culture media, pure cultures of B. thetaiotaomicron (Bt), F. prausnitzii (Fp) and cocultures. (B) Establishment of B. thetaiotaomicron and F. prausnitzii in the gastrointestinal tract (GIT) of Bt + Fp-30d rats (n = 16) was monitored weekly by enumeration in the feces; arrows represent the various inoculations with F. prausnitzii before successful colonization. (C) Cecal concentration of short chain fatty acids (SCFA) in the cecum of germfree (GF) (n = 12), Bt-30d rats (n = 19), and Bt + Fp-30d rats (n = 16). (D) Measurement of oxidoreduction potential (mV) in the cecal contents of GF (n = 11), Bt-2d (n = 13), Bt-30d (n = 19) and Bt + Fp-30d rats (n = 16). Means with different letters are significantly different (P-value <0.05).
We were unable to obtain F. prausnitzii mono-associated rats since successful implantation of F. prausnitzii required the presence of B. thetaiotaomicron. In di-associated rats, B. thetaiotaomicron produced acetate and F. prausnitzii converted it into butyrate. We then used this gnotobiotic model, where both bacteria were metabolically active, to study the effects of these two commensal bacteria on the mucus production pathway.
F. prausnitzii shifts the steady state induced by B. thetaiotaomicron in the colonic epithelium
In Bt + Fp-30d rats, colonic epithelium morphology, crypt depth and total number of cells per crypt were similar to those in Bt-30d rats (see Figure 5A and Additional file 1: Figure S1A). However, the numbers of AB- and PAS-positive cells per crypt were lower in Bt + Fp-30d than Bt-30d rats (see Figure 5B and Additional file 1: Figure S1B, C). Furthermore, the amount of KLF4 protein tended to be lower in Bt + Fp-30d than Bt-30d rats (Figure 5C). The amounts of muc2, muc4, klf4, c1galt1 and b4galt4 mRNAs were not different between Bt-30d and Bt + Fp-30d rats (Table 1). MUC2 staining was clearly lower in Bt + Fp-30d than Bt-30d rats and, indeed, was similar to that in GF rats (Figure 5D). The numbers of MUC2-positive cells tended to be lower in GF and Bt + Fp-30d rats (19.5 ± 0.6 and 21.6 ± 1.08 per crypt, respectively) than in Bt-30d rats (22.8 ± 1.7 per crypt). Chromogranin A protein was more abundant in Bt + Fp-30d than Bt-30d rats (Figure 5C). Thus, in this model with two metabolically complementary bacteria, F. prausnitzii seemed to attenuate the effects of B. thetaiotaomicron on goblet cell differentiation.
Figure 5.
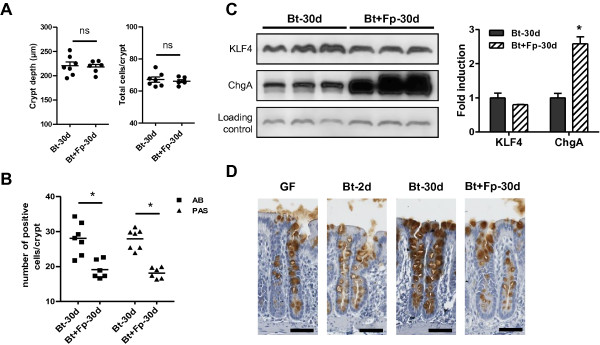
Characterization of the colonic epithelial response in B. thetaiotaomicron and F. prausnitzii di-associated rats. (A) Measurement of colonic crypt depth, total number of cells per colonic crypt and counts of (B) alcian blue- (indicated as AB) and periodic acid Schiff- (indicated as PAS) positive cells per crypt in colonic sections of Bt-30d (n = 7) and Bt + Fp-30d rats (n = 6). (C) Representative Western blot and densitometric analyses of proteins involved in the differentiation pathway of the secretory lineage, KLF4 and Chromogranin A (ChgA) in Bt-30d (n = 5) and Bt + Fp-30d rats (n = 4); protein fold induction in Bt-30d rats was used as a reference and arbitrarily defined as 1. (D) Immunostaining for MUC2 in germfree (GF) (n = 3), Bt-2d (n = 6), Bt-30d (n = 7) and Bt + Fp-30d rats (n = 6); scale bars, 50 μm. The asterisk indicates a statistical difference compared to Bt-30d rats (P-value <0.05); n.s., not significant.
Implantation of both commensal bacteria is associated with a shift in the mucin glycosylation profile
Having investigated mucus quantity by evaluating goblet cell number and mucin synthesis, we assessed expression of genes encoding host enzymes and mucus quality by studying the profile of mucin O-glycosylation. We performed analyses of sialyltransferase (st3gal4 and st6galnac3) and sulfotransferase (gal3st3 and chst7) gene expression (Table 1). In Bt-2d rats, we observed an increase in the expression of the sialyltransferase st3gal4. No modification was observed for the other genes tested. In parallel, we quantified the proportions of neutral/sulfated/sialylated oligosaccharides in the rat colonic mucins by studying the profile of mucin O-glycosylation. There were substantial changes to mucin glycosylation after colonization by B. thetaiotaomicron (see Additional file 2: Table S1). In Bt-2d rats, the proportions of sulfated (4.5% of total oligosaccharides instead of 12.9%) and neutral (40.1% instead of 52.8%) oligosaccharides were lower than in GF rats, and this was associated with higher proportions of O-glycans carrying NeuAc (24.2% instead of 18.9%) or NeuGc (31.2% instead of 15.4%) residues (Figure 6A). Only minor differences were observed between the mucins in Bt-2d and Bt-30d rats. The pattern of glycosylation of mucins from di-associated Bt + Fp-30d rats resembled that in GF rats. Interestingly, B. thetaiotaomicron increased the ratio of sialylated (NeuAc and NeuGc) to sulfated mucins in mono-associated compared to GF and di-asscociated rats (Figure 6B). Thus, as observed for goblet cell differentiation, F. prausnitzii seemed to attenuate the effects of B. thetaiotaomicron.
Figure 6.
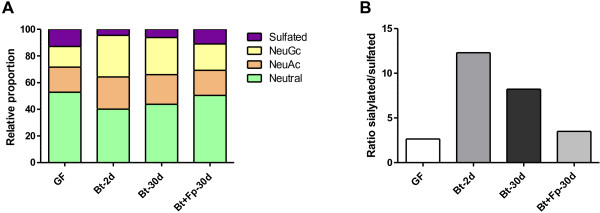
Effects of bacterial colonization on colonic mucin glycosylation. (A) Proportion of neutral and acidic oligosaccharides (carrying sulfate, N-acetyl neuraminic acid or N-glycolyl acid residues) in germfree (GF) rats (n = 3), Bt-2d (n = 2), Bt-30d (n = 3) and Bt + Fp 30d (n = 3) rats. (B) Ratio of sialylated to sulfated O-glycan chain in each group of rats.
Discussion
We report that commensal bacteria can influence goblet cells and mucin composition in the gut, providing new information about the relation among mucus, bacteria and intestinal homeostasis. B. thetaiotaomicron enhances goblet cell differentiation leading to an increase of goblet cell number and mucin gene expression in the colon of gnotobiotic rats. The presence of B. thetaiotaomicron also affects the composition of mucin O-glycans, with relative decreases in sulfated and neutral oligosaccharides in favor of sialylated oligosaccharides. B. thetaiotaomicron, therefore, appears to provoke modifications in the secretory lineage compared to GF rats, favoring mucus production and we could hypothesize that this is possibly for its own benefit. When B. thetaiotaomicron is associated with F. prausnitzii, which is an acetate consumer and a butyrate producer, the effects on both goblet cells and mucin O-glycosylation are attenuated. Using a novel gnotobiotic model, which is the first described involving F. prausnitzii, we showed that two major species of commensal bacteria can modulate the effects of the bacteria on goblet cells and thus on mucus production and mucin glycosylation.
The GIT is an environment with harsh constraints (pH, oxygen, access to nutrients) which shape the microbial composition. The genome of Bacteroides species possesses gene clusters for capsular polysaccharide synthesis [17,18] and these polysaccharides are important for bacterial colonization of the GIT: they improve survival in the GIT, and also stimulate the immune system [35-37]. Scanning electron microscopy revealed differences in the B. thetaiotaomicron cell wall in Bt-2d and Bt-30d rats. Possibly, during the first days of adaptation to the GIT, B. thetaiotaomicron produce capsular polysaccharides which collapse through a dehydration process, as previously described [38,39]. The transcriptomic profile of B. thetaiotaomicron before and after its passage through GIT shows that B. thetaiotaomicron adapts its metabolism to survive in the GIT [20]. When B. thetaiotaomicron is associated with Eubacterium rectale, a representative of the C. coccoides group, both bacteria adapt their own metabolism to the presence of the other [6]. Thus, bacteria in the GIT presumably adapt to the host, and also to other bacteria [40]. In this study, we observed that in mono-associated rats, B. thetaiotaomicron induces goblet cell differentiation leading to an increase in mucin gene expression and goblet cell number, and a parallel decrease in chromogranine produced by enteroendocrine cell. These observations are in accordance with previous work showing that the colonic epithelium also adapts to the presence of bacteria [41-43]; these adaptations are parts of the mutual interactions essential for human health.
We show that, following colonization of the GIT by B. thetaiotaomicron, the pattern of mucin glycosylation changes, with increased expression of glycans carrying sialic acid residues (either NeuAc or NeuGc), correlated with decreased expression of sulfated and neutral oligosaccharides. A previous immunohistochemical study described similar alterations in the glycosylation pattern in the mouse intestine, induced by B. thetaiotaomicron implantation or by soluble factors produced by this microorganism [44]. B. thetaiotaomicron also induces fucosylation of intestinal epithelial cells, which is linked to its ability to use fucose, a component of glycosylated mucin [45]. Thus, glycan production or distribution is modulated by B. thetaiotaomicron, and this may involve processes important for signaling and mediating the host mucosal response. According to the enhancement of the expression of st3gal4 in the presence of B. thetaiotaomicron, we can assume that there is a mechanism by which this bacterium modulates the composition of mucus chain O-glycans by impacting directly enzyme expression of the host. B. thetaiotaomicron may require high levels of expression of sialylated mucins for attachment and colonization of the GIT. Indeed, B. thetaiotaomicron may express adhesins specifically recognizing and binding to sialic acid residues and may also use sialic acid residues as host substrates. B. thetaiotaomicron possesses 28 predicted sulfatases and only 1 single predicted anaerobic sulfatase-maturating enzyme which allow bacteria to adapt to and forage on host sulfated glycans as nutrients [46]. Our observations of a substantial decrease in sulfated oligosaccharides in the rats colon two days after B. thetaiotaomicron colonization are consistent with this enzymatic potential. Presumably, B. thetaiotaomicron uses these sulfate residues rapidly not only as nutrient sources but also to gain access to other monosaccharides, such as N-acetylglucosamine (GlcNAc) and galactose (Gal), the two monosaccharides substituted by sulfate residues on mucin O-glycans. In a favorable ecosystem (that is, in the absence of other microorganisms), there is probably no need for the bacteria or the host to maintain a high level of expression of sulfated glycans. However, when B. thetaiotaomicron is associated with F. prausnitzii, the proportion of sulfated glycans is greater than that observed in the presence of B. thetaiotaomicron alone. This suggests that, in a more complex environment involving a competitive ecosystem, sulfated glycans may confer an ecological advantage on B. thetaiotaomicron.
We were unable to obtain, on a large scale and reproducibly, F. prausnitzii mono-associated rats; the implantation of F. prausnitzii required the prior presence of B. thetaiotaomicron. Furthermore, F. prausnitzii was unable to colonize the GIT early after the introduction of B. thetaiotaomicron. This may have been due to the physicochemical environment. F. prausnitzii became established in the gut only after a decrease in the oxidoreduction potential, caused by the presence of B. thetaiotaomicron, suggesting that B. thetaiotaomicron “prepares” the GIT to accommodate sustainably more oxygen-sensitive bacteria. F. prausnitzii and, to a larger extent, the C. leptum group, arrive in the GIT late after birth, presumably because they are sensitive to oxygen. Moreover, after intestinal resection, disrupting the gut structure and exposing the colon to oxygen, the C. leptum group disappears from the microbiota [47]. In the case of strict anaerobes like F. prausnitzii, their ability to colonize is governed by the physicochemical constraints of gut rather than by any lack of metabolic substrates. This contrasts with Lactobacillus bulgaricus, which is an aerobic-tolerant lactic acid bacterium, and which does not colonize the gut of GF rats in the absence of lactose [48]. That colonization by B. thetaiotaomicron precedes the implantation of F. prausnitzii gives clues about mechanisms governing microbial ecology and processes of colonization.
The two bacteria were metabolically complementary both in vitro and in vivo. F. prausnitzii consumes acetate produced by B. thetaiotaomicron and in turn produces butyrate. Short-chain fatty acids (SCFA) trigger pleiotropic signals in the host, including signals regulating mucin synthesis and secretion [33,49-51]. SCFA, in particular butyrate and to a lesser extent acetate, are reported to have inducer effects on mucin synthesis and production in vitro[33,49,51]. Butyrate also increases the expression of the transcription factor klf4 in HT29 cells [52]. On the contrary, another experiment reported an inhibitor effect of butyrate on mucin synthesis in vitro[53]. All these data show the potential role of SCFA in modulating the mucus pathway and it may be a mechanism by which they affect the goblet cell differentiation pathway in our gnotobiotic models. Using HT29-MTX, a mucus producing cell line, we showed that acetate increases KLF4 abundance. Understanding the effects of the microbiota on mucus production and goblet cell differentiation is of major importance as mucus acts as a protective barrier and disruption of this mucus layer leads to inflammation [54]; abnormalities of the mucus layer, and modulations of goblet cells and mucin secretion have been described in inflammatory bowel diseases [55]. In a murine model of DSS-induced colitis, the mucus layer is altered before the induction of any inflammation evidencing the importance of mucus layer integrity to prevent inflammation [56]. The O-glycosylation pattern of mucins has also a determinant role in health, with sulfated colonic mucins playing a protective role [57]. Indeed, mucins are poorly sulfated in patients with ulcerative colitis [58]. Butyrate induces the expression of sulfotransferases, the enzymes catalyzing sulfatation of mucin in the mouse colon [57] and also up-regulates expression of galactose-3-O-sulfotransferase 4 in human intestinal epithelial goblet cells [59]. The increase in sulfated mucins observed in our B. thetaiotaomicron and F. prausnitzii di-associated rats is consistent with these previous studies. We, therefore, suggest that metabolites, mainly acetate and butyrate, produced by B. thetaiotaomicron and F. prausnitzii, respectively, may be responsible, at least in part, for the modifications that we observed in goblet cells and mucin O-glycosylation.
Conclusions
Our study reveals relationships between intestinal microbiota and the differentiation and secretory program of colonic goblet cells. We propose that B. thetaiotaomicron and F. prausnitzii, two main members of microbiota representative of Bacteroidetes and Firmicutes, contribute to the establishment of epithelial homeostasis. These two bacteria, which are metabolically complementary, modulate in vivo the intestinal mucus barrier by modifying goblet cells and mucin glycosylation. Here we show how the balance between B. thetaiotaomicron and F. prausnitzii could be determinant for maintaining colonic epithelial homeostasis and health via their respective effects on mucus.
Methods
Bacterial strains and culture conditions
The reference strains B. thetaiotaomicron VPI-5482 (ATCC 29148) and F. prausnitzii A2-165 (DSM 17677), isolated from human fecal stool, were used. Bacteria were grown overnight at 37°C in YBHI medium (brain-heart infusion medium supplemented with 0.5% Difco yeast extract (Becton Dickinson and Company, Le Pont De Claix, France) supplemented with 1 mg/ml cellobiose, 1 mg/ml maltose and 0.5 mg/ml L-cysteine (all from Sigma-Aldrich, St-Louis, MO, USA) in an anaerobic chamber containing a gas mix of 90% N2, 5% CO2 and 5% H2. For co-culture, the 24 h pre-culture of each was added to fresh medium at 1/10 and 1/100 for F. prausnitzii and B. thetaiotaomicron, respectively.
B. thetaiotaomicron and/or F. prausnitzii were enumerated weekly in the feces of mono- and di-associated rats. Fresh feces were introduced into the anaerobic chamber, serially diluted 10-fold and plated on supplemented YBHI agar. To enumerate only F. prausnitzii in the feces of di-associated rats, 1 ml of the 10-2 dilution of the feces was treated with 0.1 ml of sodium azide (3%) for five minutes at 37°C and then serially diluted 10-fold and plated on supplemented YBHI agar with added sodium azide (0.02%).
Animals and experimental design
All procedures were carried out according to European and French guidelines for the care and use of laboratory animals (permission 78–123, granted to M.T.). GF rats (males, Fisher 344) were used. Animals were born and bred at the Institut National de la Recherche Agronomique (Jouy-en-Josas, France). All rats were housed in Trexler-type isolators (La Calhène, Vélizy, France) and fed with a standard diet, rich in polysaccharides (R03, SAFE) sterilized by gamma irradiation (45 kGy). GF rats were used as controls (n = 16). To obtain rats mono-associated with B. thetaiotaomicron, GF rats were inoculated by a single oral gavage with 1 ml of culture of B. thetaiotaomicron (107 CFU/ml) and euthanized 2 days (Bt-2d rats, n = 13) or 30 days (Bt-30d rats, n = 19) after inoculation. To obtain rats di-associated with B. thetaiotaomicron and F. prausnitzii, GF rats were orally inoculated with 1 ml of culture of B. thetaiotaomicron; 24 h later, when this bacterium was established in the GIT, 1 ml of 100× concentrated culture of F. prausnitzii (7.109 CFU/ml) was administered orally to a donor rat pretreated with 0.1 ml of sodium bicarbonate (0.2 M). This manipulation was repeated every week until F. prausnitzii was established in a donor rat. Once one rat di-associated with the two bacteria was obtained, its feces were administrated orally to additional rats pretreated with sodium bicarbonate. Rats were euthanized 30 days after F. prausnitzii implantation (Bt + Fp-30d rats, n = 16).
All rats were euthanized at the age of three months; rats were anesthetized at 9:00 a.m. with isoflurane, cecal contents were collected and colons were recovered and immediately used for cell isolation or histology.
Scanning electron microscopy
Scanning electron microscopy [60] analyses were performed on the MIMA2 platform (INRA, Massy, France). Aliquots of 0.2 g of cecal contents were suspended and fixed in 200 μl of glutaraldehyde, 3% ruthenium red for two hours in an anaerobic chamber and then stored at 4°C. Scanning electron microscopy was performed as reported previously [47].
Short chain fatty acid analysis
Acetate, propionate and butyrate concentrations were determined in cecal contents, supernatants from bacterial culture and culture media after water extraction of acidified samples using gas liquid chromatography (Nelson 1020, Perkin-Elmer, St Quentin en Yvelines, France) as described previously [61]. SCFA concentrations are expressed in mM.
Oxidoreduction potential (Eh) and pH measurement
The measurement of pH and oxidoreduction potential was as described by Martin et al., after a total immersion of the electrode in the cecal content [62]. A combined autoclavable electrode (InLAb®, Mettler-Toledo SAS, Viroflay, France) was used for all measurements.
Histology and immunohistochemisty assays
Flushed colons were opened longitudinally and cut into 2 cm sections. The samples were fixed in 4% paraformaldehyde (4 h, room temperature), dehydrated, and embedded in paraffin according to standard histological protocols. Four-micrometer thick sections of distal colon were mounted on SuperFrost® Plus slides (Thermo Fisher, Waltham, MA, USA). Paraffin-embedded sections were deparaffinized and stained with: Haematoxylin-Eosin-Safran (HES) to measure crypt depth and Alcian blue (AB) solution pH 2.5 or periodic acid-Schiff (PAS) reagent to count goblet cell number per crypt. MUC2 immunostaining was performed using the EnVision + System-HRP (Dako-Cytomation, Trappes, France) and anti-MUC2 (1:5,000; sc-15334, Santa Cruz Biotechnology, Heidelberg, Germany) as the primary antibody as previously described [12]. For all markers studied (HES, AB, PAS, MUC2), only U-shaped longitudinally cut crypts with open lumina were considered. Results reported are means obtained by analysis two distal colon sites for each rat and at least 10 crypts per site with NanozoomerDigitalPathology view software (Hamamatsu Photonics, Hamamatsu, Japan).
Western blot analysis
Colonic epithelial cells were isolated from the whole colon according to the method described by Cherbuy et al.[63]. The cell pellet from whole colon or HT29-MTX cell cultures was immediately used for protein extraction [41] and Lowry’s procedure was used for protein assays [64]. Western blot analysis was performed as previously described [41] using 12% SDS-PAGE and anti-KLF4 (1:500; IMG-6081A, Imgenex, San Diego, CA, USA), anti-Chromogranin A (1:500; ab15160, Abcam, Cambridge, MA, USA), or anti-P21 (1:200; sc-397, Santa Cruz Biotechnology) with appropriate peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). For each Western blot, protein loads were determined using anti-cullin1 antibodies (1:400; sc-17775, Santa Cruz Biotechnology). Signals detected on autoradiographic films were quantified by scanning densitometry with Biovision 1000 and Bio-1D software (Vilber Lourmat, France).
Total RNA extraction and real-time quantitative PCR analysis
Total RNA was extracted from isolated colonic epithelial cells by the guanidinium thiocyanate method [65]. RNA concentration and purity were determined by absorbance measurement using a nanodrop ND-1000 (Thermo Fisher Scientific, Illkirch, France) and RNA Integrity Number (RIN) was checked using the RNA 6000 Nano LabChip® kit (Agilent Technologies, Santa Clara, CA, USA) and the Agilent 2100 bioanalyzer at ICE platform (INRA, Jouy-en-Josas, France). All RNA had a RIN between 8 and 10, indicating a high RNA quality in all samples. Reverse transcription was performed with 7 μg of RNA of each sample using the High-Capacity cDNA Archive Kit (Applied Biosystems by Life Technologies SAS, Saint Aubin, France) according to the manufacturer’s instructions. Each cDNA sample was tested for PCR inhibition with the TaqMan® Exogenous Internal Positive Control (Applied Biosystems) and no inhibition was detected for any sample. The cDNA products were analyzed in triplicate by RT-qPCR with an ABI PRISM 7000 Sequence Detection System and the 7000 system software version 1.2.3 (Applied Biosystems). Several genes were analyzed using TaqMan Gene Expression Assays (Applied Biosystems): klf4 (Rn00821506_g1), muc2 (Rn01498197_m1), muc4 (Rn01475265_m1), c1galt1 (Rn01455817_m1), b4galt4 (Rn01439430503812_m1), st3gal4 (Rn01786289_m1), st6galnac3 (Rn00569406_m1), gal3st3 (Rn01233548_m1) and chst7 (Rn01430599_m1). 18S rRNA (Hs99999901_s1) was used as a reference. Results obtained were normalized to the value for 18S rRNA and compared with the mean target gene expression in GF rats as the calibrator sample. The following formula was used: fold change = 2-ΔΔCt, where ΔΔCt threshold cycle (Ct) equals (target Ct - reference Ct) of sample minus (target Ct - reference Ct) of the calibrator.
Cell culture
The mucus-secreting cell line HT29-MTX was obtained from Thecla Lesuffleur (INSERM UMR S 938, Paris, France) and used from passages 27 to 36 [31]. Cells were routinely grown in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) with 4.5 g/L glucose (Lonza, Verviers, Belgium), supplemented with 10% (v/v) fetal calf serum (FCS) inactivated by incubation for one hour at 56°C (Lonza, Verviers, Belgium), with 1% (v/v) L-Glutamine 200 mM (Lonza). The experimental design involved cells being grown for seven days in six-well tissue culture plates at a density of 12.104 cells per ml. Cells were maintained at 37°C in a 10% CO2:90% air atmosphere. The culture medium was changed daily. Undifferentiated cells were obtained and incubated with 0, 10, 20 mM acetate or 0, 5, 10 mM propionate or 0, 1, 5 mM butyrate for 17 h. To avoid a pH effect, when SCFA were added in culture medium, the pH value was checked and adjusted to those of culture medium. Each condition was tested in triplicate and experiments were repeated three times. Cells were scraped off and immediately used for protein extraction.
Analysis of mucin glycosylation
Rat colonic mucosa was scraped and mucins were purified as previously described [66]. Briefly, mucins were solubilized in 4 M guanidine chloride solution containing 5 mM EDTA, 10 mM benzamidine, 5 mM N-ethylmaleimide, 0.1 mg/mL soy bean trypsin inhibitor and 1 mM PMSF (phenylmethanesulfonyl fluoride). Bacteria were removed by centrifugation at 3,000 g for 15 minutes. Supernatant was collected and CsCl was added to give a density of 1.4 g/mL and mucins were purified by isopycnic density-gradient centrifugation (Beckman Coulter LE80K ultracentrifuge; 70.1 Ti rotor, 58,000 rpm at 15°C for 72 h; Beckman Coulter France S.A.S, Villepinte, France). The mucin-containing fractions were pooled, dialyzed against water and lyophilized. Mucins were then subjected to β-elimination under reducing conditions (0.1 M NaOH, 1 M KBH4 for 24 h at 45°C). The mixtures of oligosaccharide alditols were permethylated by the sodium hydroxide procedure. After derivation, the reaction products were dissolved in 200 μl of methanol and further purified on a C18 Sep-Pak (Waters Corporation, Milford, MA, USA). Permethylated oligosaccharides were analyzed by MALDI TOF mass spectrometry in positive ion reflective mode as [M + Na]+. The relative proportions of each oligosaccharide were calculated by integration of peaks on MS spectra, and are expressed as percentages of the total.
Statistical analyses
Data are reported as means ± SEM for the number of animals indicated. Group data were compared using one-way analysis of variance (ANOVA) followed by a Wilcoxon test (jmp version 7) when the ANOVA revealed differences between groups. Differences with P-value <0.05 were considered to be statistically significant.
Abbreviations
AB: Alcian blue; Ct: Threshold cycle; DMEM: Dulbecco’s modified Eagle’s minimal essential medium; FCS: Fetal calf serum; Gal: galactose; GF: germfree; GIT: Gastrointestinal tract; GlcNAc: N-acetylglucosamine; HES: Haematoxylin-Eosin-Safran; PAS: Periodic-acid Schiff; RIN: RNA Integrity Number; SCFA: Short-chain fatty acid.
Competing interests
The authors have no conflicting financial interests.
Authors’ contributions
LW, SM, CC, PL and MT designed in vitro and in vivo experiments and LW, SM, MLN, SB, MJCC, VR, CP, CB and CRM performed these experiments. LW, SM, CRM, PL and MT wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Characterization of the colonic epithelial response in B. thetaiotaomicron and F. prausnitzii di-associated rats. (A) Representative pictures of colonic sections stained with HES in GF, Bt-2d, Bt-30d and Bt + Fp-30d rats. Representative pictures showing goblet cells staining with (B) alcian blue (indicated as AB) and (C) periodic acid Schiff (indicated as PAS) in Bt-30d rats and Bt + Fp 30d (n = 3) rats. Scale bars, 50 μm.
Deduced monosaccharide composition of mucin O-glycans from GF, Bt-2d, Bt-30d and Bt + Fp 30d rats, as identified by MALDI-TOF mass spectrometry in positive ion mode. The relative proportions of each oligosaccharide were calculated by integration of peaks on MS spectra. Mucus O-glycans types were separated into neutral oligosaccharides (blue), oligosaccharides carrying sialic acid (orange) and sulfated residues (green). Results are presented as means ± SD of the percentage of each oligosaccharide.
Contributor Information
Laura Wrzosek, Email: laura.wrzosek@jouy.inra.fr.
Sylvie Miquel, Email: sylvie.miquel@jouy.inra.fr.
Marie-Louise Noordine, Email: marie-louise.noordine@jouy.inra.fr.
Stephan Bouet, Email: stephan.bouet@jouy.inra.fr.
Marie Joncquel Chevalier-Curt, Email: marie.chev@hotmail.fr.
Véronique Robert, Email: veronique.robert@jouy.inra.fr.
Catherine Philippe, Email: catherine.philippe@jouy.inra.fr.
Chantal Bridonneau, Email: chantal.bridonneau@jouy.inra.fr.
Claire Cherbuy, Email: claire.cherbuy@jouy.inra.fr.
Catherine Robbe-Masselot, Email: Catherine.Robbe@univ-lille1.fr.
Philippe Langella, Email: philippe.langella@jouy.inra.fr.
Muriel Thomas, Email: muriel.thomas@jouy.inra.fr.
Acknowledgements
We thank Alhosna Benjdia, Muriel Mercier-Bonin, Harry Sokol and Julie Tomas for fruitful discussions and critical reading of the manuscript and the Animalerie Axénique de MICALIS (ANAXEM) platform (Microbiologie de l’Alimentation au Service de la Santé (MICALIS), Jouy-en-Josas, France) for animal facilities. We gratefully acknowledge Thierry Meylheuc for scanning electron microscopy (MIMA2 platform, INRA, Massy, France) and Rémi Cachon for technical assistance with oxidoreduction potential (Eh) analysis (GPMA, Dijon, France). LW was the recipient of a PhD grant from the French Ministry of Education and Research. This study was a part of FPARIS collaborative project selected and supported by the Vitagora Competitive Cluster and funded by the French FUI (Fond Unique Interministériel; FUI: n°F1010012D), the FEDER (Fonds Européen de Développement Régional; Bourgogne: 34606), the Burgundy Region, the Conseil Général 21 and the Grand Dijon. This work was also supported by Merck Médication Familiale (Dijon, France) and Biovitis (Saint Etienne de Chomeil, France). SM received a salary from the same grants.
References
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. Erratum in: Science 2008, 322:1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Munoz-Tamayo R, Paslier DL, Nalin R, Dore J, Leclerc M. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52:2141–2146. doi: 10.1099/ijs.0.02241-0. [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli EM, Simmering R, Faure M, Donnicola D, Mansourian R, Rochat F, Corthesy-Theulaz I, Cherbut C. Multifaceted transcriptional regulation of the murine intestinal mucus layer by endogenous microbiota. Genomics. 2008;91:70–77. doi: 10.1016/j.ygeno.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kandori H, Hirayama K, Takeda M, Doi K. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim. 1996;45:155–160. doi: 10.1538/expanim.45.155. [DOI] [PubMed] [Google Scholar]
- Tomas J, Wrzosek L, Bouznad N, Bouet S, Mayeur C, Noordine ML, Honvo-Houeto E, Langella P, Thomas M, Cherbuy C. Primocolonization is associated with colonic epithelial maturation during conventionalization. FASEB J. 2013;27:645–655. doi: 10.1096/fj.12-216861. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksik P, Sokol H, Lepage P, Vasquez N, Manichanh C, Mangin I, Pochart P, Dore J, Marteau P. Review article: the role of bacteria in onset and perpetuation of inflammatory bowel disease. Aliment Pharmacol Ther. 2006;24:11–18. doi: 10.1111/j.1365-2036.2006.03053.x. [DOI] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong ML, Xu A, Chavakis T, Bornstein AB, Ehrhart-Bornstein M, Lamounier-Zepter V, Lohmann T, Wolf T, Bornstein SR. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2012. [Epub ahead of print.] [DOI] [PubMed]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuffleur T, Porchet N, Aubert JP, Swallow D, Gum JR, Kim YS, Real FX, Zweibaum A. Differential expression of the human mucin genes MUC1 to MUC5 in relation to growth and differentiation of different mucus-secreting HT-29 cell subpopulations. J Cell Sci. 1993;106:771–783. doi: 10.1242/jcs.106.3.771. [DOI] [PubMed] [Google Scholar]
- Archer SY, Meng S, Shei A, Hodin RA. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci U S A. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420:211–219. doi: 10.1042/BJ20082222. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol. 2011;45:S120–S127. doi: 10.1097/MCG.0b013e31822fecfe. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A. 2008;105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Evrard B, Balestrino D, Dosgilbert A, Bouya-Gachancard JL, Charbonnel N, Forestier C, Tridon A. Roles of capsule and lipopolysaccharide O antigen in interactions of human monocyte-derived dendritic cells and Klebsiella pneumoniae. Infect Immun. 2010;78:210–219. doi: 10.1128/IAI.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany SC, Levery SB, Kim JS, Reuhs BL, Lion LW, Ghiorse WC. Structure and carbohydrate analysis of the exopolysaccharide capsule of Pseudomonas putida G7. Environ Microbiol. 2001;3:774–784. doi: 10.1046/j.1462-2920.2001.00248.x. [DOI] [PubMed] [Google Scholar]
- Turpin W, Humblot C, Noordine ML, Wrzosek L, Tomas J, Mayeur C, Cherbuy C, Guyot JP, Thomas M. Behavior of lactobacilli isolated from fermented slurry (ben-saalga) in gnotobiotic rats. PLoS One. 2013;8:e57711. doi: 10.1371/journal.pone.0057711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbuy C, Honvo-Houeto E, Bruneau A, Bridonneau C, Mayeur C, Duee PH, Langella P, Thomas M. Microbiota matures colonic epithelium through a coordinated induction of cell cycle-related proteins in gnotobiotic rat. Am J Physiol Gastrointest Liver Physiol. 2010;299:G348–G357. doi: 10.1152/ajpgi.00384.2009. [DOI] [PubMed] [Google Scholar]
- Rul F, Ben-Yahia L, Chegdani F, Wrzosek L, Thomas S, Noordine ML, Gitton C, Cherbuy C, Langella P, Thomas M. Impact of the metabolic activity of Streptococcus thermophilus on the colon epithelium of gnotobiotic rats. J Biol Chem. 2011;286:10288–10296. doi: 10.1074/jbc.M110.168666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Wrzosek L, Ben-Yahia L, Noordine ML, Gitton C, Chevret D, Langella P, Mayeur C, Cherbuy C, Rul F. Carbohydrate metabolism is essential for the colonization of Streptococcus thermophilus in the digestive tract of gnotobiotic rats. PLoS One. 2011;6:e28789. doi: 10.1371/journal.pone.0028789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas M, Axelsson LG, Cayuela C, Midtvedt T, Trugnan G. Indigenous microbes and their soluble factors differentially modulate intestinal glycosylation steps in vivo. Use of a “lectin assay” to survey in vivo glycosylation changes. Histochem Cell Biol. 2005;124:423–433. doi: 10.1007/s00418-005-0004-1. [DOI] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont Bacteroides thetaiotaomicron. J Biol Chem. 2011;286:25973–25982. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly F, Mayeur C, Bruneau A, Noordine ML, Meylheuc T, Langella P, Messing B, Duee PH, Cherbuy C, Thomas M. Drastic changes in fecal and mucosa-associated microbiota in adult patients with short bowel syndrome. Biochimie. 2010;92:753–761. doi: 10.1016/j.biochi.2010.02.015. [DOI] [PubMed] [Google Scholar]
- Ben-Yahia L, Mayeur C, Rul F, Thomas M. Growth advantage of Streptococcus thermophilus over Lactobacillus bulgaricus in vitro and in the gastrointestinal tract of gnotobiotic rats. Benef Microbes. 2012;3:211–219. doi: 10.3920/BM2012.0012. [DOI] [PubMed] [Google Scholar]
- Gaudier E, Jarry A, Blottiere HM, de Coppet P, Buisine MP, Aubert JP, Laboisse C, Cherbut C, Hoebler C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1168–G1174. doi: 10.1152/ajpgi.00219.2004. [DOI] [PubMed] [Google Scholar]
- Gaudier E, Rival M, Buisine MP, Robineau I, Hoebler C. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol Res. 2009;58:111–119. doi: 10.33549/physiolres.931271. [DOI] [PubMed] [Google Scholar]
- Hatayama H, Iwashita J, Kuwajima A, Abe T. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun. 2007;356:599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Rex S, Tseng CC. Kruppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–798. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- Augenlicht L, Shi L, Mariadason J, Laboisse C, Velcich A. Repression of MUC2 gene expression by butyrate, a physiological regulator of intestinal cell maturation. Oncogene. 2003;22:4983–4992. doi: 10.1038/sj.onc.1206521. [DOI] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, Einerhand AW. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Gersemann M, Becker S, Kubler I, Koslowski M, Wang G, Herrlinger KR, Griger J, Fritz P, Fellermann K, Schwab M, Wehkamp J, Stange EF. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, Sjovall H, Hansson GC. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS One. 2010;5:e12238. doi: 10.1371/journal.pone.0012238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobisawa Y, Imai Y, Fukuda M, Kawashima H. Sulfation of colonic mucins by N-acetylglucosamine 6-O-sulfotransferase-2 and its protective function in experimental colitis in mice. J Biol Chem. 2010;285:6750–6760. doi: 10.1074/jbc.M109.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf AH, Tsai HH, Parker N, Hoffman J, Walker RJ, Rhodes JM. Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin Sci (Lond) 1992;83:623–626. doi: 10.1042/cs0830623. [DOI] [PubMed] [Google Scholar]
- Gaudier E, Forestier L, Gouyer V, Huet G, Julien R, Hoebler C. Butyrate regulation of glycosylation-related gene expression: evidence for galectin-1 upregulation in human intestinal epithelial goblet cells. Biochem Biophys Res Commun. 2004;325:1044–1051. doi: 10.1016/j.bbrc.2004.10.141. [DOI] [PubMed] [Google Scholar]
- Dallo SF, Kannan TR, Blaylock MW, Baseman JB. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol Microbiol. 2002;46:1041–1051. doi: 10.1046/j.1365-2958.2002.03207.x. [DOI] [PubMed] [Google Scholar]
- Lan A, Bruneau A, Bensaada M, Philippe C, Bellaud P, Rabot S, Jan G. Increased induction of apoptosis by Propionibacterium freudenreichii TL133 in colonic mucosal crypts of human microbiota-associated rats treated with 1,2-dimethylhydrazine. Br J Nutr. 2008;100:1251–1259. doi: 10.1017/S0007114508978284. [DOI] [PubMed] [Google Scholar]
- Martin F, Cachon R, Pernin K, De Coninck J, Gervais P, Guichard E, Cayot N. Effect of oxidoreduction potential on aroma biosynthesis by lactic acid bacteria in nonfat yogurt. J Dairy Sci. 2011;94:614–622. doi: 10.3168/jds.2010-3372. [DOI] [PubMed] [Google Scholar]
- Cherbuy C, Darcy-Vrillon B, Morel MT, Pegorier JP, Duee PH. Effect of germfree state on the capacities of isolated rat colonocytes to metabolize n-butyrate, glucose, and glutamine. Gastroenterology. 1995;109:1890–1899. doi: 10.1016/0016-5085(95)90756-4. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Rossez Y, Maes E, Lefebvre Darroman T, Gosset P, Ecobichon C, Joncquel Chevalier Curt M, Boneca IG, Michalski JC, Robbe-Masselot C. Almost all human gastric mucin O-glycans harbor blood group A, B or H antigens and are potential binding sites for Helicobacter pylori. Glycobiology. 2012;22:1193–1206. doi: 10.1093/glycob/cws072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the colonic epithelial response in B. thetaiotaomicron and F. prausnitzii di-associated rats. (A) Representative pictures of colonic sections stained with HES in GF, Bt-2d, Bt-30d and Bt + Fp-30d rats. Representative pictures showing goblet cells staining with (B) alcian blue (indicated as AB) and (C) periodic acid Schiff (indicated as PAS) in Bt-30d rats and Bt + Fp 30d (n = 3) rats. Scale bars, 50 μm.
Deduced monosaccharide composition of mucin O-glycans from GF, Bt-2d, Bt-30d and Bt + Fp 30d rats, as identified by MALDI-TOF mass spectrometry in positive ion mode. The relative proportions of each oligosaccharide were calculated by integration of peaks on MS spectra. Mucus O-glycans types were separated into neutral oligosaccharides (blue), oligosaccharides carrying sialic acid (orange) and sulfated residues (green). Results are presented as means ± SD of the percentage of each oligosaccharide.