Abstract
Molecular signatures of melanoma have propelled new approaches to early diagnosis, monitoring of treatment response, and targeted therapy. This review discusses messenger RNA (mRNA), genomic and epigenomic melanoma biomarkers in blood and tissue specimens. The major focus is on tissue-based molecular assays to upstage sentinel lymph nodes and blood-based assays to detect melanoma progression by monitoring levels of circulating tumor cells and circulating DNA.
Keywords: biomarkers, melanoma, circulating tumor cells, microRNA, lymph node, epigenetics
INTRODUCTION
Although molecular alterations have been investigated as potential biomarkers of cancer progression or outcome, only a handful of prognostic biomarkers have been validated in cutaneous melanoma. Here, we report some of the biomarker technical platforms and assays that have been developed by our group for diagnostic and/or prognostic assessment of patients with cutaneous melanoma. When validated in the phase III setting, these biomarker assays may facilitate accurate diagnosis, stratification for treatment, and monitoring of treatment response.
Molecular biomarkers can be categorized based on the molecular component that is assessed for the tumor-related alterations. The first biomarker category comprises messenger RNA (mRNA) biomarkers, which are being validated for clinical use in multiple studies. The detection of these mRNA biomarkers is sensitive, specific, and robust using available molecular technology. Several clinical trials have investigated the use of mRNA biomarkers for upstaging melanoma-draining sentinel lymph nodes; other studies are using mRNA biomarkers to detect circulating tumor cells (CTC) in patients receiving systemic treatment for advanced melanoma.
The other biomarker categories comprise genomic and epigenomic biomarkers. Genomic biomarkers such as mutation (mt), single nucleotide polymorphism (SNP), and loss of heterozygosity (LOH) have been found in high frequency in melanoma. Most recently, epigenetic aberrations such as gene promoter region methylation of CpG islands and microRNA(miR) have taken center stage in the hunt for biomarkers. Several of these genomic and epigenomic biomarkers show promise for prognostic assessment of primary cutaneous melanoma. These different types of molecular biomarkers (Table 1) will be discussed in more detail along with important studies that demonstrate potential use in clinical settings.
Table 1. Melanoma Molecular Biomarkers.
| MARKER | CHARACTERISTICS | CLINICAL RELEVANCE | REF |
|---|---|---|---|
| mRNA BM | |||
| MART-1 | a frequent melanoma associated antigen specific for melanoma | Diagnostic/Prognostic: Combination of these biomarkers can be used in the diagnosis of SLN to upstage melanoma patients and for detection of CTC during treatment or follow-up. | 1, 5-8, 10, 11, 46, 48-50, 59 |
| MAGE-A3 | cancer-testis antigen not found in normal tissues except testis and placenta | 5-8, 10, 14, 49, 50, 53, 56, 59 | |
| GalNac-T | key enzyme involved in gangliosides GM2 and GD2 synthesis | 5-10, 15, 16, 50 | |
| PAX3 | involved in the regulation of melanin synthesis, migration and anti-apoptosis well-expressed in melanomas | 5, 7, 8, 10, 50 | |
| MITF | essential for the development and postnatal survival of melanocytes | 7, 19, 48, 59 | |
| HMW-MAA | melanoma chondroitin sulfate proteoglycan | Diagnostic: Improve desmoplastic melanoma diagnosis | 21 |
| FABP-7 | involved in lipid-metabolism | Prognostic: independent poor-prognostic factor for DFS and OS if found in tumor | 25, 26 |
| Survivin | inhibitor of apoptosis protein family | Prognostic: expression in tumor is correlated to good prognosis among stage IV patients who received postoperative immunotherapy | 27 |
| CXCR4 | chemokine receptor | Tumor characterization: the most common chemokine receptor expressed in PE liver melanoma metastases | 38 |
| CCR-9 | chemokine receptor | Tumor characterization: Expression in tumor may facilitate metastasis to the small intestine | 40 |
| Genomic BM | |||
| BRAF | a component of the Ras-MAPK-ERK pathway | Diagnostic/Prognostic: V600E mutation detected in patient serum can predict disease outcome and therapeutic response | 59, 64, 91 |
| RET | receptor tyrosine kinase | Diagnostic: G691S polymorphism improves desmoplastic melanoma diagnosis | 74 |
| Apaf-1 | tumor-suppressor gene mediating p53-induced apoptosis | Prognostic: LOH detection in tumor and serum associated with poor prognosis in patient | 77, 92 |
| FABP7 | Lipid-metabolizing capacity associated with fatty acids | Prognostic: LOH detection in tumor is associated with poor prognosis in patient | 26 |
| RASSF1A/RARb(beta) | tumor suppressor gene | Prognostic: Detection of hypermethylated RASSF1A in patient serum is associated with worse survival in patients receiving biochemotherapy | 6, 79, 83, 95 |
| RARβ | tumor suppressor gene | 6, 79, 83, 95 | |
| estrogen recepter-α | sex hormone receptor | Prognostic: Hypermethylation of ER-αfound in serum is associated with poor prognosis | 94 |
| MINT31 | multiple noncoding, methylated-in-tumor loci | Prognostic: Hypermethylation of MINT31 found in tumor is linked to good prognosis in stage III melanoma | 79 |
| DNMT3 | DNA methyltransferase | Prognostic: high level of DNMT3 in LN metastatic tumor is associated with poor prognosis in patients | 80 |
| miR-29c | microRNA | Prognostic: high level of miR-29c in LN metastatic tumor is associated with better prognosis in patients | 80 |
| miR-532-5p | microRNA | Tumor characterization: may contribute to melanoma progression by downregulation of RUNX3 expression | 85 |
mRNA BIOMARKERS
Specific genes with functional roles in tumor progression are of particular interest as mRNA biomarkers in both tissue and blood. These include genes from the melanogenesis pathway, such as tyrosinase, gp100, TRP-1, TRP-2, and MART-1 [1,2]. However, some mRNA biomarkers also can be expressed in low levels in normal melanocytes and nevi, producing false-positives [3,4]. As described below, our group has extensively investigated many prognostic mRNA biomarkers for upstaging lymph node status and identifying circulating tumor cells (CTC) [1,5-12].
Candidate mRNA Biomarkers
Some of the candidate mRNA biomarkers we have extensively investigated are described below. We have divided them into melanoma-related, apoptosis-related, and chemokine receptor biomarkers (Table 1).
MAGE-A3 is a melanoma-associated antigen that is not found in normal tissues except testis and placenta [13,14]. This gene is a member of the MAGE family of testis-related antigens that are highly specific in cancer tissues including melanoma. Their function is not well understood, limiting their potential utility. These so-called testis-related genes may be just non-specific activation of genes of limited function in cancer.
Cell surface tumor-related gangliosides of melanoma such as GM2 and GD2 are well defined oncofetal melanoma-related antigens usually found in aggressive melanomas [15,16]. β1→4-N-acetylgalactosaminyltransferase (GalNAc-T) is a key enzyme involved in synthesis of gangliosides GM2 and GD2 from GM3 and GD3, respectively [9,15,16]. GM2 and GD2 are not expressed in melanocytes or nevi. Our data indicate that elevated GalNAc-T mRNA expression in melanoma cells appears to be a biomarker for aggressive melanoma [9].
PAX3 (paired-box homeotic gene transcription factor 3) is involved in the regulation of melanin synthesis, migration and anti-apoptosis [17,18]. PAX3 has been considered as a stem cell marker and is well-expressed in melanomas and not in normal skin melanocytes or benign nevi by reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) and in situ hybridization [18]. Microphthalmia-associated transcription factor (MITF) plays an important role in melanocyte biology and in melanoma progression. PAX3 is considered as a regulator of MITF, which has demonstrated utility for detecting CTC [19].
HMW-MAA, also known as melanoma chondroitin sulfate proteoglycan [20], is a sensitive mRNA biomarker for primary desmoplastic melanoma (DM) [21]. Because metastatic DM is very difficult to diagnose by immunohistochemical staining (IHC) such as with anti-MART-1 and anti-HMB-45 antibodies, we used HMW-MAA RT-qPCR to assess 40 primaries and 23 metastases of DM. Results showed that 25 (63%) DM primaries and 16 (70%) DM metastases expressed HMW-MAA mRNA, whereas MART-1 was expressed in 9 (23%) primaries and 5 (22%) metastases in the same melanoma specimens. HMW-MAA mRNA was expressed in 8 (57%) of 14 nodal metastases, whereas MART-1 mRNA was expressed in 3 (21%) of 14 nodal metastases.
A new biomarker recently found in melanoma is FABP7 (Fatty acid binding protein 7). FABP7 was reported to have lipid-metabolizing capacity associated with fatty acids [22,23]. In the brain, it has been linked to cell proliferation and tissue differentiation [24]. FABP7 can be regulated by protein kinase C (PKC) and the MAPK/ERK1/2 pathway through independent mechanisms in melanoma cell lines. Furthermore, in vitro studies demonstrated that FABP7 is involved in cell proliferation and invasion [25]. We reported that FABP7 mRNA was detected in 60 of 87 (69%) AJCC Stage I-III melanomas. However, it was detected only in 13 of 68(19%) AJCC Stage III-IV metastatic melanomas. Analyzing of 37 paired primary and metastatic melanomas by IHC assessment, FABP7 was detected in 27 of 37 (73%) primary melanomas and in 10 of 37(27%) metastatic melanomas[26]. FABP7 detection in metastatic tissues was inversely correlated with disease-free and overall survival and was a significant independent prognostic factor for survival (Figure 1).
Figure 1.
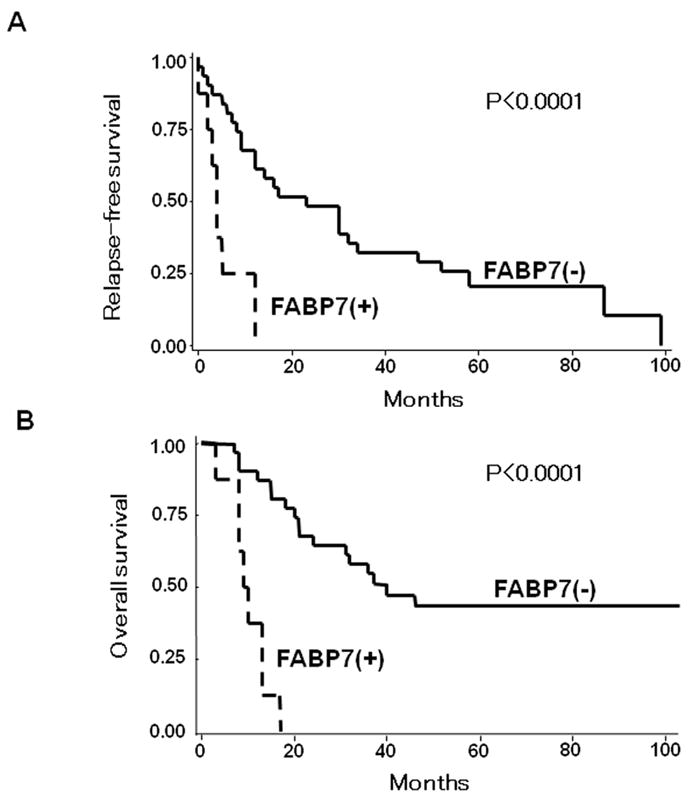
Kaplan–Meier curves of relapse-free survival (A) and overall survival (B) based on FABP7 mRNA detection in melanoma metastases. Reprinted with permission from Goto Y et al., Aberrant Fatty Acid-Binding Protein 7 Gene Expression In Cutaneous Malignant Melanoma, J Invest Dermatol, 130:221-9, 2010.
Survivin is a member of anti-apoptosis molecular inhibitors(IAP), which often promote various cancers. Survivin is a protein known to be significantly expressed in highly malignant cutaneous melanomas. Lower expression of survivin was correlated with good prognosis among stage IV patients who received postoperative vaccine immunotherapy [27].
Chemokines are small secreted chemotactic cytokines involved in cell trafficking and specific organ site methylation [28]. Chemokines and their receptors have been identified as key factors that control the migration of tumor cells to specific organ sites [29]. The expression of several chemokines and their receptors is upregulated during melanoma progression. In melanoma, several chemokines and chemokine receptors have been linked to tumor growth and specific organ metastasis [30-34]. Two of these chemokine receptors expressed in melanoma are CXCR4 and CCR7 [35-37]. In 2005, Scala et al. reported that CXCR4 expression was detected in 31 of 71(43.6%) primary cutaneous melanomas and associated with poor prognosis [36]. We assessed CXCR4 expression in resected tumor tissue from patients who underwent hepatic surgery for melanoma liver metastasis [38]. We identified CXCR4 as the most common chemokine receptor expressed in paraffin-embedded liver metastases. RT-qPCR demonstrated CXCR4 expression in 24 of 27 (89%) metastases. In vitro treatment of melanoma cells with CXCL12 (CXCR4-specific ligand) significantly increased cell migration (P<0.001). CXCL12 is highly expressed by liver cells and supports the attraction of CXCR4-positive melanoma cells.
CCR9-CCL25 interaction has been implicated as being critical for the migration of peripheral T-cells to inflammatory small intestine [39]. We demonstrated CCR9 expression in 88 of 102 small intestine metastatic melanomas, in 8 of 8 melanoma lines derived from small intestine metastases, and in 0 of 96 metastatic melanomas to other organ sites [40]. In vitro migration and invasion studies on CCR9(+) melanoma lines showed migration in response to CCL25. CCR9 expression by small intestine metastases and concomitant α4 and β1 integrin expression were confirmed by flow cytometry. These findings demonstrate the importance of the CCR9-CCL25 axis in preferential metastasis to small intestine. This is a significant metastasis event because metastasis of any cancer type to the small intestine is uncommon. This demonstrated that chemokine-receptor axis can promote site-specific metastasis to the small intestine from a primary tumor located at any anatomical location, without a direct anatomical blood drainage pattern.
Recently, using human melanoma xenografted in athymic nude SCID mice for melanoma brain metastasis, Izraely et al. [41] showed that CCR4 expression was significantly higher in brain metastatic variants than in corresponding local variants. CCR4 is suggested to be associated with brain metastasis in human melanoma and may be an important factor for identifying melanoma cells likely to metastasize to the brain [41]. In general, chemokine receptors are not diagnostic but provide valuable information of potential metastasis ability and site of metastasis. These need to be further explored.
Molecular Upstaging of Sentinel Lymph Nodes
Lymph node metastasis of melanoma is one of the most significant prognostic determinants. Sentinel lymphadenectomy is standard for surgical staging of clinically localized melanoma [42]. Accurate assessment of the sentinel lymph node (SLN) is important in determining disease staging and prognosis. However, the detection of micrometastasis in SLNs is not always accurate based on hematoxylin and eosin (H&E) and IHC staining. Although IHC using anti-S100p, HMB45, and MART-1 antibodies is standard, a significant number of patients with histopathology-negative SLNs subsequently develop recurrent disease. Improved techniques for detecting clinically significant micrometastases in melanoma-draining SLNs are needed to reduce the subjectivity of current detection methods. Also, better biomarkers are needed for prognosis.
Our group developed multimarker RT-PCR assays to detect occult metastasis in SLNs. These assays, initially applied to frozen specimens, have since been validated for paraffin-embedded (PE) tissues [11,12,43-49]. We performed RT-PCR on archived PE SLNs from 215 clinically SLN(-) patients who underwent lymphatic mapping and SLND for melanoma and were followed up for >8 yrs. PE SLNs (n=308) from these patients were sectioned and assessed by qPCR for four melanoma-associated biomarkers: MART-1, MAGE-A3, GalNAcT, and PAX3. Fifty three (25%) patients had histopathology-positive SLNs by H&E and/or IHC [10]. Forty-eight (30%) of the 162 patients with histopathology-negative SLNs had SLNs which expressed >1 mRNA biomarker, and Cox proportional hazards model analysis showed a significantly increased risk of disease recurrence of these 48 patients (P<0.0001). The presence of >1 biomarker in histopathology-negative SLNs was significantly associated with lower survival rate by multivariate analysis (P <0.0002). This study was recently updated with longer median follow-up (>11 yrs) and results remain highly significant (Figure 2; [50]). The study demonstrates the prognostic utility of molecular upstaging of SLNs with specific mRNA biomarkers. It is very clear that selection of mRNA biomarkers, sampling of SLN, and molecular assays used are highly important. The study demonstrates that specific molecular upstaging of SLNs has more prognostic value than IHC positivity in longer-term follow-up. Previous studies reporting no correlation between RT-PCR upstaging of SLN and clinical outcome had flaws in specimen sampling, mRNA biomarkers, assay sensitivity, assay specificity, and/or patient cohorts.
Figure 2.
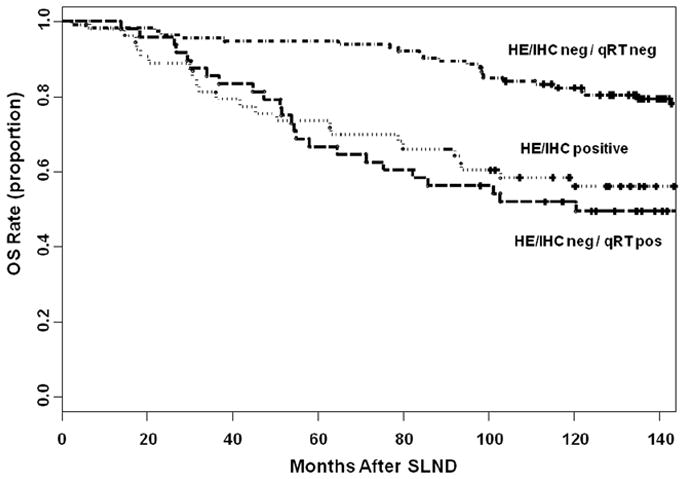
Kaplan–Meier curves of overall survival based on histopathologic (H&E/IHC) and molecular (qRT) status of the sentinel lymph node (SLN). Reprinted with permission from Nicholl MB et al., Molecular upstaging based on paraffin-embedded sentinel lymph nodes: ten-year follow-up confirms prognostic utility in melanoma patients, Ann Surg, 253:116-22, 2011.
A recent systematic meta-analysis [51] assessed multimarker RT-qPCR upstaging of SLNs in 22 studies that were conducted in years 1998-2006 and enrolled 4,019 patients with clinical stage I/II cutaneous melanoma. RT-qPCR status was associated with TNM stage and with overall and disease-free survival. Currently, RT-qPCR upstaging of histopathology-negative SLNs is being investigated in the phase III Multicenter Selective Lymphadenectomy Trial-II (MSLT-II). Over 1900 patients have been entered into this multicenter international trial to date. Results of the molecular studies are expected to validate results of phase II RT-qPCR studies and determine the clinical relevance of molecular upstaging in SLN specimens.
Landmark Blood Molecular Biomarker Studies
During tumor progression, malignant melanoma cells invade lymphatic and blood vessels and shed cells that circulate in the peripheral bloodstream. Thus molecular biomarkers in blood are promising surrogates for monitoring tumor progression [44,52-55]. RT-qPCR detection of CTC in blood of melanoma patients has been associated with disease stage and clinical outcome [53]. RT-qPCR assay can detect a few CTC among millions of peripheral blood leukocytes (PBL) [56]. Investigations have used a single-biomarker assay for detection of CTC, but single-biomarker assays are limited by known heterogeneous expression, particularly in advanced disease [1,3,56]. Hoon et al. [56] previously reported that a combination of CTC mRNA biomarkers is necessary to compensate for heterogeneous biomarker expression in melanoma patients; multiple biomarkers can increase the sensitivity of CTC detection and reduce false-negative results.
The efficacy of a multimarker assay depends on the careful selection of CTC biomarkers [57], serial rather than single-point assessment [7,8], and quantification of biomarker expression to compensate for ectopic and background mRNA [58]. Based on these strategies, Koyanagi et al. [5,19] have developed a multimarker RT-qPCR assay to detect CTC in blood of melanoma patients. Koyanagi et al. demonstrated the utility of the RT-qPCR CTC assays in monitoring patients treated with immuno and chemotherapy [6-8]. Recently, Kitago et al. described monoclonal antibody and immunomagnetic bead capture assay to isolate melanoma CTC expressing HMW-MAA; isolated CTC were assessed by multimarker RT-qPCR assay (mRNA biomarkers) and by qPCR assay (BRAFmt) [59]. The bead assay validated the direct RT-qPCR, demonstrating presence of CTC. The direct assay is logistically more feasible and can be easily performed in a multi-institutional clinical trial setting.
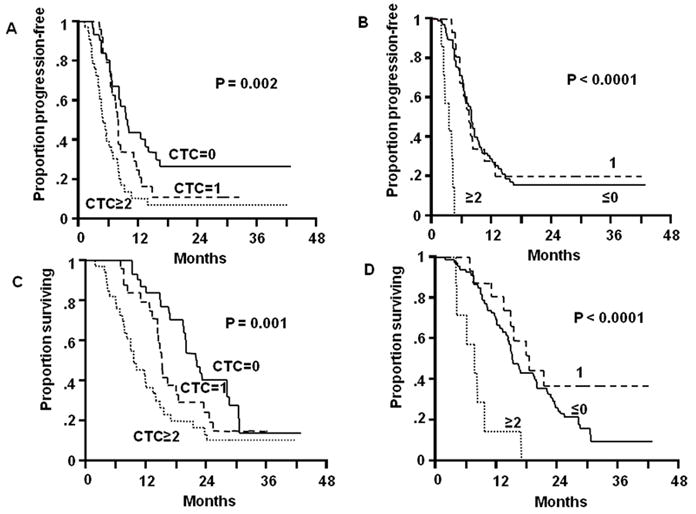
Tissue-based prognostic assessment is a static (single-point) measurement that does not directly reflect ongoing disease progression, particularly subclinical systemic metastasis. By contrast, blood-based assessment can be performed as serial measurements, which may be particularly valuable to manage melanoma patients receiving systemic chemo- and/or immunotherapy [60]. Koyanagi et al. assessed serial blood specimens from patients enrolled in a prospective phase II multicenter neoadjuvant clinical trial of biochemotherapy before and after surgical treatment of AJCC stage III melanoma. They found that changes in CTC detection were significantly correlated with disease progression and overall survival (Figure 3A) [8]. Separately, they also assessed serial blood specimens collected from 87 patients before and during induction biochemotherapy and maintenance biotherapy for stage IV melanoma; changes in CTC detection were significantly correlated with treatment response, progression-free survival, and overall survival (Figure 3B) [7]. Recently, a large prospective multicenter clinical trial demonstrated the prognostic utility of CTC in stage IV melanoma patients [61] and stage III patients (D. Hoon, unpublished data).
Figure 3.
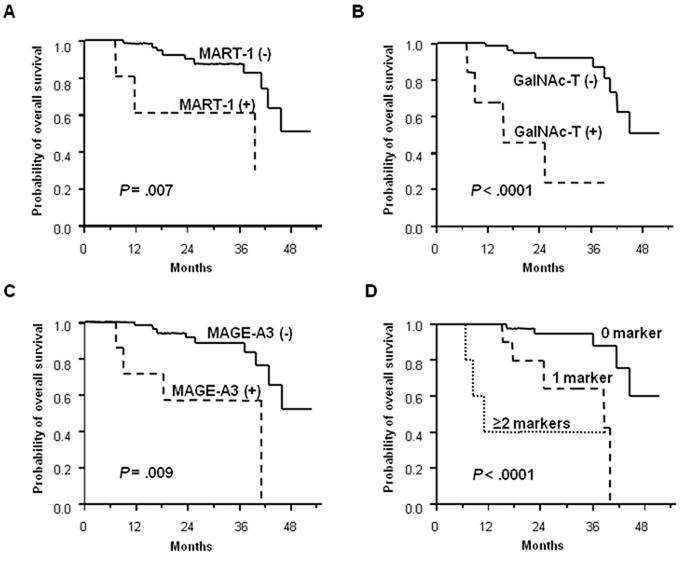
A. (A) Kaplan-Meier curves of OS based on MART-1 detection after treatment. (B) Kaplan-Meier curves of OS based on GalNAc-T detection after treatment. (C) Kaplan-Meier curves of OS based on MAGE-A3 detection after treatment. (D) Kaplan-Meier curves of OS based on CTC BM detection after treatment. Reprinted with permission from Koyanagi K et al., Serial monitoring of circulating melanoma cells during neoadjuvant biochemotherapy for stage III melanoma: outcome prediction in a multicenter trial, J Clin Oncol. 23:8057-64, 2005. License Number: 2510280678967
B. (A) Kaplan-Meier curves of PFS based on CTC BM detection after two cycles of induction BC. (B) Kaplan-Meier curves of PFS based on changes in CTC BM detection during two cycles of induction BC. (C) Kaplan-Meier curves of OS based on CTC BM detection after two cycles of induction BCT. (D) Kaplan-Meier curves of OS based on changes in CTC BM detection during two cycles of induction BC. In each panel, the solid line corresponds to no CTC BMs, the broken line is 1 CTC BM, and the dotted line is ≥2 CTC BMs. Reprinted with permission from Koyanagi K et al., Serial monitoring of circulating melanoma cells during neoadjuvant biochemotherapy for stage III melanoma: outcome prediction in a multicenter trial, J Clin Oncol. 23:8057-64, 2005. License Number: 2510280678967.
CTC detection in serial blood specimens in melanoma may similarly be employed to determine which component of the treatment is most effective and which needs to be improved. As treatment regimens for advanced melanoma become multimodal and multiphasic, the development of a tool to identify high-risk patients and to monitor response to systemic therapy is urgently needed. Serial RT-qPCR assay can assess CTC changes during different phases of treatment, and this makes RT-qPCR detection of CTC a promising method to evaluate treatment efficacy. Identification of CTC with specific gene expression allows us to better screen high-risk CTC likely to establish distant metastasis. Individual CTC do not have equal metastasis potential. The measurement of numbers of CTC may not be prognostic particularly in early stages of cancer.
GENOMIC AND EPIGENOMIC BIOMARKERS
Sequential genetic aberrations have been correlated with the development and progression of various cancers. Several somatic gene alterations in melanoma have been reported, such as CDKN2A and CDK4 [62]. Recently, epigenetic alterations have become a hot topic in melanoma. In 2001, Wu et al [63] defined epigenetics as “the study of changes in gene function that are mitotically and/or meiotically heritable and that do not entail a change in DNA sequence”. Today this definition is globally accepted. The major players in epigenetic gene regulation to date are gene promoter methylation, histone modifications, and short non-coding mRNAs. It will be important to investigate genomic, transcriptome, and epigenetic biomarkers together in relation to tumor progression and disease outcome.
Genomic biomarkers
BRAF kinase has become a key target of interest in melanoma because of its high frequency of mutation. BRAF kinase is a component of the Ras-MAPK-ERK pathway; BRAF mutation occurs frequently in exons 11 V600E. We found BRAF V600E mutation (BRAFmt) frequency was significantly (P<0.0024) higher in metastatic tumors (n=68) than in primary melanomas (n=59) [64]. This suggested that the BRAFmt may be acquired during melanoma progression to distant metastasis. This also suggested BRAFmt does not always occur at initiation of melanoma, supporting the concept that it is not always the cause of melanoma. In reviewing the literature, there is variability in reporting the actual frequency of BRAFmt in primary melanoma. Assessment of melanoma BRAFmt has gained importance because of the effectiveness of PLX4032 [65] and GSK2118436 [66], new agents that target BRAFmt.
RET (rearranged during transfection) proto-oncogene encodes a receptor tyrosine kinase [67-69] containing four cadherin-related motifs and a cysteine-rich region in the extracellular domain [67,70]. Glial cell line-derived neurotrophic factor (GDNF) family members bind the extracellular domain of RET through a complex formed with glycosyl-phosphatidylinositol-anchored coreceptor (GFRα1–4), a member of the GDNF receptor family [71]. Activated RET induces signaling through RAS-BRAF-ERK, phosphatidylinositol 3-kinase (PI3K)-Akt and p38 mitogen-activated protein kinase (MAPK) pathways [70]. Activation of both RET-RAS-BRAF-MEK-ERK and RET-PI3K-Akt pathways leads to cell proliferation and survival, whereas the RET-PI3K pathway is frequently involved in cell motility [70,71]. Several germline mutations of RET play an important role in development of multiple endocrine neoplasia (MEN) syndromes MEN2A, MEN2B, and familial medullary thyroid carcinoma [71,72].
G691S RET polymorphism (RETp) is a single nucleotide germline polymorphism in exon 11 of the juxtamembrane region of RET, which enhances the response of RET to GDNF in pancreatic cancer [73]. In cutaneous malignant melanomas, particularly desmoplastic cutaneous melanomas, which are highly neurotropic, RETp reportedly plays an important role in enhancing malignant behavior [74]. RETp was shown to enhance and prolong phosphorylation of RET signaling pathways including ERK1/2 and AKT after GDNF stimulation, leading to vigorous cell proliferation and migration in melanoma cells (Figure 4). RETp-induced enhancement of cell proliferation and migration was not affected by BRAFmt, which is found frequently in metastatic melanomas. We demonstrated that RETp could be acquired during melanoma progression. This would be a significant advantage for melanoma survival and invasion. If RETp plays a central role in inducing primary tumor neurotropism in melanomas, then targeting tyrosine kinase inhibitors against RETp might be a useful approach to melanoma therapy.
Figure 4.
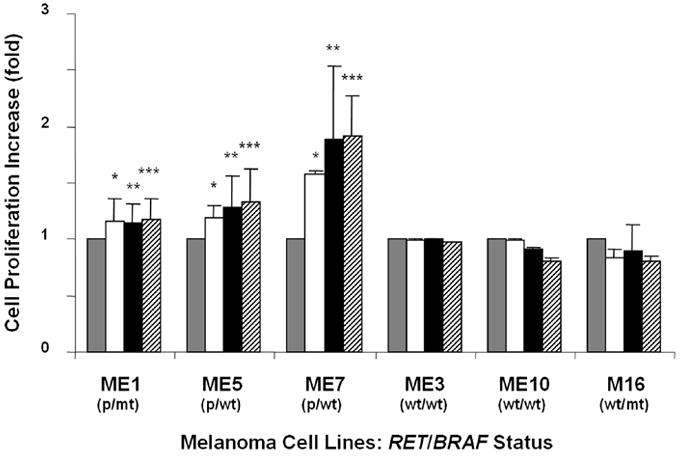
GDNF-induced cell proliferation and migration in melanoma cells bearing G691S RET polymorphism and/or V600E BRAF mutation. p; G691S RET polymorphism, mt; V600E BRAF mutation, wt; wild type. Proliferation of melanoma cells was analyzed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay 48h after GDNF stimulation. GDNF □ 5 ng/ml, ■ 25 ng/ml or ▨ 50 ng/ml.
 Delivery agent-treated control. Bars±s.d. show fold increase over each control cell. *,**,*** P<0.05 versus each control. Bars±s.d. without asterisks show no significant change compared to each control. Reprinted with permission from Narita N et al., Functional RET G691S Polymorphism in Cutaneous Malignant Melanoma, Oncogene, 28:3058-3068, 2009.
Delivery agent-treated control. Bars±s.d. show fold increase over each control cell. *,**,*** P<0.05 versus each control. Bars±s.d. without asterisks show no significant change compared to each control. Reprinted with permission from Narita N et al., Functional RET G691S Polymorphism in Cutaneous Malignant Melanoma, Oncogene, 28:3058-3068, 2009.
The frequency of loss of heterozygosity (LOH) in tumors, along with specific gene function in tumor cells, suggests that LOH may play a significant role in regulating tumor-suppressor genes and oncogenes. Frequent LOH of DNA microsatellites on specific chromosomal regions has been reported in cutaneous melanoma [75,76]. These LOH biomarkers have prognostic potential. For example, apoptotic protease activating factor-1 (APAF-1) is a tumor-suppressor gene that mediates apoptosis [77]. We found that LOH of microsatellites covering the APAF-1 locus (12q22-23) was significantly more common in metastatic tumors (36 of 98 specimens; 37%) than in primary melanomas (10 of 54 specimens; 19%). In metastatic melanomas, APAF-1 loss significantly correlated with a worse prognosis. More recently, we reported the frequency of LOH in the region covering FABP7 [26], which is a tumor-related gene involved in proliferation and invasion of melanoma cells [25]. LOH was identified in 10 of 20 (50%) metastatic melanomas at the 6q22.31 region containing the FABP7 gene, and 0 of 14 primary melanomas. FABP7 expression is significantly decreased in metastases of melanoma due to LOH, and its decrease is associated with significantly poorer disease outcome.
Overall, there are many genomic biomarkers identified in melanoma, but few have high frequency and functional utility with the exception of BRAF V600Emt. Continued genomic deep-sequencing will probably reveal genomic aberrations that are functionally important in specific melanoma subsets.
Epigenomic biomarkers
DNA methylation plays one of the most important roles in regulating gene expression and chromatin architecture. CpG island methylation can result in suppression of gene expression, and contribute to tumorigenesis and cancer progression [78]. Epigenetic suppression can occur by methylation of specific CpG islands in the promoter region, histone methylation or acetylation and/or miR activation [79-81]. In melanoma, more than 50 genes have been reported to demonstrate aberrant hypermethylation of promoter CpG islands [82].
We were the first group to identify and verify the inactivation of RAS association domain family protein 1A, RASSF1A, which is a human tumor suppressor gene in melanoma [83]. Hypermethylation of two regions in the RASSF1A CpG island was investigated in metastatic cutaneous melanomas. Methylation of the RASSF1A CpG island was detected in 57% of tumors. No methylation was detected in normal skin tissues or lymphocytes. Hypermethylation of CpG regions correlated with no expression of the RASSF1A gene. RASSF1A is key gene in regulating mitosis and methylation; thus, it is highly important in controlling tumor invasion and metastasis. RASSF1A gene suppression by promoter methylation is strongly correlated with melanoma progression and outcome [79,80].
The CpG island methylator phenotype (CIMP) may be associated with development of malignancy through coordinated inactivation of tumor suppressor and tumor-related genes (TRGs) and methylation of multiple noncoding, methylated-in-tumor (MINT) loci. These epigenetic changes create a distinct CIMP pattern that has been linked to progression and disease outcome in gastrointestinal cancers [84]. The existence of a clinically significant CIMP in cutaneous melanoma progression was recently demonstrated by our group [79]. We showed an increase in hypermethylation of several TRGs (WIF1, TFPI2, RASSF1A, and SOCS1) with advancing clinical tumor stage. Furthermore, we reported a significant positive association between the methylation status of MINT17, MINT31, and specific TRGs. These findings demonstrated that a CIMP pattern is significant for melanoma progression. We recently demonstrated that DNMT3(a/b), which plays a significant role in methylation of TRGs, was significantly upregulated during melanoma progression [80]. The regulation of DNMT3 was controlled by another epigenetic factor, miR29c [80]. These events strongly indicate that specific epigenetic aberrations in melanoma progression are very significant, thus new potential targets.
Recently, we investigated the expression of RUNX3 and its regulatory factor, microRNA miR-532-5p [85]. RUNX3 is a tumor-suppressor gene [86,87]. Expression of RUNX3 mRNA and human miR-532-5p was assessed in cell lines and in primary and metastatic melanomas. RUNX3 expression was down-regulated in all 11 (100%) melanoma lines relative to normal melanocytes. In primary and metastatic melanomas, RUNX3 had reduced expression relative to normal skin. Evidence of RUNX3 promoter region methylation was demonstrated in 5 of 17 (29%) melanoma cell lines, 2 of 52 (4%) primary melanomas, and 5 of 30 (17%) metastatic melanomas. miR-532-5p expression was upregulated in melanoma lines and metastatic melanomas relative to normal melanocytes and primary melanomas, respectively. To investigate the relationship between RUNX3 and miR-532-5p, we transfected anti-miR-532-5p into melanoma lines and evaluated RUNX3 expression. Inhibition of miR-532-5p activated RUNX3 mRNA and protein expression. miR-532-5p may contribute to melanoma progression by downregulation of RUNX3 expression. In the future, we will likely identify miR that play a significant role in melanoma progression as seen on other cancer systems.
Landmark Studies Using Serum Specimens
Cell-free tumor-specific DNA has been detected in plasma and serum from cancer patients [88-90]. This observation has been expanded to develop blood biomarkers. We have demonstrated prognostic utility for circulating DNA microsatellites, mutation, and methylation. In one study, we assessed nine microsatellites to examine allelic instability (AI) in serum specimens obtained from 41 stage IV melanoma patients before the initiation of biochemotherapy [91]. AI was detected in 12 of 41 (29%) patients. The response rate of these 12 patients was 17%, whereas that of the 29 patients without AI was 72%. The presence of AI was statistically significant and independently associated with disease progression.
AI encompassing the APAF-1 locus (12q22-23) is found frequently in metastatic melanoma [77]. When we evaluated 12q22-23 AI status as a surrogate biomarker to predict response to biochemotherapy, we found that AI of the 12q22-23 region was significantly lower in responders (5 of 24, 21%) compared with nonresponders (11 of 20, 55%). As expected, nonresponders to biochemotherapy (AI-positive group) had a significantly worse survival than responders (AI-negative group) [92].
Also, we showed the presence and predictive utility of circulating BRAFmt in DNA from melanoma patients [93]. Overall survival was significantly lower in 20 patients with the BRAFmt before biochemotherapy compared with those that did not have the BRAFmt. After biochemotherapy, BRAFmt was lower in the responder group (1 of 10, 10%) than in the nonresponder group (7 of 10, 70%). The BRAF V600Emt can be useful for monitoring melanoma patients receiving biochemotherapy.
Methylation detected in serum DNA can predict disease outcome and therapeutic response in patients receiving concurrent biochemotherapy for metastatic melanoma [94,95]. We developed multiple methylated serum biomarkers based on frequently methylated tumor-related genes in melanoma tissues. In a study of RASSF1A, RAR-β2 and MGMT, we found that circulating methylated RASSF1A was significantly less frequent in biochemotherapy responders (3 of 23, 13%) than nonresponders (10 of 24, 42%), and it was significantly correlated with overall survival [95] (Figure 5). Patients with RASSF1A, RAR-β2, or at least one of the three biomarkers had significantly worse overall survival than patients with no biomarkers. In a separate study, we demonstrated serum estrogen receptor α hypermethylation was detected more frequently in advanced melanomas than localized melanomas and was the only factor predicting progression-free and overall survival in patients receiving biochemotherapy [94].
Figure 5.
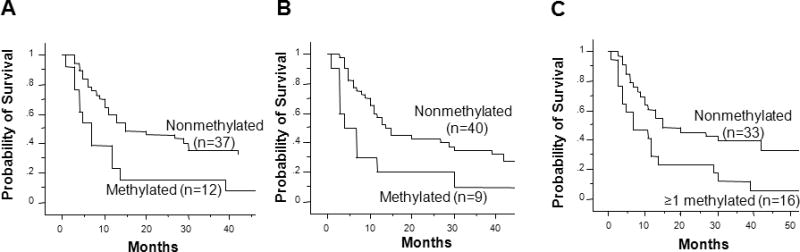
Kaplan-Meier curves of overall survival based on pre-biochemotherapy methylation status of three serum biomarkers: RASSF1A, RAR-β2 and MGMT. (A) Survival probability according to RASSF1A methylation status (log-rank test, P =.013). (B) Survival probability according to RAR-β2 methylation status (log-rank test, P = .02). (C) Survival probability according to methylation of at least one of three markers (log-rank test, P = .01). Reprinted with permission from Mori T et al., Predictive Utility of Circulating Methylated DNA in Serumof Melanoma Patients Receiving Biochemotherapy, J Clin Oncol. 23:9351-58, 2005. License Number: 2587200686801.
Our group was among the first to report the prognostic potential for combined assessment of CTC and methylated blood DNA biomarkers [6]. We assessed matched pairs of PBL and serum specimens from 50 AJCC stage IV melanoma patients before administration of biochemotherapy. PBL were analyzed for three mRNA CTC biomarkers: MART-1, GalNAc-T, and MAGE-A3. Sera were analyzed for two methylated DNA biomarkers: RASSF1A and RAR-β2. CTC were detected in 13 of 15 (86%) patients with serum tumor-related methylated DNA, and in 13 of 35 (37%) patients without methylated DNA. The number of CTC biomarkers detected was significantly associated with methylated DNA. Patients with both CTC and methylated DNA showed significantly poorer response to biochemotherapy and poorer progression. Findings indicate that these two assays in combination may be a very useful determinant of disease status and may improve efficacy of monitoring melanoma progression.
CONCLUSION
The above review covers only some of the highlights of our approaches in molecular biomarkers applied to both tissues and blood for diagnosis and prognosis. The technology available for development and validation of molecular biomarkers has significantly improved in the last decade; as a result, a number of molecular biomarkers have been investigated in a multitude of platforms. In the near future molecular biomarkers will play a significant role in the diagnosis and management of melanoma. Also, a validated molecular signature for melanoma may eventually allow highly efficient, tailored treatments for this cancer. Molecular oncology has and will continue to impact clinical practice and be at the forefront of melanoma translational research. As molecular technology and sensitivity towards specific targets improves, it is inevitable that melanoma will be better classified based on molecular characteristics.
Acknowledgments
This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, Melanoma Research Alliance, as well as the Award Number P0 CA029605, P0 CA012582, and R-33 CA100314 from the National Institutes of Health, NCI. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
Footnotes
Disclosure: All authors disclose no financial/commercial conflict of interest except that Dr. D. Hoon has licensing agreement with Abraxis Healthcare on several of the biomarkers discussed in this manuscript.
References
- 1.Takeuchi H, Kuo C, Morton DL, et al. Expression of differentiation melanoma-associated antigen genes is associated with favorable disease outcome in advanced-stage melanomas. Cancer Res. 2003;63:441–448. [PubMed] [Google Scholar]
- 2.Kawakami Y, Eliyahu S, Delgado CH, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci U S A. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser R, Rass K, Seiter S, et al. Detection of circulating melanoma cells by specific amplification of tyrosinase complementary DNA is not a reliable tumor marker in melanoma patients: a clinical two-center study. J Clin Oncol. 1997;15:2818–2825. doi: 10.1200/JCO.1997.15.8.2818. [DOI] [PubMed] [Google Scholar]
- 4.Jung FA, Buzaid AC, Ross MI, et al. Evaluation of tyrosinase mRNA as a tumor marker in the blood of melanoma patients. J Clin Oncol. 1997;15:2826–2831. doi: 10.1200/JCO.1997.15.8.2826. [DOI] [PubMed] [Google Scholar]
- 5.Koyanagi K, Kuo C, Nakagawa T, et al. Multimarker quantitative real-time PCR detection of circulating melanoma cells in peripheral blood: relation to disease stage in melanoma patients. Clin Chem. 2005;51:981–988. doi: 10.1373/clinchem.2004.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koyanagi K, Mori T, O’Day SJ, et al. Association of circulating tumor cells with serum tumor-related methylated DNA in peripheral blood of melanoma patients. Cancer Res. 2006;66:6111–6117. doi: 10.1158/0008-5472.CAN-05-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyanagi K, O’Day SJ, Boasberg P, et al. Serial monitoring of circulating tumor cells predicts outcome of induction biochemotherapy plus maintenance biotherapy for metastatic melanoma. Clin Cancer Res. 2010;16:2402–2408. doi: 10.1158/1078-0432.CCR-10-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyanagi K, O’Day SJ, Gonzalez R, et al. Serial monitoring of circulating melanoma cells during neoadjuvant biochemotherapy for stage III melanoma: outcome prediction in a multicenter trial. J Clin Oncol. 2005;23:8057–8064. doi: 10.1200/JCO.2005.02.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo CT, Bostick PJ, Irie RF, et al. Assessment of messenger RNA of beta 1-->4-N-acetylgalactosaminyl-transferase as a molecular marker for metastatic melanoma. Clin Cancer Res. 1998;4:411–418. [PubMed] [Google Scholar]
- 10.Takeuchi H, Morton DL, Kuo C, et al. Prognostic significance of molecular upstaging of paraffin-embedded sentinel lymph nodes in melanoma patients. J Clin Oncol. 2004;22:2671–2680. doi: 10.1200/JCO.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeuchi H, Taback B, Kuo C, Hoon DS. Clinicopathological utility of molecular staging for melanoma patients undergoing sentinel lymphadenectomy. Ann Surg Oncol. 2004;11:152S–155S. doi: 10.1007/BF02523620. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi H, Wascher RA, Kuo C, et al. Molecular diagnosis of micrometastasis in the sentinel lymph node. Cancer Treat Res. 2005;127:221–252. doi: 10.1007/0-387-23604-x_12. [DOI] [PubMed] [Google Scholar]
- 13.Gaugler B, Van den Eynde B, van der Bruggen P, et al. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyashiro I, Kuo C, Huynh K, et al. Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem. 2001;47:505–512. [PubMed] [Google Scholar]
- 15.Tsuchida T, Saxton RE, Irie RF. Gangliosides of human melanoma: GM2 and tumorigenicity. J Natl Cancer Inst. 1987;78:55–60. doi: 10.1093/jnci/78.1.55. [DOI] [PubMed] [Google Scholar]
- 16.Tsuchida T, Saxton RE, Morton DL, Irie RF. Gangliosides of human melanoma. J Natl Cancer Inst. 1987;78:45–54. doi: 10.1093/jnci/78.1.45. [DOI] [PubMed] [Google Scholar]
- 17.Maschhoff KL, Baldwin HS. Molecular determinants of neural crest migration. Am J Med Genet. 2000;97:280–288. doi: 10.1002/1096-8628(200024)97:4<280::aid-ajmg1278>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 18.Scholl FA, Kamarashev J, Murmann OV, et al. PAX3 is expressed in human melanomas and contributes to tumor cell survival. Cancer Res. 2001;61:823–826. [PubMed] [Google Scholar]
- 19.Koyanagi K, O’Day SJ, Gonzalez R, et al. Microphthalmia transcription factor as a molecular marker for circulating tumor cell detection in blood of melanoma patients. Clin Cancer Res. 2006;12:1137–1143. doi: 10.1158/1078-0432.CCR-05-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campoli MR, Chang CC, Kageshita T, et al. Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit Rev Immunol. 2004;24:267–296. doi: 10.1615/critrevimmunol.v24.i4.40. [DOI] [PubMed] [Google Scholar]
- 21.Goto Y, Arigami T, Murali R, et al. High molecular weight-melanoma-associated antigen as a biomarker of desmoplastic melanoma. Pigment Cell Melanoma Res. 2010;23:137–140. doi: 10.1111/j.1755-148X.2009.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anthony TE, Mason HA, Gridley T, et al. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arai Y, Funatsu N, Numayama-Tsuruta K, et al. Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J Neurosci. 2005;25:9752–9761. doi: 10.1523/JNEUROSCI.2512-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haunerland NH, Spener F. Fatty acid-binding proteins--insights from genetic manipulations. Prog Lipid Res. 2004;43:328–349. doi: 10.1016/j.plipres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Goto Y, Matsuzaki Y, Kurihara S, et al. A new melanoma antigen fatty acid-binding protein 7, involved in proliferation and invasion, is a potential target for immunotherapy and molecular target therapy. Cancer Res. 2006;66:4443–4449. doi: 10.1158/0008-5472.CAN-05-2505. [DOI] [PubMed] [Google Scholar]
- 26.Goto Y, Koyanagi K, Narita N, et al. Aberrant Fatty Acid-Binding Protein 7 Gene Expression In Cutaneous Malignant Melanoma. J Invest Dermatol. 2010;130:221–229. doi: 10.1038/jid.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi H, Morton DL, Elashoff D, Hoon DS. Survivin expression by metastatic melanoma predicts poor disease outcome in patients receiving adjuvant polyvalent vaccine. Int J Cancer. 2005;117:1032–1038. doi: 10.1002/ijc.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnik A. Chemokines and cancer. Int J Cancer. 2006;119:2026–2029. doi: 10.1002/ijc.22024. [DOI] [PubMed] [Google Scholar]
- 30.Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J Clin Pathol. 2007;60:596–599. doi: 10.1136/jcp.2005.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarini-Meury AA, Conrad C. Melanoma and innate immunity--aActive inflammation or just erroneous attraction? Melanoma as the source of leukocyte-attracting chemokines. Semin Cancer Biol. 2009;19:84–91. doi: 10.1016/j.semcancer.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 32.Raman D, Baugher PJ, Thu YM, Richmond A. Role of chemokines in tumor growth. Cancer Lett. 2007;256:137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richmond A, Yang J, Su Y. The good and the bad of chemokines/chemokine receptors in melanoma. Pigment Cell Melanoma Res. 2009;22:175–186. doi: 10.1111/j.1755-148X.2009.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi H, Kitago M, Hoon DS. Effects of chemokines on tumor metastasis. Cancer Treat Res. 2007;135:177–184. doi: 10.1007/978-0-387-69219-7_13. [DOI] [PubMed] [Google Scholar]
- 35.Hoon DS, Kitago M, Kim J, et al. Molecular mechanisms of metastasis. Cancer Metastasis Rev. 2006;25:203–220. doi: 10.1007/s10555-006-8500-x. [DOI] [PubMed] [Google Scholar]
- 36.Scala S, Ottaiano A, Ascierto PA, et al. Expression of CXCR4 predicts poor prognosis in patients with malignant melanoma. Clin Cancer Res. 2005;11:1835–1841. doi: 10.1158/1078-0432.CCR-04-1887. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi H, Fujimoto A, Tanaka M, et al. CCL21 chemokine regulates chemokine receptor CCR7 bearing malignant melanoma cells. Clin Cancer Res. 2004;10:2351–2358. doi: 10.1158/1078-0432.ccr-03-0195. [DOI] [PubMed] [Google Scholar]
- 38.Kim J, Mori T, Chen SL, et al. Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann Surg. 2006;244:113–120. doi: 10.1097/01.sla.0000217690.65909.9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mora JR, Bono MR, Manjunath N, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- 40.Amersi FF, Terando AM, Goto Y, et al. Activation of CCR9/CCL25 in cutaneous melanoma mediates preferential metastasis to the small intestine. Clin Cancer Res. 2008;14:638–645. doi: 10.1158/1078-0432.CCR-07-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izraely S, Klein A, Sagi-Assif O, et al. Chemokine-chemokine receptor axes in melanoma brain metastasis. Immunol Lett. 2010;130:107–114. doi: 10.1016/j.imlet.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Chen SL, Iddings DM, Scheri RP, Bilchik AJ. Lymphatic mapping and sentinel node analysis: current concepts and applications. CA Cancer J Clin. 2006;56:292–309. doi: 10.3322/canjclin.56.5.292. quiz 316-297. [DOI] [PubMed] [Google Scholar]
- 43.Amersi F, Giuliano AE, Hoon DS. Molecular assessment of sentinel lymph nodes. In: Mariani G, Giuliano AE, Strauss HW, editors. Radioguided Surgery. New York: Springer; 2008. pp. 206–217. [Google Scholar]
- 44.Grimm EA, Hoon DS, Duncan LM. Biomarkers for Cutaneous Melanoma. In: Balch CM, Houghton AN, Sober AJ, et al., editors. Cutaneous Melanoma. St. Louis: Quality Medical Publishing, Inc.; 2008. pp. 883–897. [Google Scholar]
- 45.Karim RZ, Scolyer RA, Li W, et al. False negative sentinel lymph node biopsies in melanoma may result from deficiencies in nuclear medicine, surgery, or pathology. Ann Surg. 2008;247:1003–1010. doi: 10.1097/SLA.0b013e3181724f5e. [DOI] [PubMed] [Google Scholar]
- 46.Kuo CT, Hoon DS, Takeuchi H, et al. Prediction of disease outcome in melanoma patients by molecular analysis of paraffin-embedded sentinel lymph nodes. J Clin Oncol. 2003;21:3566–3572. doi: 10.1200/JCO.2003.01.063. [DOI] [PubMed] [Google Scholar]
- 47.Martinez SR, Mori T, Hoon DS. Molecular upstaging of sentinel lymph nodes in melanoma: where are we now? Surg Oncol Clin N Am. 2006;15:331–340. doi: 10.1016/j.soc.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Morton DL, Hoon DS, Cochran AJ, et al. Lymphatic mapping and sentinel lymphadenectomy for early-stage melanoma: therapeutic utility and implications of nodal microanatomy and molecular staging for improving the accuracy of detection of nodal micrometastases. Ann Surg. 2003;238:538–549. doi: 10.1097/01.sla.0000086543.45557.cb. discussion 549-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bostick PJ, Morton DL, Turner RR, et al. Prognostic significance of occult metastases detected by sentinel lymphadenectomy and reverse transcriptase-polymerase chain reaction in early-stage melanoma patients. J Clin Oncol. 1999;17:3238–3244. doi: 10.1200/JCO.1999.17.10.3238. [DOI] [PubMed] [Google Scholar]
- 50.Nicholl MB, Elashoff D, Takeuchi H, et al. Molecular upstaging based on paraffin-embedded sentinel lymph nodes: ten-year follow-up confirms prognostic utility in melanoma patients. Ann Surg. 2011;253:116–122. doi: 10.1097/SLA.0b013e3181fca894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mocellin S, Hoon DS, Pilati P, et al. Sentinel lymph node molecular ultrastaging in patients with melanoma: a systematic review and meta-analysis of prognosis. J Clin Oncol. 2007;25:1588–1595. doi: 10.1200/JCO.2006.09.4573. [DOI] [PubMed] [Google Scholar]
- 52.Hoon DS. Are circulating tumor cells an independent prognostic factor in patients with high-risk melanoma? Nat Clin Pract Oncol. 2004;1:74–75. doi: 10.1038/ncponc0041. [DOI] [PubMed] [Google Scholar]
- 53.Hoon DS, Bostick P, Kuo C, et al. Molecular markers in blood as surrogate prognostic indicators of melanoma recurrence. Cancer Res. 2000;60:2253–2257. [PubMed] [Google Scholar]
- 54.Mocellin S, Hoon D, Ambrosi A, et al. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res. 2006;12:4605–4613. doi: 10.1158/1078-0432.CCR-06-0823. [DOI] [PubMed] [Google Scholar]
- 55.Voit C, Kron M, Rademaker J, et al. Molecular staging in stage II and III melanoma patients and its effect on long-term survival. J Clin Oncol. 2005;23:1218–1227. doi: 10.1200/JCO.2005.04.098. [DOI] [PubMed] [Google Scholar]
- 56.Hoon DS, Wang Y, Dale PS, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–2116. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- 57.Xi L, Nicastri DG, El-Hefnawy T, et al. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007;53:1206–1215. doi: 10.1373/clinchem.2006.081828. [DOI] [PubMed] [Google Scholar]
- 58.de Graaf H, Maelandsmo GM, Ruud P, et al. Ectopic expression of target genes may represent an inherent limitation of RT-PCR assays used for micrometastasis detection: studies on the epithelial glycoprotein gene EGP-2. Int J Cancer. 1997;72:191–196. doi: 10.1002/(sici)1097-0215(19970703)72:1<191::aid-ijc27>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 59.Kitago M, Koyanagi K, Nakamura T, et al. mRNA expression and BRAF mutation in circulating melanoma cells isolated from peripheral blood with high molecular weight melanoma-associated antigen-specific monoclonal antibody beads. Clin Chem. 2009;55:757–764. doi: 10.1373/clinchem.2008.116467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siena S, Sartore-Bianchi A, Di Nicolantonio F, et al. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoshimoto S, Shingai T, Wang H, Elashoff RM, Morton DL, Hoon DS. MMAIT Clinical Trials Group.: Validation of a multimarker blood assay for postoperative assessment of stage IV melanoma patients in a prospective international phase III trial. J Clin Oncol. 2009;27 Abstract 9045. [Google Scholar]
- 62.Udayakumar D, Tsao H. Melanoma genetics: an update on risk-associated genes. Hematol Oncol Clin North Am. 2009;23:415–429. vii. doi: 10.1016/j.hoc.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 63.Wu C, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science. 2001;293:1103–1105. doi: 10.1126/science.293.5532.1103. [DOI] [PubMed] [Google Scholar]
- 64.Shinozaki M, Fujimoto A, Morton DL, Hoon DS. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10:1753–1757. doi: 10.1158/1078-0432.ccr-1169-3. [DOI] [PubMed] [Google Scholar]
- 65.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kefford R, Arkenau H, Brown MP, et al. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol. 2010;28 Abstr 8503. [Google Scholar]
- 67.Iwamoto T, Taniguchi M, Asai N, et al. cDNA cloning of mouse ret proto-oncogene and its sequence similarity to the cadherin superfamily. Oncogene. 1993;8:1087–1091. [PubMed] [Google Scholar]
- 68.Takahashi M, Buma Y, Iwamoto T, et al. Cloning and expression of the ret proto-oncogene encoding a tyrosine kinase with two potential transmembrane domains. Oncogene. 1988;3:571–578. [PubMed] [Google Scholar]
- 69.Zbuk KM, Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nat Rev Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi M. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 2001;12:361–373. doi: 10.1016/s1359-6101(01)00012-0. [DOI] [PubMed] [Google Scholar]
- 71.Kodama Y, Asai N, Kawai K, et al. The RET proto-oncogene: a molecular therapeutic target in thyroid cancer. Cancer Sci. 2005;96:143–148. doi: 10.1111/j.1349-7006.2005.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wells SA, Jr, Santoro M. Targeting the RET pathway in thyroid cancer. Clin Cancer Res. 2009;15:7119–7123. doi: 10.1158/1078-0432.CCR-08-2742. [DOI] [PubMed] [Google Scholar]
- 73.Sawai H, Okada Y, Kazanjian K, et al. The G691S RET polymorphism increases glial cell line-derived neurotrophic factor-induced pancreatic cancer cell invasion by amplifying mitogen-activated protein kinase signaling. Cancer Res. 2005;65:11536–11544. doi: 10.1158/0008-5472.CAN-05-2843. [DOI] [PubMed] [Google Scholar]
- 74.Narita N, Tanemura A, Murali R, et al. Functional RET G691S polymorphism in cutaneous malignant melanoma. Oncogene. 2009;28:3058–3068. doi: 10.1038/onc.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Healy E, Belgaid C, Takata M, et al. Prognostic significance of allelic losses in primary melanoma. Oncogene. 1998;16:2213–2218. doi: 10.1038/sj.onc.1200203. [DOI] [PubMed] [Google Scholar]
- 76.Shirasaki F, Takata M, Hatta N, Takehara K. Loss of expression of the metastasis suppressor gene KiSS1 during melanoma progression and its association with LOH of chromosome 6q16.3-q23. Cancer Res. 2001;61:7422–7425. [PubMed] [Google Scholar]
- 77.Fujimoto A, Takeuchi H, Taback B, et al. Allelic imbalance of 12q22-23 associated with APAF-1 locus correlates with poor disease outcome in cutaneous melanoma. Cancer Res. 2004;64:2245–2250. doi: 10.1158/0008-5472.can-03-2932. [DOI] [PubMed] [Google Scholar]
- 78.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 79.Tanemura A, Terando AM, Sim MS, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15:1801–1807. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen T, Kuo C, Nicholl MB, et al. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2010;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 82.Howell PM, Jr, Liu S, Ren S, et al. Epigenetics in human melanoma. Cancer Control. 2009;16:200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- 83.Hoon DS, Spugnardi M, Kuo C, et al. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Maat MF, van de Velde CJ, van der Werff MP, et al. Quantitative analysis of methylation of genomic loci in early-stage rectal cancer predicts distant recurrence. J Clin Oncol. 2008;26:2327–2335. doi: 10.1200/JCO.2007.14.0723. [DOI] [PubMed] [Google Scholar]
- 85.Kitago M, Martinez SR, Nakamura T, et al. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma. Clin Cancer Res. 2009;15:2988–2994. doi: 10.1158/1078-0432.CCR-08-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ito Y. Oncogenic potential of the RUNX gene family: ‘overview’. Oncogene. 2004;23:4198–4208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- 87.Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29:2605–2615. doi: 10.1038/onc.2010.88. [DOI] [PubMed] [Google Scholar]
- 88.Chen XQ, Stroun M, Magnenat JL, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 89.Nawroz H, Koch W, Anker P, et al. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- 90.Yamada T, Nakamori S, Ohzato H, et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: correlation with clinicopathological features. Clin Cancer Res. 1998;4:1527–1532. [PubMed] [Google Scholar]
- 91.Taback B, O’Day SJ, Boasberg PD, et al. Circulating DNA microsatellites: molecular determinants of response to biochemotherapy in patients with metastatic melanoma. J Natl Cancer Inst. 2004;96:152–156. doi: 10.1093/jnci/djh011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fujimoto A, O’Day SJ, Taback B, et al. Allelic imbalance on 12q22-23 in serum circulating DNA of melanoma patients predicts disease outcome. Cancer Res. 2004;64:4085–4088. doi: 10.1158/0008-5472.CAN-04-0957. [DOI] [PubMed] [Google Scholar]
- 93.Shinozaki M, O’Day SJ, Kitago M, et al. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13:2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mori T, Martinez SR, O’Day SJ, et al. Estrogen receptor-alpha methylation predicts melanoma progression. Cancer Res. 2006;66:6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mori T, O’Day SJ, Umetani N, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol. 2005;23:9351–9358. doi: 10.1200/JCO.2005.02.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]


