Abstract
The tumor necrosis factor (TNF) antagonist infliximab was recently found to reduce depressive symptoms in patients with increased baseline inflammation as reflected by a plasma C-reactive protein concentration >5mg/L. To further explore predictors and targets of response to infliximab, differential gene expression was examined in peripheral blood mononuclear cells from infliximab responders (n=13) versus non-responders (n=14) compared to placebo at baseline and 6hr, 24hr, and 2 weeks after the first infliximab infusion. Treatment response was defined as 50% reduction in depressive symptoms at any point during the 12-week trial. One-hundred-forty-eight gene transcripts were significantly associated (1.2 fold, adjusted p≤0.01) with response to infliximab and were distinct from placebo responders. Transcripts predictive of infliximab response were associated with gluconeogenesis and cholesterol transport, and were enriched in a network regulated by hepatocyte nuclear factor (HNF)4-alpha, a transcription factor involved in gluconeogenesis and cholesterol and lipid homeostasis. Of the 148 transcripts differentially expressed at baseline, 48% were significantly regulated over time in infliximab responders, including genes related to gluconeogenesis and the HNF4-alpha network, indicating that these predictive genes were responsive to infliximab. Responders also demonstrated inhibition of genes related to apoptosis through TNF signaling at 6hr and 24hr after infusion. Transcripts down-regulated in responders 2 weeks after infliximab were related to innate immune signaling and nuclear factor-kappa B. Thus, baseline transcriptional signatures reflective of alterations in glucose and lipid metabolism predicted antidepressant response to infliximab, and infliximab response involved regulation of metabolic genes and inhibition of genes related to innate immune activation.
Keywords: depression, infliximab, tumor necrosis factor, gene expression, gluconeogenesis
INTRODUCTION
Previous studies have suggested that activation of the peripheral inflammatory response and the release of inflammatory cytokines may contribute to depressive symptoms in a subpopulation of patients with major depression (Miller et al. 2009). Indeed, patients with major depression have been shown to exhibit increases in plasma concentrations and peripheral blood gene expression of inflammatory cytokines including tumor necrosis factor (TNF), interleukin (IL)-1 and IL-6 (Dhabhar et al. 2009; Dowlati et al. 2010; Hepgul et al. 2013; Howren et al. 2009), as well as decreases in anti-inflammatory cytokines such as IL-10 (Dhabhar et al. 2009). Moreover, reproducible increases in peripheral blood concentrations of the acute phase protein C-reactive protein (CRP) have been reported (Howren et al. 2009). Administration of inflammatory cytokines such as interferon (IFN)-alpha or inflammatory stimuli such as typhoid vaccination or endotoxin have also been shown to induce depressive symptoms (Capuron et al. 2002; Eisenberger et al. 2010; Harrison et al. 2009; Maddock et al. 2005; Raison et al. 2006; Reichenberg et al. 2001). In addition, inflammatory cytokines interact with virtually every pathophysiologic pathway relevant to depression including neurotransmitter metabolism, neuroendocrine function, synaptic plasticity and regional brain activity (Haroon et al. 2012; Miller et al. 2009). Finally, data suggest that blocking inflammatory cytokines or their signaling pathways may have antidepressant properties in patients with inflammatory disorders and depression (Monk et al. 2006; Raison et al. 2013; Tyring et al. 2006).
Increased inflammation in depressed patients appears to have a special relationship with treatment resistance. Patients with treatment resistant depression (TRD) have been shown to exhibit increased inflammatory markers, and increased inflammatory markers including TNF gene expression have been found to predict treatment non-response (Cattaneo et al. 2013; Eller et al. 2008; Lanquillon et al. 2000; Raison et al. 2013; Sluzewska et al. 1997). Moreover, clinical factors associated with non-response to antidepressants including obesity, childhood maltreatment, and anxiety disorders have all been associated with increased inflammation (Danese et al. 2008; Danese et al. 2007; Nanni et al. 2012; Oskooilar et al. 2009; Rush et al. 2008). Inflammatory cytokines can also sabotage and circumvent the mechanisms of action of antidepressant medications. For example, by increasing the activity and expression of monoamine transporters as well as decreasing monoamine synthesis, inflammatory cytokines can undermine the ability of antidepressants to block monoamine reuptake and increase monoamine availability in the synapse (Haroon et al. 2012; Miller In Press). Inflammatory cytokines can also inhibit neurogenesis, an important component of the salutary effects of antidepressants (Perera et al. 2011). Finally, inflammatory cytokines reduce the expression of the glutamate transporters in astrocytes and stimulate astrocytic release of glutamate, a neurotransmitter which is not a primary target of conventional antidepressant medications (Haroon et al. 2012; Miller et al. 2009).
Given the relationship between increased inflammation and treatment non-response, we recently conducted a double-blind, placebo-controlled clinical trial to determine whether the TNF antagonist infliximab would lead to a greater reduction in depressive symptoms than placebo in patients with TRD (Raison et al. 2013). Surprisingly, no differences in clinical response were found between infliximab and placebo in the group as a whole. However, there was a significant interaction between baseline inflammation as measured by CRP and treatment response, with patients exhibiting high inflammation (CRP >5mg/L) performing clinically better (greater reduction in depressive symptoms) when administered infliximab versus placebo. In addition, patients who responded to infliximab exhibited significantly higher plasma concentrations of TNF and its soluble receptors compared to infliximab non-responders (Raison et al. 2013). Taken together, these data indicate that cytokine antagonism may have therapeutic efficacy in TRD, but only in patients with increased inflammatory markers at baseline.
To further examine additional predictors and targets of response to infliximab in patients with TRD, in the current study, we endeavored to explore differential gene expression as a function of response to infliximab versus placebo in peripheral blood mononuclear cells (PBMCs) of TRD patients from our previous study.
MATERIALS AND METHODS
Study procedures
The study was a randomized, double-blind trial of infliximab versus placebo in individuals with major depression or bipolar disorder, depressed who were non-responsive to antidepressant treatment as determined by a score ≥2 on the Massachusetts General Hospital Staging (MGH-S) method in the current episode (Raison et al. 2013). Participants were recruited via local television, radio and newspaper advertisements and included males and females aged 25–60 years who were on a consistent antidepressant regimen or off antidepressant therapy for at least 4 weeks prior to baseline. To balance the number of male and female participants and to equalize levels of inflammation across groups, treatment assignment was stratified by sex and CRP (≥2mg/L versus <2mg/L). To be enrolled, subjects had to exhibit moderate severity of depression as determined at screening by a score ≥14 on the 16 item Quick Inventory of Depressive Symptomatology Self Report (Trivedi et al. 2004). Antidepressant regimens were required to remain stable throughout the study and could include conventional antidepressant medications, mood stabilizers, antipsychotic medication, stimulants and benzodiazepines. Exclusion criteria included any autoimmune disorder (confirmed by laboratory testing); history of tuberculosis (confirmed by chest X-ray, skin and blood testing) or high risk of tuberculosis exposure; hepatitis B or C or human immunodeficiency virus infection (confirmed by laboratory testing); evidence of active fungal infection; history of recurrent viral or bacterial infections; history of cancer excluding basal cell or squamous cell carcinoma of the skin (fully excised with no recurrence); unstable cardiovascular, endocrinologic, hematologic, hepatic, renal, or neurologic disease (determined by physical examination and laboratory testing); history of schizophrenia (determined by Structured Clinical Interview for DSM-IV - SCID)(First et al. 1997); active psychotic symptoms of any type; substance abuse/dependence within the past 6 months (determined by SCID); active suicidal ideation determined by a score ≥3 on item #3 on the17-item Hamilton Depression Rating Scale (HAM-D-17)(Hamilton 1960) and/or a score <28 on the Mini-Mental State Exam (Folstein et al. 1975). All subjects provided written informed consent to the study, and all study procedures were approved by the Emory University Institutional Review Board. The study was registered in ClinicalTrials.gov, Identifier: NCT00463580.
Clinical Assessments
The diagnosis of depression was made using the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV), as assessed by the SCID (First et al. 1997). Depression severity was measured by the HAM-D-17 at baseline and at Weeks 1, 2, 3, 4, 6, 8, 10, and 12 (Hamilton 1960). Treatment response was defined as a 50% reduction in the HAM-D-17 at any point during the 12 week study.
Infliximab and Placebo Infusion Procedures
All subjects were administered either infliximab (5mg/kg, n=30) or placebo (n=30) through an indwelling catheter at the Emory Division of Digestive Diseases at 3 separate time points (baseline, 2 weeks and 6 weeks). Infliximab infusion dosing protocol and scheduling were matched to the standard intravenous induction routine for the treatment of inflammatory bowel disease. Infliximab or placebo was dispensed by independent pharmacists in a 250ml saline bag according to a computer-generated randomization list, blocked in units of 4 and provided by a study statistician. The saline-dissolved placebo was matched to infliximab in color and consistency. Infliximab and placebo were provided free-of-charge by Centocor Ortho Biotech Services (Horsham, PA). For subjects exhibiting evidence of infection, infusions were delayed until symptoms resolved and/or appropriate treatment was initiated. Patients were not allowed to take non-steroidal or steroidal anti-inflammatory medications during the study except for aspirin 81mg per day if medically indicated. Medications for hypertension, diabetes, hypothyroidism, allergies, infections and other medical conditions were allowed as dictated by the patients’ treating physicians. All study staff remained blinded to group assignment throughout the trial.
Biological Samples
Whole blood for gene expression analysis was collected in Tempus Blood RNA Tubes (Applied Biosystems, Carlsbad, CA) at baseline (9 am), 6 hours after the first infusion, 24 hours after the first infusion and immediately prior to the second infusion on Week 2. RNA tubes were stored at −80C until RNA extraction using RNeasy kits (QIAGEN, Valencia, CA) according to manufacturer instructions. RNA sample concentrations and the A260/280 ratio were determined using the MBA 2000 System (Perkin-Elmer, Shelton, CT, USA). Each sample was linearly amplified by WT-Ovation RNA amplification system (NuGEN) and then submitted to the Emory Cancer Genomics Core for microarray analysis. After hybridization to Illumina Human HT-12 Expression BeadChips (Illumina, San Diego, CA), BeadChips were scanned on the Illumina BeadArray Reader to determine raw probe fluorescence intensity.
Statistical Analysis
The socio-demographic and clinical variables across the groups were compared using t-tests and chi-square analyses/Fisher’s Exact Test. Raw expression data were exported from Illumina Genome Studio into R (http://www.R-project.org) for subsequent analysis. The data was normalized and log transformed using vsn (Huber et al. 2002) and corrected for hybridization batch effects using an Empirical Bayes estimation (Johnson et al. 2007). A total of 16,962 probes passing the filter criteria of Illumina probe detection p-value of < 0.01 in at least 10% of the samples were used for subsequent analyses, and data were deposited in NCBI Gene Expression Omnibus as series GSE45468. The association between treatment response and gene expression was assessed using generalized linear models in R, while adjusting for the effects of age, gender, body mass index (BMI) and medication status. Gene transcripts differentially expressed in responders compared to non-responders were defined as having a ≥20% difference (1.2 fold change) corresponding to a <10% FDR (Cole et al. 2003; Miller et al. 2008; Torres et al. 2013) and cut-off of p≤0.01 (Haider et al. 2008), and were therefore highly predictive based on both statistical significance (p value) as well as biological effect size (fold change). Mixed-effects linear models were used to assess differences in gene expression across time points in R (p<0.05). Corrections for multiple testing were performed using 10,000 permutations for each test as previously described (Mehta et al. 2011). Predictions were performed using nearest shrunken centroids (NSC) method (pamr) with a leave out one 100-fold cross validation scheme to ensure robustness of the prediction (Tibshirani et al. 2002). This NSC method computes an average expression vector for each class, which is then shrunken towards the overall expression mean across the classes in order to avoid over-fitting. The samples were repeatedly split into training and test sets (cross-validation) to allow selection of the optimal number of transcripts for the classifier and for accuracy estimations (Witten et al. 2010). Functional annotation of transcripts within pathways was assessed using MetaCore (GeneGo Inc., St. Joseph, MI) or the Wikipathways tool via the Webgestalt interface (Pico et al. 2008). Differentially expressed genes were also analyzed using a network-based transcription factor analysis in MetaCore (Ekins et al. 2007). The MetaCore transription factor analysis uses a transcriptional regulation algorithm to query a manually-curated database, and has been determined to be both accurate and comprehensive in identifying transcriptional regulatory pathways and target genes (Shmelkov et al. 2011). Transcript origin analysis was performed to identify the cellular source of differentially expressed transcripts in whole blood based on log transformed cell diagnostic scores (Cole et al. 2011). The transcripts were tested for enrichment relative to the null hypothesis (genome-wide) average scores using single sample t-tests (Felger et al. 2011). All statistical tests were two-sided with a significance level of p<0.05.
RESULTS
Demographics and Clinical Variables
Gene expression profiles were available from a total of 57 individuals at baseline, comprising 27 patients treated with infliximab (n=13 responders and n=14 non-responders) and 30 treated with placebo (n=15 responders and n=15 non-responders). Gene expression profiles were available for 53 individuals at the remaining time points, 24 were treated with infliximab (n=12 responders and n=12 non-responders) and 29 with placebo (n=15 responders and n=14 non-responders). Three infliximab-treated patients, including 2 responders, did not have gene expression data available at any time point. Demographic and clinical variables between the intervention and control groups were well-matched with no significant differences in the sample as a whole (n=60)(Raison et al. 2013) or in the 57 patients included in the present study (data not shown). Distributions and comparisons of the demographic and clinical variables between treatment responders and non-responders for both the infliximab and placebo-treated groups are shown in Table 1. No differences in gender, ethnicity, BMI, CRP, baseline HAM-D-17 depression scores, or the number of individuals on current antidepressants, mood stabilizers, antipsychotic medicines and psychotropic medicines were observed between the responders and non-responders in either treatment group (all p>0.05). Responders were significantly older than non-responders in the infliximab-treated group.
Table 1.
Demographic and clinical characteristics of responders versus non-responders to placebo or infliximab infusion.
| Infliximab Non-responders | Infliximab Responders | P-value | Placebo Non-responders | Placebo Responders | P-value | |
|---|---|---|---|---|---|---|
|
| ||||||
| (N = 14) | (N = 13) | (N = 15) | (N = 15) | |||
|
| ||||||
| Gender | 1 | 1 | ||||
| Female | 9 [64.3%] | 8 [61.5%] | 10 [66.7%] | 10 [66.7%] | ||
| Male | 5 [35.7%] | 5 [38.5%] | 5 [33.3%] | 5 [33.3%] | ||
|
| ||||||
| Race | 0.33 | 0.89 | ||||
| Caucasian | 9 [64.3%] | 12 [92.3%] | 10 [71.4%] | 12 [80%] | ||
| African American | 5 [35.7%] | 1 [7.7%] | 3 [21.4%] | 2 [13.3%] | ||
| Other | 1 [7.1%] | 1 [6.7%] | ||||
|
| ||||||
| Age | 38.9 [7.6] | 46.0 [8.0] | 0.03 | 42.1 [10.0] | 46.6 [8.6] | 0.19 |
|
| ||||||
| BMI | 30.9 [9.1] | 31.0 [4.4] | 0.97 | 32.4 [8.2] | 33.1 [8.1] | 0.83 |
|
| ||||||
| Baseline CRP (mg/L) | 6.0 [7.6] | 7.4 [11.2] | 0.71 | 7.8 [11.0] | 3.1 [2.6] | 0.12 |
|
| ||||||
| Baseline HAM-D-17 | 24.1 [3.6] | 23.6 [4.6] | 0.74 | 23.9 [4.1] | 23.3 [3.6] | 0.67 |
|
| ||||||
| Week 12 HAM-D-17 | 21.9 [7.6] | 12.7 [6.8] | <0.005 | 20.2 [6.7] | 8.1 [3.8] | <0.001 |
|
| ||||||
| Current antidepressant | 0.45 | 0.7 | ||||
| Yes | 5 [35.7%] | 7 [53.8%] | 9 [60%] | 11 [73.3%] | ||
| No | 9 [64.3%] | 6 [46.2%] | 6 [40%] | 4 [26.7%] | ||
|
| ||||||
| Current antipsychotic | 0.65 | 1 | ||||
| Yes | 2 [14.3%] | 3 [23%] | 2 [13.3%] | 2 [13.3%] | ||
| No | 12 [85.7%] | 10 [77%] | 13 [86.7%] | 13 [86.7%] | ||
|
| ||||||
| Current mood stabilizer | 1 | 1 | ||||
| Yes | 2 [14.3%] | 1 [7.7%] | 2 [13.3%] | 1 [7.1%] | ||
| No | 12 [85.7%] | 12 [92.3%] | 13 [86.7%] | 14 [92.9%] | ||
|
| ||||||
| Current psychotropic | 0.45 | 0.43 | ||||
| Yes | 6 [42.9%] | 8 [61.5%] | 9 [60%] | 12 [80%] | ||
| No | 8 [57.1%] | 5 [38.5%] | 6 [40%] | 3 [20%] | ||
Results expressed as number [percent] or mean [SD]. BMI: body mass index; CRP: C-reactive protein; HAM-D-17: 17-item Hamilton Depression Rating Scale
Baseline Gene Expression Prediction of Infliximab Treatment-Response
To identify transcripts whose baseline expression was associated with infliximab treatment response, baseline gene expression profiles were regressed against the treatment responder status for the 27 individuals who were treated with infliximab, adjusting for gender, ethnicity, age, BMI and current medication status. Expression levels of 148 transcripts were significantly associated (1.2 fold change, adjusted p≤0.01) with treatment response. The predictive values of these 148 transcripts differentially expressed between responders and non-responders at baseline were then interrogated. The expression profiles were subjected to the nearest shrunken centroid classification while randomly splitting the samples into test and training sets and performing a 10-fold cross validation of the predictions. To ensure robustness of the classification, the predictions were further repeated 100 times using a leave-out-one cross validation scheme. Using baseline gene expression profiles of the 148 transcripts allowed a 100% prediction of the infliximab responder status with 100% specificity and 100% sensitivity. These transcripts and their relative expression levels at baseline in infliximab responders vs. non-responders are listed in Supplemental Table 1. To confirm whether the differentially expressed transcripts were specific to infliximab treatment, we compared the results to transcripts associated with response to placebo treatment. None of the transcripts significantly predicting response to infliximab treatment (n=148 transcripts) overlapped with transcripts significantly associated with clinical response in the placebo-treated individuals (n=12 transcripts), indicating that the predictive transcripts were specific to infliximab response. To identify the genes that had the most sensitivity or specificity for predicting response to infliximab, we examined the 10 transcripts most significantly different between responders and non-responders at baseline, and from these determined the top 2 named genes that exhibited the greatest fold increase and the greatest fold decrease. The top 2 upregulated genes were TGM2 (ILMN_1705750) and PGAM4 (ILMN_1706841), and the top 2 downregulated genes were DTNBP1 (ILMN_1783806) and LOC100134241 (ILMN_3248948).
The transcripts encoding three inflammatory genes recently reported to be predictive of response to the antidepressants escitalopram or nortriptyline (Cattaneo et al. 2013) were also examined independently to determine whether these genes were differentially expressed between responders and non-responders to infliximab at baseline. The transcript for TNF (ILMN_1728106) was significantly higher at baseline in infliximab responders compared to non responders using a reduced statistical stringency, p<0.05 (fold change = 1.3), whereas IL1B (ILMN_1775501, p=0.78, fold change=−1.1) and MIF (ILMN_1807044, p=0.26, fold change= 1.08) were not differentially expressed in infliximab responders versus non-responders at baseline.
Biological Significance of the Infliximab Treatment-Response Baseline Predictors
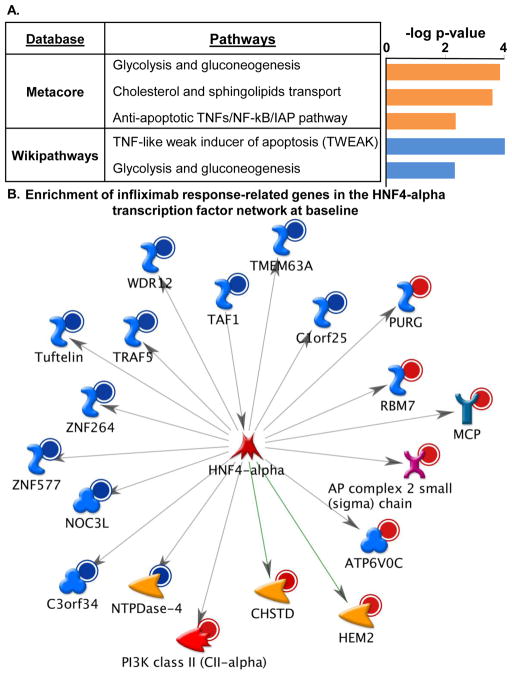
To investigate the biological significance of the infliximab treatment response predictors, functional annotation of the 148 differentially expressed transcripts was performed using Metacore pathways analysis and Wikipathways tool in Webgestalt. The top three pathways represented by Metacore analysis and the two significant pathways from Wikipathways are presented in Figure 1A. These databases produced similar results that included pathways relevant to glycolysis and gluconeogenesis, and to apoptosis through TNF-related signaling and the TNF-like weak inducer of apoptosis (TWEAK), which is encoded by the gene TNFSF12. The Metacore analysis also revealed differential regulation of genes relevant to cholesterol transport. Transcription factor network analysis in Metacore revealed a significant over-representation of genes regulated by several transcription factors, the top two being SP-1 (32/148 genes; z=128.32; p = 1.56e-88) and hepatocyte nuclear factor (HNF) 4-alpha (20/148 genes; z=97.86; p = 6.63e-52), a transcription factor that has been linked to both gluconeogenesis and cholesterol and lipid homeostasis (see Figure 1B for associated genes). Of note, there were no significant differences in the presence of diabetes or hypercholesterolemia between the infliximab responder and non-responder groups (diabetes 2=1.18, df=1, p=0.29; hypercholesterolemia 2=1.36, df=1, p=0.24). To identify which specific immune cell subtypes contributed to the differentially expressed gene expression profiles, we performed a transcript origin analysis (TOA) of the predictive transcripts (Cole et al. 2011; Felger et al. 2011). Of the 148 predictive transcripts, 100 were RefSeq genes. Of these, TOA analysis was performed on 72 transcripts (72% of total RefSeq transcripts) with available gene expression profiles for immune cell subtypes (monocytes, CD4+ and CD8+ T cells, B cells, NK cells and plasmacytoid dendritic cells). Based on cell type diagnostic scores, the predictive transcripts were significantly enriched for transcripts originating from plasmacytoid dendritic cells (p=0.022) and B cells (p=0.043).
Figure 1. Pathways and transcription factor networks significantly represented in differentially regulated gene transcripts at baseline that were predictive of the antidepressant response to infliximab.
Infliximab (5mg/kg, n=30) was administered over 12 weeks through an indwelling catheter at baseline, 2 weeks and 6 weeks, and antidepressant response was determined as a 50% reduction in depression severity as measured by the 17-item HAM-D (measured at baseline and 1, 2, 3, 4, 6, 8, 10, and 12 weeks) at any time point during the study. Genes at baseline that were significantly predictive of treatment response (n=148, 1.2 fold change, p 0.01) were involved in pathways related to glycolysis and gluconeogenesis, cholesterol and sphingolipid transport, and apoptosis through tumor necrosis factor (TNF)-related signaling pathway as assessed using Metacore and Wikipathways (A). The most significant transcription factors represented were SP-1 (not shown) and a 19-gene network regulated by hepatocyte nuclear factor (HNF) 4-alpha (B). Red circle= up-regulated genes; Blue circle= down-regulated genes; orange arrowhead= enzyme; red arrowhead= lipid kinase; pink X= transporter; red star= transcription factor; turquoise Y= receptor; blue S =binding protein; blue clover =protein.
Effects of a Single Infusion of Infliximab on Predictive Baseline Transcripts over Time
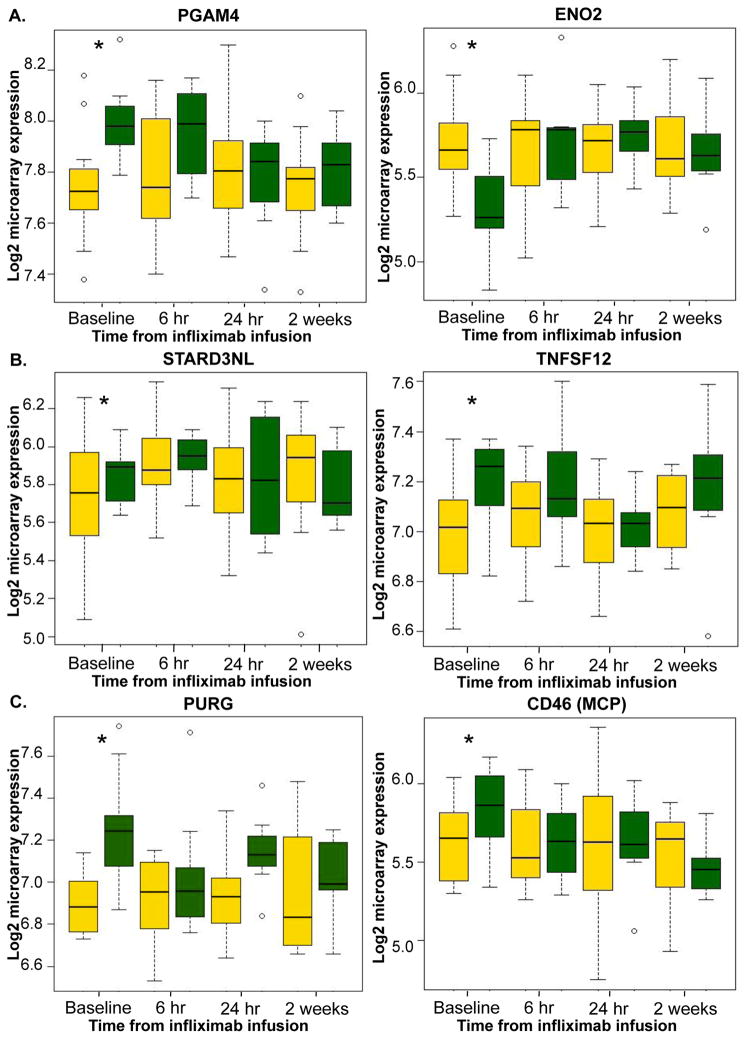
Changes in expression of the 148 baseline predictive transcripts were examined over time to determine whether the initial infusion of infliximab regulated genes that were differentially expressed at baseline in responders. Of the 148 predictive transcripts, 71 transcripts (47.9%) were significantly regulated by infliximab (adjusted p<0.05) in the responders at any time point following infliximab infusion (6 hr, 24 hr, or 2 weeks). In contrast, only 13 of the 148 (8.78%) transcripts were significantly regulated across time among individuals that did not respond to infliximab, and only 11 of the 148 (7.4%) transcripts were significantly regulated across time among individuals that responded to placebo treatment, indicating that the majority of predictive transcripts were changed over time only in infliximab responders. Genes from the significantly represented Metacore pathways and the HNF4-alpha transcription factor network that were differentially expressed at baseline were then examined over time to determine whether these genes were significantly regulated by infliximab in responders. Interestingly, 2 of the 3 genes represented in the glycolysis and gluconeogenesis pathway (PGAM4, ENO2) at baseline were differentially regulated in response to the initial injection of infliximab in responders but not non-responders (Figure 2A). Furthermore, 1 of the 3 genes represented in the pathway related to cholesterol transport (STARD3NL) and 1 of the 3 genes involved in apoptosis through TNF-related signaling (TNFSF12/TWEAK) (Figure 2B), as well as 13 of the 19 genes in the HNF4-alpha transcription factor network (e.g. PURG and CD46/MCP) (Figure 2C), were significantly regulated over time in responders compared to non-responders following the initial infliximab infusion (p<0.05 at any time). In addition to PGAM4, of the four most predictive transcripts, DTNBP1, but not TGM2 or LOC100134241, was included among the transcripts significantly regulated by infliximab over time in responders.
Figure 2. Gene transcripts that were predictive of antidepressant response at baseline were altered in responders following the initial infliximab infusion.
Infliximab (5mg/kg, n=30) was administered over 12 weeks through an indwelling catheter at baseline, 2 weeks and 6 weeks, and antidepressant response was determined as a 50% reduction in depression severity as measured by the 17-item HAM-D at any time point during the study. Gene expression was assessed at baseline, and 6 hr, 24 hr, and 2 weeks following the first infusion. Transcripts at baseline that were significantly predictive of treatment response and involved in the (A) glycolysis and gluconeogenesis pathway (PGAM4, ENO2), (B) cholesterol and sphingolipid transport pathway (STARD3NL) or apoptosis through tumor necrosis factor (TNF)-related signaling pathway (TNFSF12), and (C) hepatocyte nuclear factor (HNF) 4-alpha transcription factor network (e.g. PURG, CD46/MCP), were differentially regulated over time by infliximab in responders (green boxes) but not in non-responders (yellow boxes) at any point over time (p<0.05). Expression values differed between responders and non-responders only at baseline. *p<0.05 responders compared to non-responders
Differential Regulation of Gene Expression in Infliximab Responders versus Non-responders at Specific Time Points
To examine how inhibition of TNF regulated gene expression at different time points in infliximab responders versus non-responders, we compared gene expression between these 2 groups following the first infliximab infusion at each of the time points separately. Using the same criteria as above (1.2 fold change, adjusted p≤0.01), transcripts differentially regulated in responders compared to non-responders were identified at 6 hr (n=145), 24 hr (n=157), and 2 weeks (n=56) (see Supplementary Table 2). Of note, of the 148 transcripts that differentiated infliximab responders versus non-responders at baseline, only 18 (12.16%) of these transcripts still differentiated the groups at 6 hr and only 6 (4.05%) of these transcripts differentiated the groups at 24 hr and at 2 weeks.
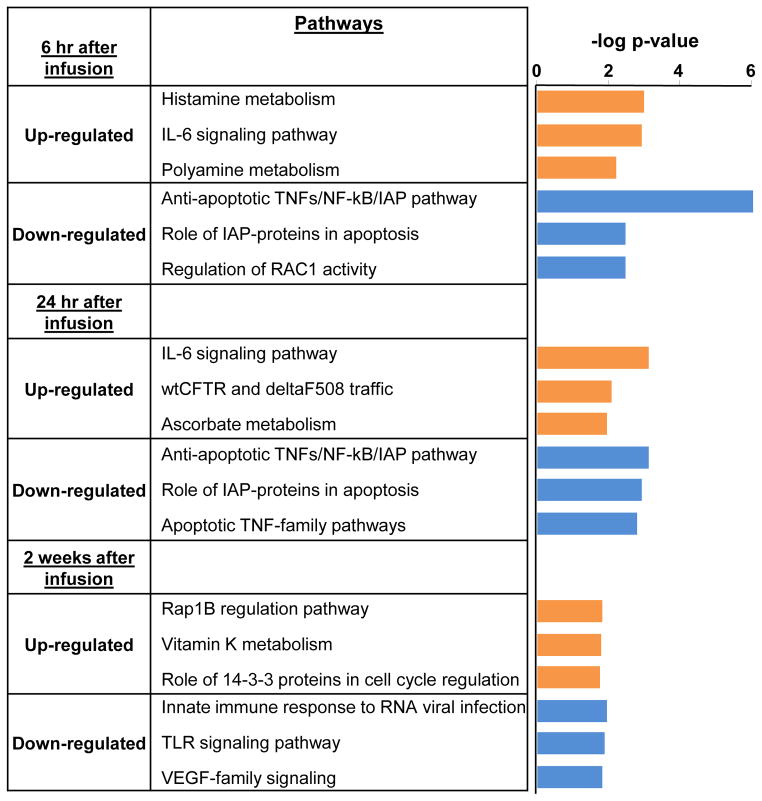
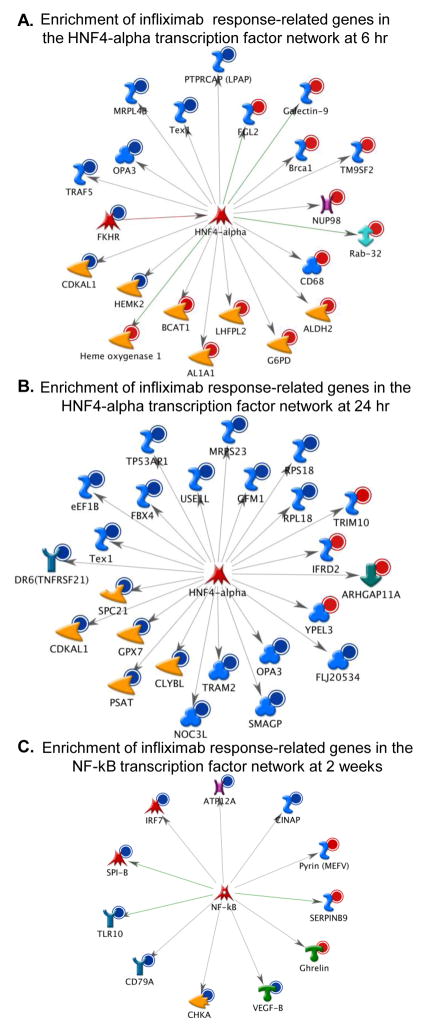
To examine the biological significance of transcripts that were differentially regulated over time in responders compared to non-responders, up-regulated genes and down-regulated genes were analyzed separately at each time point to identify pathways that were significantly increased or decreased in response to infliximab in responders versus non-responders (Figure 3). Metacore analysis revealed that, in general, at the earlier time points (6 and 24 hr) responders exhibited increased expression of genes related to IL-6 signaling, and decreased expression of genes related to apoptosis through TNF-related signaling pathways. Interestingly, at 2 weeks following the initial infliximab infusion, gene expression related to innate immune responses (antiviral and TLR signaling) were decreased in the responders with respect to non-responders. Transcription factor networks that may drive differential gene expression patterns during infliximab treatment were also examined. At 6 hr SP-1 (31/145 genes; z=121.49; p=1.72e-84) and HNF4-alpha (21/145 genes; z=99.24; p=1.03e-56) remained the two most significant transcription factors represented, whereas at 24 hr, HNF4-alpha was the most significant transcription factor represented (24/157 genes; z=105.54, p=8.35e-65). Interestingly, there was a redistribution of the transcripts regulated by HNF4-alpha that was associated with treatment response across time compared to those at baseline (Figure 1B), and these transcripts were primarily up-regulated at 6hr (Figure 4A) and then down-regulated at 24hr (Figure 4B). At 2 weeks following the initial infusion of infliximab, aside from SP-1 (16/56 genes; z=134.44; p=6.13e-50), nuclear factor-kappa B (NF-kB) (11/56 genes; z=110.01, p=4.88e-34) was the most significantly represented transcription factor network, comprised mostly of genes down-regulated in responders with respect to non-responders (8/11) (Figure 4C).
Figure 3. Genes down-regulated by infliximab over time in responders were related to apoptosis through tumor necrosis factor (TNF) and innate immune activation.
Infliximab (5mg/kg, n=30) was administered over 12 weeks through an indwelling catheter at baseline, 2 weeks and 6 weeks, and antidepressant response was determined as a 50% reduction in depression severity as measured by the 17-item HAM-D at any time point during the study. Genes that were up-regulated and down-regulated in responders compared to non-responders were analyzed separately at 6 hr, 24 hr, and 2 weeks to identify significant pathways that were increased or inhibited in response to infliximab that may be related to treatment response. At the earlier time points (6 and 24 hr), responders exhibited increased expression of genes related to IL-6 signaling, and decreased expression of genes related to apoptosis through TNF-related signaling pathways. At 2 weeks, gene expression profiles related to innate immune responses (antiviral and TLR signaling) were decreased in responders with respect to non-responders.
Figure 4. Genes differentially expressed between responders and non-responders were enriched in transcriptional networks of hepatocyte nuclear factor (HNF) 4-alpha and nuclear factor-kappaB (NF-kB), early and late following the initial infliximab infusion, respectively.
Infliximab (5mg/kg, n=30) was administered over 12 weeks through an indwelling catheter at baseline, 2 weeks and 6 weeks, and antidepressant response was determined as a 50% reduction in depression severity as measured by the 17-item HAM-D at any time point during the study. Genes that were differentially regulated in responders compared to non-responders were analyzed at 6 hr, 24 hr, and 2 weeks. 6 hr after infliximab, the transcriptional network for HNF4-alpha was composed of mostly of genes up-regulated (red circles, 13/21 genes) in responders compared to non-responders (A), whereas at 24 hr, the transcriptional network for HNF4-alpha was comprised mostly of genes down-regulated in responders (blue circle, 20/24 genes) (B). 2 weeks after the initial infliximab infusion, the genes differentially regulated in responders versus non-responders were enriched in the NF-kB transcriptional network and comprised mostly of down-regulated genes (blue circle, 8/11 genes) (C). Red circle= up-regulated genes; Blue circle= down-regulated genes; orange arrowhead= enzyme; red arrowhead= lipid kinase; pink X= transporter; red star= transcription factor; turquoise Y= receptor; blue S =binding protein; blue clover =protein, green double-headed arrow=RAS superfamily; pink and brown H=channel; green down arrow=regulator; orange arrowhead with hole=protease; green T= receptor ligand.
Because the analyses at 6 hr, 24 hr, and 2 weeks were conducted in only 53 of 57 patients, we verified that these subjects had a similar pattern of differential gene expression as the 57 patients examined at baseline. Indeed, these 53 patients exhibited differential expression of all 148 transcripts at baseline described above, as well as an additional 6 transcripts, indicating that this subset of 53 patients was highly representative of the group as a whole.
DISCUSSION
Examination of gene transcripts at baseline that were predictive of an antidepressant response to infliximab revealed enrichment of genes involved in pathways and a transcriptional control network (HNF4-alpha) related to both glycolysis and gluconeogenesis and to lipid and cholesterol homeostasis (Rhee et al. 2003; Yin et al. 2011). These genes were specific to infliximab response and distinct from placebo treatment. Interestingly, response to infliximab was associated with alterations in expression of many of the transcripts in these pathways as well as the HNF4-alpha network over time, indicating that regulation of these genes may be involved in infliximab-induced transcriptional alterations that mediate response. In addition, there was a shift in transcriptional activity within the HNF4-alpha network early in the response to infliximab, evinced by activation at 6 hr followed by suppression at 24 hr of a number of genes regulated by this transcription factor. Moreover, infliximab responders compared to non-responders demonstrated greater early (6 and 24 hr) inhibition of genes related to apoptotic pathways through TNF signaling, suggesting that TNF pathways showed greater inhibition in subjects exhibiting a treatment response. These changes were followed by decreased expression of gene transcripts related to innate immune signaling and to NF-kB transcriptional control 2 weeks after infliximab infusion in responders compared to non-responders. Together, these data indicate that patients that responded to infliximab exhibited gene expression profiles consistent with alterations in metabolic processes involving glucose metabolism and cholesterol homeostasis that were regulated by infliximab early following the initial infusion, and that these patients also exhibited a more exaggerated anti-inflammatory response to infliximab compared to non-responders that persisted at two weeks following a single infliximab infusion.
Gene expression was examined in this study at 6 and 24 hr following the first infliximab injection to capture early effects of infliximab on gene expression, and then to determine whether changes in gene expression were evident 2 weeks after a single infusion. It is interesting to note that a number of genes were significantly differentially expressed between responders and non-responders at baseline (n=148) and early in response to infliximab (6 hr n=145; 24 hr n=157), whereas a smaller number (n=56) of differentially expressed genes, and only ~4% of the original 148 predictive transcripts, were observed at 2 weeks. This is consistent with findings that the initial infliximab infusion regulated many of the predictive gene transcripts that were differentially expressed at baseline, but only in responders. These predictive transcripts were related to glycolysis and gluconeogensis, and to cholesterol homeostasis and apoptotic signaling through TNF-related pathways. Although genes in the network of apoptosis related to TNF signaling were decreased in responders early in response to infliximab infusion, gene expression differences between responders and non-responders related to glucose metabolism and cholesterol homeostasis were not present after infliximab infusion. This is not to say that the response to infliximab was mediated by a lack of differential gene expression between responders and non-responders at 2 weeks, but rather that many of the genes that were predictive of a response to infliximab at baseline, particularly those genes related to glycolysis and gluconeogensis and the HNF4-alpha transcriptional network, were also affected by inflximab, but only in patients that demonstrated an antidepressant response. Therefore, many of the genes differentially expressed at baseline appeared not only to predict the response to infliximab, but were also involved in transcriptional changes associated with this response.
The association of genes predictive of infliximab response with pathways and transcriptional control related to glucose metabolism and cholesterol homeostasis is interesting given the recent attention paid to the relationship among obesity, metabolic syndrome, inflammation and depression (Shelton and Miller 2010). Numerous studies have reported co-morbidity of depression and metabolic disorders such as diabetes (Rustad et al. 2011). Moreover, recent studies have shown a dose response relationship between body weight and antidepressant treatment response (Oskooilar et al. 2009). In addition, there is a strong relationship between BMI and multiple inflammatory markers, the higher the BMI, the greater the inflammation (Shelton and Miller 2010; Shoelson et al. 2007). In terms of transcriptional control of genes relevant to metabolic disorders that were observed in the infliximab responders, the HNF4-alpha transcription factor has not only been linked to diabetes (Hansen et al. 2005; Yamagata et al. 1996), but has also been demonstrated to be important for both gluconeogenesis and cholesterol homeostasis (Rhee et al. 2003; Yin et al. 2011). Therefore, a signature of differential gene expression at baseline that is related to alterations in glucose metabolism and HNF4-alpha transcription is consistent with the idea that patients that responded to infliximab may have altered glucose and lipid metabolism. Interestingly, however, only 1 patient out of 13 infliximab responders was diagnosed with diabetes and 3 of 13 had hypercholesterolemia (compared to 1 out of 14 infliximab non-responders). Thus, the relationship between altered glucose and lipid metabolism and response to infliximab cannot be explained by the presence of manifest disease, and may represent a more incipient process that contributes to treatment resistance, inflammation and ultimately responsiveness to anti-inflammatory treatment strategies.
The gene expression pattern that emerged as significantly different between responders and non-responders at 2 weeks following infliximab infusion was the suppression of a subset of genes related to innate immune responses, which is interesting in light of the previous findings from this trial indicating that subjects with increased inflammation as reflected by a CRP >5 mg/L as well as increased TNF and its soluble receptors at baseline were more likely to respond to infliximab (Raison et al. 2013). Of note, 2 weeks is a time point when mean depression scores were significantly reduced in subjects that responded to infliximab (t=4.21, df=12, p<0.001). Therefore, transcriptional signatures of treatment response to infliximab appear to involve a regulation of existing metabolic imbalances accompanied by an increased anti-inflammatory response to TNF inhibition that may be beneficial in subjects that display increased inflammation and/or altered metabolic function.
The results from this study are interesting in conjunction with a recent study that conducted a targeted analysis of the expression of genes related to inflammatory cytokines in depressed subjects before and after antidepressant treatment with escitalopram or nortriptyline (Cattaneo et al. 2013). This study found that patients resistant to antidepressant treatment exhibited increased inflammatory genes including TNF compared to those that responded. In contrast, in the current study, increased TNF expression at baseline was predictive of response to infliximab. These data are consistent with the notion that increased inflammation prior to treatment predicts non-response to conventional antidepressant therapy while potentially predicting a successful response to immune-targeted treatment.
Although the infliximab responder group as a whole did not exhibit increased CRP at baseline compared to non-responders, patients with CRP >5mg/ml favored response to infliximab, and infliximab responders exhibited higher baseline concentrations of circulating TNF and its soluble receptors (Raison et al. 2013) and increased TNF family (TNF and TNFSF12) gene expression, further supporting the presence of low grade inflammation in the responder group. In relation to metabolic disorders, obesity is associated with chronic activation of the innate immune system that is thought to be driven by adipose tissue, which can subsequently lead to insulin resistance, impaired glucose tolerance and diabetes (Osborn and Olefsky 2012; Shoelson et al. 2007). The presence of both a transcriptional signature of altered metabolic function coupled with increased inflammation would indicate a possible association between treatment response and obesity, yet infliximab responders did not exhibit increased BMI compared to non-responders at baseline. Therefore, the association between metabolic-related transcripts and response to infliximab may be independent of adiposity, however, BMI is only one measure of adiposity and other measures (e.g. waist-hip ratio and body fat percentage) were not included in this study. The regulation of these metabolic transcripts by anti-inflammatory infliximab treatment suggests that this low-grade inflammation may drive changes in metabolism, however more work is necessary to fully characterize the metabolic function in TRD and its relationship with adiposity and inflammation.
Of the other patient characteristics of infliximab responders that may have contributed to the observed differences in genes expression, infliximab responders were slightly older than non-responders. Aging has been associated with increased production of inflammatory cytokines (Fagiolo et al. 1993), as well as exaggerated inflammatory responses in the brain and exacerbated depressive-like behavior in response to activation of the immune system in the periphery (Godbout and Johnson 2009; Godbout et al. 2008). Nevertheless, it should be noted that the gene expression analyses were adjusted for age among other clinical variables.
When considering the immune cell subsets that were represented in the TOA by the differentially expressed transcripts at baseline, plasmacytoid dendritic cells exert multiple functions, including the production of IFN-alpha and other inflammatory cytokines and regulation of T-cell responses that can contribute to inflammation (Jahnsen et al. 2002). Interestingly, B cells can affect glucose metabolism by activating proinflammatory macrophages and T cells in adipose tissue (Winer et al. 2011). Nevertheless, it is important to note that the use of gene expression profiles in peripheral blood immune cells to extrapolate pathways and processes in other tissues has limitations. Although gene expression related to gluconeogensis and cholesterol homeostasis may appear to be indicative of general metabolic alterations in infliximab responders, the findings might also be indicative of changes in the energy demands of the immune cells themselves. For example, glucose metabolism is highly regulated in lymphocytes, and lymphocyte activation requires dramatically more glucose to meet energy demands for proliferation (Maciver et al. 2008). Furthermore, glucose deficiency can lead to cell death (Maciver et al. 2008; Rathmell et al. 2003).
In addition to the identification of a set of predictive transcripts related to metabolic function (including the highly predictive PGAM4), HNF4-alpha transcriptional control, and TWEAK signaling, we also identified two other highly predictive transcripts for genes related to neuronal functioning that have been associated with neurodegenerative disorders and psychiatric illness, increased expression of TGM2 and decreased expression of DTNBP1. TGM2 encodes a tissue glutaminase, a family of enzymes that catalyze post-translational modifications of proteins, including cross-linking, and may play a role in the pathology of neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease (Andringa et al. 2004; Wang et al. 2008). Moreover, the DTNBP1 gene, which encodes dysbindin, a protein involved in signal transduction and thought to regulate glutamate transmission, exhibits genetic variation that has been associated with both schizophrenia and major depression (Kim et al. 2008; Numakawa et al. 2004), and predicts early (4 week) responses to antidepressants (Pae et al. 2007). Interestingly, like PGAM4 and other metabolic-related gene transcripts, DTNBP1 expression was significantly regulated by infliximab over time in responders but not non-responders, and there was no difference in DTNBP1 expression between responders and non-responders at any time point after infliximab infusion, indicating that changes in expression of this gene may be related to the response to infliximab.
In addition to the aforementioned limitations of this study (lack of control subjects, BMI as the only measure of adiposity, increased age of the infliximab responders), several additional methodological issues warrant consideration. The comparison between infliximab responders and non-responders was made on a relatively small sample size <30, which may have affected the ability to detect group differences. Nevertheless, a sizeable number of highly significant transcripts (n=148) were identified that predicted response to infliximab. This number of differentially expressed transcripts is consistent with (and in fact exceeds) the number of transcripts differentially expressed in a previous study comparing responders to non-responders in patients receiving infliximab for rheumatoid arthritis (Lequerre et al. 2006). Furthermore, although treatment assignment was randomized and patient characteristics were balanced in the placebo and infliximab groups, we were unable to control for demographic variables, such as age, in infliximab responders versus non-responders. Moreover, because stringent exclusion criteria were required due to the use of infliximab which carries significant liabilities in terms of infectious diseases and autoimmune disorders, the results may be difficult to generalize to other populations of patients with TRD who may exhibit even greater activation of inflammatory pathways due to co-morbid conditions that were exclusionary in our trial. Finally, a number of methods are available for indentifying biological significance of differentially expressed genes. However, similar results were obtained for the pathway analysis of genes differentially expressed at baseline using two frequently published databases (Metacore and Wikipathways), indicating the validity of the conclusions drawn from this data set. Nevertheless, future studies will be necessary to confirm the reliability of these findings by exploring the association between antidepressant response to infliximab in TRD patients with both metabolism-related gene expression and clinical metabolic assessments, such as fasting glucose or cholesterol, that were not explored in the present study.
In sum, this study identified a number of significantly expressed transcripts associated with gluconeogensis and cholesterol transport that predicted the antidepressant response to infliximab in patients with TRD. Interestingly, these predictive transcripts were regulated over time by infliximab in the responders, and appeared to be involved in the response. This regulation of metabolic-related genes during treatment response was accompanied by an increase in anti-inflammatory responses to inhibition of TNF by infliximab. Together these findings suggest that depressed patients with increased peripheral inflammation in association with evidence of biological processes indicative of a metabolic disorder may benefit from novel anti-inflammatory treatments for depression, such as infliximab. Nevertheless, more work is necessary to determine the relationship between glucose and cholesterol metabolism, TRD, and inflammation.
Supplementary Material
Acknowledgments
This work was supported by funds from the National Institute of Mental Health (R21MH0771172) and Centocor Ortho Biotec Services LLC. In addition, the study was supported by PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program and PHS Grant M01 RR0039 from the General Clinical Research Center program, National Institutes of Health, National Center for Research Resources.
Footnotes
Financial Disclosure
All authors declare that there are no conflicts of interest, and all financial disclosures are listed for each author: Charles L. Raison has served on the advisory board for, consulted for, developed continuing medical education material for, or received travel funds from Pamlab, Lilly, North American Center for Continuing Education, Pfizer, Johnson & Johnson, and CME Incite; Andrew H. Miller has served as a consultant for Abbott Laboratories, AstraZeneca, GlaxoSmithKline, Lundbeck Research USA, F. Hoffmann-La Roche Ltd., Schering-Plough Research Institute and Wyeth/Pfizer Inc. and has received research support from Centocor Inc., GlaxoSmithKline, and Schering-Plough Research Institute; Divya Mehta, Bobbi J. Woolwine, Ebrahim Haroon, Elisabeth B. Binder, and Jennifer C. Felger have nothing to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andringa G, Lam KY, Chegary M, Wang X, Chase TN, Bennett MC. Tissue transglutaminase catalyzes the formation of alpha-synuclein crosslinks in Parkinson’s disease. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2004;18(7):932–934. doi: 10.1096/fj.03-0829fje. [DOI] [PubMed] [Google Scholar]
- Capuron L, Gumnick JF, Musselman DL, Lawson DH, Reemsnyder A, Nemeroff CB, Miller AH. Neurobehavioral effects of interferon-alpha in cancer patients: phenomenology and paroxetine responsiveness of symptom dimensions. Neuropsychopharmacology. 2002;26(5):643–652. doi: 10.1016/S0893-133X(01)00407-9. [DOI] [PubMed] [Google Scholar]
- Cattaneo A, Gennarelli M, Uher R, Breen G, Farmer A, Aitchison KJ, Craig IW, Anacker C, Zunsztain PA, McGuffin P, et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: differentiating between baseline ‘predictors’ and longitudinal ‘targets’. Neuropsychopharmacology. 2013;38(3):377–385. doi: 10.1038/npp.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 2003;19(14):1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108(7):3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese A, Moffitt TE, Pariante CM, Ambler A, Poulton R, Caspi A. Elevated inflammation levels in depressed adults with a history of childhood maltreatment. Arch Gen Psychiatry. 2008;65(4):409–415. doi: 10.1001/archpsyc.65.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese A, Pariante CM, Caspi A, Taylor A, Poulton R. Childhood maltreatment predicts adult inflammation in a life-course study. Proc Natl Acad Sci U S A. 2007;104(4):1319–1324. doi: 10.1073/pnas.0610362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhabhar FS, Burke HM, Epel ES, Mellon SH, Rosser R, Reus VI, Wolkowitz OM. Low serum IL-10 concentrations and loss of regulatory association between IL-6 and IL-10 in adults with major depression. Journal of psychiatric research. 2009;43(11):962–969. doi: 10.1016/j.jpsychires.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Dowlati Y, Herrmann N, Swardfager W, Liu H, Sham L, Reim EK, Lanctot KL. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67(5):446–457. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- Eisenberger NI, Berkman ET, Inagaki TK, Rameson LT, Mashal NM, Irwin MR. Inflammation-induced anhedonia: endotoxin reduces ventral striatum responses to reward. Biol Psychiatry. 2010;68(8):748–754. doi: 10.1016/j.biopsych.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- Eller T, Vasar V, Shlik J, Maron E. Pro-inflammatory cytokines and treatment response to escitalopram in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(2):445–450. doi: 10.1016/j.pnpbp.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Fagiolo U, Cossarizza A, Scala E, Fanales-Belasio E, Ortolani C, Cozzi E, Monti D, Franceschi C, Paganelli R. Increased cytokine production in mononuclear cells of healthy elderly people. Eur J Immunol. 1993;23(9):2375–2378. doi: 10.1002/eji.1830230950. [DOI] [PubMed] [Google Scholar]
- Felger JC, Cole SW, Pace TW, Hu F, Woolwine BJ, Doho GH, Raison CL, Miller AH. Molecular signatures of peripheral blood mononuclear cells during chronic interferon-alpha treatment: relationship with depression and fatigue. Psychol Med. 2011:1–13. doi: 10.1017/S0033291711002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV. Washington DC: American Psychiatric Press; 1997. [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. Journal of psychiatric research. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Godbout JP, Johnson RW. Age and neuroinflammation: a lifetime of psychoneuroimmune consequences. Immunology and allergy clinics of North America. 2009;29(2):321–337. doi: 10.1016/j.iac.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Godbout JP, Moreau M, Lestage J, Chen J, Sparkman NL, JOC, Castanon N, Kelley KW, Dantzer R, Johnson RW. Aging exacerbates depressive-like behavior in mice in response to activation of the peripheral innate immune system. Neuropsychopharmacology. 2008;33(10):2341–2351. doi: 10.1038/sj.npp.1301649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider AS, Cohen J, Fei J, Zaba LC, Cardinale I, Toyoko K, Ott J, Krueger JG. Insights into gene modulation by therapeutic TNF and IFNgamma antibodies: TNF regulates IFNgamma production by T cells and TNF-regulated genes linked to psoriasis transcriptome. The Journal of investigative dermatology. 2008;128(3):655–666. doi: 10.1038/sj.jid.5701064. [DOI] [PubMed] [Google Scholar]
- Hamilton M. A rating scale for depression. Journal of neurology, neurosurgery, and psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SK, Rose CS, Glumer C, Drivsholm T, Borch-Johnsen K, Jorgensen T, Pedersen O, Hansen T. Variation near the hepatocyte nuclear factor (HNF)-4alpha gene associates with type 2 diabetes in the Danish population. Diabetologia. 2005;48(3):452–458. doi: 10.1007/s00125-005-1671-0. [DOI] [PubMed] [Google Scholar]
- Haroon E, Raison CL, Miller AH. Psychoneuroimmunology meets neuropsychopharmacology: translational implications of the impact of inflammation on behavior. Neuropsychopharmacology. 2012;37(1):137–162. doi: 10.1038/npp.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison NA, Brydon L, Walker C, Gray MA, Steptoe A, Critchley HD. Inflammation causes mood changes through alterations in subgenual cingulate activity and mesolimbic connectivity. Biol Psychiatry. 2009;66(5):407–414. doi: 10.1016/j.biopsych.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepgul N, Cattaneo A, Zunszain PA, Pariante CM. Depression pathogenesis and treatment: what can we learn from blood mRNA expression? BMC Med. 2013;11(1):28. doi: 10.1186/1741-7015-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howren MB, Lamkin DM, Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: a meta-analysis. Psychosom Med. 2009;71(2):171–186. doi: 10.1097/PSY.0b013e3181907c1b. [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Jahnsen FL, Farkas L, Lund-Johansen F, Brandtzaeg P. Involvement of plasmacytoid dendritic cells in human diseases. Hum Immunol. 2002;63(12):1201–1205. doi: 10.1016/s0198-8859(02)00759-0. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Mandelli L, Pae CU, De Ronchi D, Jun TY, Lee C, Paik IH, Patkar AA, Steffens D, Serretti A, et al. Is there protective haplotype of dysbindin gene (DTNBP1) 3 polymorphisms for major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(2):375–379. doi: 10.1016/j.pnpbp.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Lanquillon S, Krieg JC, Bening-Abu-Shach U, Vedder H. Cytokine production and treatment response in major depressive disorder. Neuropsychopharmacology. 2000;22(4):370–379. doi: 10.1016/S0893-133X(99)00134-7. [DOI] [PubMed] [Google Scholar]
- Lequerre T, Gauthier-Jauneau AC, Bansard C, Derambure C, Hiron M, Vittecoq O, Daveau M, Mejjad O, Daragon A, Tron F, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis research & therapy. 2006;8(4):R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. Journal of leukocyte biology. 2008;84(4):949–957. doi: 10.1189/jlb.0108024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock C, Landau S, Barry K, Maulayah P, Hotopf M, Cleare AJ, Norris S, Pariante CM. Psychopathological symptoms during interferon-alpha and ribavirin treatment: effects on virologic response. Mol Psychiatry. 2005;10(4):332–333. doi: 10.1038/sj.mp.4001634. [DOI] [PubMed] [Google Scholar]
- Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, Mercer KB, Putz B, Bradley B, Holsboer F, et al. Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: evidence from endocrine and gene expression studies. Arch Gen Psychiatry. 2011;68(9):901–910. doi: 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AH, Maletic V, Raison CL. Inflammation and its discontents: the role of cytokines in the pathophysiology of major depression. Biol Psychiatry. 2009;65(9):732–741. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AHHE, Raison CL, Felger JC. Cytokine Targets in the Brain: Impact on Neurotransmitters and Neurocircuits. Depression and Anxiety. doi: 10.1002/da.22084. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JM, Doll R, Ma R, Cole SW. A functional genomic fingerprint of chronic stress in humans: blunted glucocorticoid and increased NF-kappaB signaling. Biol Psychiatry. 2008;64(4):266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk JP, Phillips G, Waite R, Kuhn J, Schaaf LJ, Otterson GA, Guttridge D, Rhoades C, Shah M, Criswell T, et al. Assessment of tumor necrosis factor alpha blockade as an intervention to improve tolerability of dose-intensive chemotherapy in cancer patients. J Clin Oncol. 2006;24(12):1852–1859. doi: 10.1200/JCO.2005.04.2838. [DOI] [PubMed] [Google Scholar]
- Nanni V, Uher R, Danese A. Childhood maltreatment predicts unfavorable course of illness and treatment outcome in depression: a meta-analysis. Am J Psychiatry. 2012;169(2):141–151. doi: 10.1176/appi.ajp.2011.11020335. [DOI] [PubMed] [Google Scholar]
- Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, Iwata N, Ozaki N, Taguchi T, Tatsumi M, Kamijima K, et al. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Human molecular genetics. 2004;13(21):2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nature medicine. 2012;18(3):363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- Oskooilar N, Wilcox CS, Tong ML, Grosz DE. Body mass index and response to antidepressants in depressed research subjects. J Clin Psychiatry. 2009;70(11):1609–1610. doi: 10.4088/JCP.09l05226blu. [DOI] [PubMed] [Google Scholar]
- Pae CU, Serretti A, Mandelli L, De Ronchi D, Patkar AA, Jun TY, Kim JJ, Lee CU, Lee SJ, Lee C, et al. Dysbindin associated with selective serotonin reuptake inhibitor antidepressant efficacy. Pharmacogenetics and genomics. 2007;17(1):69–75. doi: 10.1097/01.fpc.0000236330.03681.6d. [DOI] [PubMed] [Google Scholar]
- Perera TD, Dwork AJ, Keegan KA, Thirumangalakudi L, Lipira CM, Joyce N, Lange C, Higley JD, Rosoklija G, Hen R, et al. Necessity of hippocampal neurogenesis for the therapeutic action of antidepressants in adult nonhuman primates. PLoS One. 2011;6(4):e17600. doi: 10.1371/journal.pone.0017600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6(7):e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends Immunol. 2006;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF, Haroon E, Miller AH. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA psychiatry. 2013;70(1):31–41. doi: 10.1001/2013.jamapsychiatry.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Molecular and cellular biology. 2003;23(20):7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberg A, Yirmiya R, Schuld A, Kraus T, Haack M, Morag A, Pollmacher T. Cytokine-associated emotional and cognitive disturbances in humans. Arch Gen Psychiatry. 2001;58(5):445–452. doi: 10.1001/archpsyc.58.5.445. [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100(7):4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush AJ, Wisniewski SR, Warden D, Luther JF, Davis LL, Fava M, Nierenberg AA, Trivedi MH. Selecting among second-step antidepressant medication monotherapies: predictive value of clinical, demographic, or first-step treatment features. Arch Gen Psychiatry. 2008;65(8):870–880. doi: 10.1001/archpsyc.65.8.870. [DOI] [PubMed] [Google Scholar]
- Rustad JK, Musselman DL, Nemeroff CB. The relationship of depression and diabetes: pathophysiological and treatment implications. Psychoneuroendocrinology. 2011;36(9):1276–1286. doi: 10.1016/j.psyneuen.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Shelton RC, Miller AH. Eating ourselves to death (and despair): the contribution of adiposity and inflammation to depression. Prog Neurobiol. 2010;91(4):275–299. doi: 10.1016/j.pneurobio.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmelkov E, Tang Z, Aifantis I, Statnikov A. Assessing quality and completeness of human transcriptional regulatory pathways on a genome-wide scale. Biology direct. 2011;6:15. doi: 10.1186/1745-6150-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132(6):2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- Sluzewska A, Sobieska M, Rybakowski JK. Changes in acute-phase proteins during lithium potentiation of antidepressants in refractory depression. Neuropsychobiology. 1997;35(3):123–127. doi: 10.1159/000119332. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Pace TW, Liu T, Felger JC, Mister D, Doho GH, Kohn JN, Barsevick AM, Long Q, Miller AH. Predictors of depression in breast cancer patients treated with radiation: Role of prior chemotherapy and nuclear factor kappa B. Cancer. 2013 doi: 10.1002/cncr.28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi MH, Rush AJ, Ibrahim HM, Carmody TJ, Biggs MM, Suppes T, Crismon ML, Shores-Wilson K, Toprac MG, Dennehy EB, et al. The Inventory of Depressive Symptomatology, Clinician Rating (IDS-C) and Self-Report (IDS-SR), and the Quick Inventory of Depressive Symptomatology, Clinician Rating (QIDS-C) and Self-Report (QIDS-SR) in public sector patients with mood disorders: a psychometric evaluation. Psychol Med. 2004;34(1):73–82. doi: 10.1017/s0033291703001107. [DOI] [PubMed] [Google Scholar]
- Tyring S, Gottlieb A, Papp K, Gordon K, Leonardi C, Wang A, Lalla D, Woolley M, Jahreis A, Zitnik R, et al. Etanercept and clinical outcomes, fatigue, and depression in psoriasis: double-blind placebo-controlled randomised phase III trial. Lancet. 2006;367(9504):29–35. doi: 10.1016/S0140-6736(05)67763-X. [DOI] [PubMed] [Google Scholar]
- Wang DS, Dickson DW, Malter JS. Tissue transglutaminase, protein cross-linking and Alzheimer’s disease: review and views. International journal of clinical and experimental pathology. 2008;1(1):5–18. [PMC free article] [PubMed] [Google Scholar]
- Winer DA, Winer S, Shen L, Wadia PP, Yantha J, Paltser G, Tsui H, Wu P, Davidson MG, Alonso MN, et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nature medicine. 2011;17(5):610–617. doi: 10.1038/nm.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten D, Tibshirani R, Gu SG, Fire A, Lui WO. Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol. 2010;8:58. doi: 10.1186/1741-7007-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384(6608):458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- Yin L, Ma H, Ge X, Edwards PA, Zhang Y. Hepatic hepatocyte nuclear factor 4alpha is essential for maintaining triglyceride and cholesterol homeostasis. Arteriosclerosis, thrombosis, and vascular biology. 2011;31(2):328–336. doi: 10.1161/ATVBAHA.110.217828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.