Abstract
Background
Glycosaminoglycans, such as hyaluronic acid, heparin, and chondroitin sulfate, are among the top ranked products in industrial biotechnology for biomedical applications, with a growing world market of billion dollars per year. Recently a remarkable progress has been made in the development of tailor-made strains as sources for the manufacturing of such products. The genetic modification of E. coli K4, a natural producer of chondroitin sulfate precursor, is challenging considering the lack of detailed information on its genome, as well as its mobilome. Chondroitin sulfate is currently used as nutraceutical for the treatment of osteoarthritis, and several new therapeutic applications, spanning from the development of skin substitutes to live attenuated vaccines, are under evaluation.
Results
E. coli K4 was used as host for the overexpression of RfaH, a positive regulator that controls expression of the polysaccharide biosynthesis genes and other genes necessary for the virulence of E. coli K4. Various engineering strategies were compared to investigate different types of expression systems (plasmid vs integrative cassettes) and integration sites (genome vs endogenous mobile element). All strains analysed in shake flasks on different media showed a capsular polysaccharide production improved by 40 to 140%, compared to the wild type, with respect to the final product titer. A DO-stat fed-batch process on the 2L scale was also developed for the best performing integrative strain, EcK4r3, yielding 5.3 g∙L-1 of K4 polysaccharide. The effect of rfaH overexpression in EcK4r3 affected the production of lipopolysaccharide and the expression of genes involved in the polysaccharide biosynthesis pathway (kfoC and kfoA), as expected. An alteration of cellular metabolism was revealed by changes of intracellular pools of UDP-sugars which are used as precursors for polysaccharide biosynthesis.
Conclusions
The present study describes the identification of a gene target and the application of a successful metabolic engineering strategy to the unconventional host E. coli K4 demonstrating the feasibility of using the recombinant strain as stable cell factory for further process implementations.
Keywords: E. coli K4, Capsular polysaccharide, rfaH, Antiterminator, Chondroitin sulfate, UDP-precursors
Introduction
Chondroitin sulfate (CS) is a natural linear polysaccharide formed by disaccharide units of N-acetyl-D-galactosamine (GalNAc) and Glucuronic acid (GlcA) β 1:4 and β 1:3 linked. It is a ubiquitous component of the extracellular matrix of vertebrates where it exerts its established chondroprotective properties by improving the biosynthesis of connective tissue components and increasing the viscosity of synovial fluid at disease sites [1,2]. CS also inhibits cartilage degradative enzymes therefore it is widely used for the treatment of osteoarthrithis. Moreover, recent novel studies suggest its potential applicability for cancer prevention, formulation of skin substitutes, and vaccine development [3-5].
The external coating, defined as capsule, of Escherichia coli K4 is composed of fructosylated chondroitin and therefore represents an unlimited source of precursor supply for the production of CS. Moreover the use of microbial polymer eliminates the risk of interspecies viral and prionic contaminations.
Several thriving research efforts regarded the set up of fermentation strategies targeting the maximization of capsular polysaccharide (CPS) production from E. coli K4 followed by recovery of a 90% pure polysaccharide and final chemical sulfation resulting in a polymer that is equivalent to the extractive counterpart [6-8]. Productivity was further increased by overexpressing the enzyme responsible for polymer assembly in the wild type (wt) strain using an inducible plasmid as expression system. However, although a 100% improvement in the final CPS titer was achieved, these experiments also revealed a great genetic instability of the recombinant strain, that limited process scale-up [9].
The K4 CPS belongs to group II K antigens. Capsules produced from E. coli K5 and K1 are well studied members of this group [10]. The structure of the cluster responsible for CPS biosynthesis is organised into three regions. Regions 1 and 3 contain the kps genes common to all group II members and mainly involved in the transport of the polymer in the periplasm and outside the cell wall [11,12]. The genes coding for enzymes that direct the synthesis and assembly of the final polysaccharide are found in region 2, that is the serotype specific. Besides the IS2 insertion sequence, region 2 in E. coli K4 comprises three genes of unknown function (kfoB, kfoG, kfoD), kfoE responsible for the addition of fructose residues [13], kfoA coding for a UDP-glucose 4-epimerase, kfoF coding for a UDP-glucose dehydrogenase and the well characterised kfoC gene coding for chondroitin polymerase [14,15].
One of the factors regulating capsule expression is the transcriptional activator RfaH that carries out an antitermination process, that also requires the cis-acting operon polarity suppressor element (ops) located just upstream of a JUMP start (just upstream of many polysaccharide gene starts). The role of the ops element is to recruit RfaH, and therefore its presence on the nascent transcript is crucial. Deletion of the ops element results in fact in the lack of capsule expression in E. coli K5 [16]. The JUMP start sequence is also required for RfaH to function; indeed it is conserved and present at the 5′ end of RfaH-regulated operons, and it might be necessary to fold the mRNA into stem-loop structures that mediate these processes [17]. RfaH plays several roles in E. coli affecting the biosynthesis of lipopolysaccharide (LPS), O-antigen, and haemolysin, and the production of the F-factor sex pilus [17]. Its function is to control promoter distal gene expression by preventing the termination of transcripts and promoting transcription over long distances. In E. coli K5 RfaH seems to prevent the termination of region 3 transcripts allowing read through transcription, and thereby also regulating the transcription of region 2 [16]. E. coli K5 strains with mutations in the rfaH gene showed undetectable levels of expression of region 2 genes, and of the corresponding encoded proteins. The regulation of transcription of group II capsule gene cluster is quite complex, also depending on the activity of other regulator proteins namely H-NS (nucleoid associated protein) and Bip-A (tyrosine phosphorilated GTPase) that are involved in the thermal regulation of region 1 and 3 promoters [18].
In the present study an integrative expression cassette comprising the rfaH gene under the control of part of the constitutive promoter of the gapA gene, coding for glyceraldehyde 3-phosphate dehydrogenase, was constructed and introduced into wild type E. coli K4. The recombinant strains overexpressing the regulator demonstrated a significant increase of the CPS yield and proved suitable for the scale up of the fermentation process.
Results
Strain construction
The host E. coli K4 was modified by the addition of extra, plasmidic or integrative, copies of the rfaH gene. E. coli K4-pTrcrfaH possesses additional inducible copies of rfaH on the pTrc plasmid, under the control of the hybrid promoter consisting of the -10 region from lacUV5 and the -35 region from trpB. Protein profiles of this strain grown in shake flask showed an overexpressed band of about 18 KDa by SDS-Page 3h after IPTG induction and the intensity of this band slightly increased after 8 and 24h of growth (data not shown). The sequence of the rfaH PCR product cloned in the pTrc plasmid was compared to that of E. coli K12 and silent point mutations, three transitions and three transversions, were identified (Additional file 1).
EcK4r1 and EcK4r3 were generated by the integration of a fragment containing the rfaH gene under the control of the P1 portion of the gapA promoter, targeting the endogenous plasmid pK4EC05 and the lacZ gene, respectively. The expression cassette also comprised the kanamycin resistance gene flanked by FRT sites that allowed the removal of the selection marker by means of a consecutive pop-out step, resulting in resistance-free strains.
Shake flask experiments
Direct comparison of the three recombinant strains was performed in shake flask conditions on the control medium [19] to investigate physiological differences resulting from (i) an increased copy number of the rfaH gene (ii) the different expression systems used, and (iii) integration targeted to endogenous plasmid or genome (strains EcK4r1 and EcK4r3, respectively). All recombinant strains demonstrated a significant increase of CPS production after 24h of growth together with a higher YK4/X, thereby clearly demonstrating the positive effect of a higher availability of RfaH on CPS production (Table 1). E. coli K4-pTrcrfaH was used in this study only as a tool to evaluate whether rfaH overexpression could boost CPS production, and after 24h of growth a 2.4-fold higher concentration of polysaccharide was in fact obtained with a 155% increase of the YK4/X; a slightly lower growth rate and biomass production compared to the wild type strain was probably due to the higher metabolic burden.
Table 1.
Summary of growth, K4 CPS production and relative yields obtained in shake flask experiments from the reference E.coli K4 and the recombinant strains
|
Strain |
Carbon/Nitrogen |
Biomass |
μmax |
K4 |
YK4/X |
YK4/S |
|---|---|---|---|---|---|---|
| (gcdw∙L-1) | (h-1) | (mg∙L-1) | (mg∙gcdw-1) | (mg∙g-1) | ||
|
E.coli K4 |
Gly/Soy |
2.24 |
0.88 |
124 |
55 |
11 |
|
E.coli K4-pTrcrfaH |
Gly/Soy |
2.08 |
0.67 |
294 |
140 |
- |
| EcK4r1 |
Gly/Soy |
2.49 |
0.76 |
174 |
70 |
- |
| EcK4r3 |
Gly/Soy |
2.39 |
0.77 |
212 |
88 |
20 |
|
E.coli K4 |
Glu/Ye |
2.89 |
1.03 |
190 |
66 |
18 |
| EcK4r3 | Glu/Ye | 2.83 | 1.05 | 283 | 100 | 27 |
The concentration of capsular polysaccharide was measured in the broth at the end of the experiment after 24 h of growth. YK4/X indicates mg of K4 CPS produced per g of dry cells. YK4/S indicates mg of K4 CPS produced per g of carbon source consumed. μmax indicates the maximum specific growth rate in h-1. The values are averages of at least 4 separate experiments with a standard deviation below 10%. Abbreviations: Glu (Glucose), Gly (Glycerol), Ye (Yeast extract).
The performance of integrative recombinants EcK4r1 vs. EcK4r3 was quite different in terms of final total polysaccharide produced and yield over biomass. As shown in Table 1 the percentage of increase of both polymer and YK4/X at the end of the growth, compared to the wild type strain, was almost two fold higher for EcK4r1 versus EcK4r3.
When testing E. coli K4 and EcK4r3 on a new medium containing glucose and yeast extract, an increase of biomass and K4 CPS was observed. Furthermore, also the yield of K4 CPS per gram biomass increased on the new medium for both strains, indicating a decoupling of polysaccharide and biomass formation (Table 1). EcK4r3 cultivations also showed the decrease of production of fructosylated biopolymer from 14 to 1% and the drastic reduction of the pH24h from 6.4±0.28 (on the standard medium) to 5.2±0.14. In order to verify that the additional copies of rfaH did not modify the aggressiveness of EcK4r3, a MIC test was performed. In particular the antimicrobial resistance of EcKr3 was compared to that of the wild type strain by using a commercial kit. Results did not highlight any difference between the two strains.
Hydrolysis of the K4 CPS
A partial defructosylation of the K4 CPS was obtained after the incubation at pH=5 and pH=6 in conditions simulating growth in shake flasks but without cells (Table 2). In particular at pH=5 the percentage of defructosylated polymer increased from about 12.5% after 3h of incubation to 25% at the end of the process. Defructosylation was less pronounced when incubating the polymer at pH=6.
Table 2.
Hydrolysis of the K4 CPS produced during shake flask experiments
| pH=5 | pH=6 | |||
|---|---|---|---|---|
|
Time |
K4 |
Defructosylated |
K4 |
Defructosylated |
|
(h) |
(mg∙L-1) |
(mg∙L-1) |
(mg∙L-1) |
(mg∙L-1) |
|
3 |
189 |
27 |
199 |
17 |
|
6 |
181 |
35 |
198 |
18 |
| 16 | 162 | 54 | 192 | 24 |
The polysaccharide (216 mg∙L-1) was incubated in the standard growth medium at pH=5 and pH=6 without cells for 16h. The temperature was kept constant at 37°C and agitation was set to 200 rpm. Samples were collected after 3, 6 and 16h of incubation to analyse defructosylation over time.
Analysis of gene expression
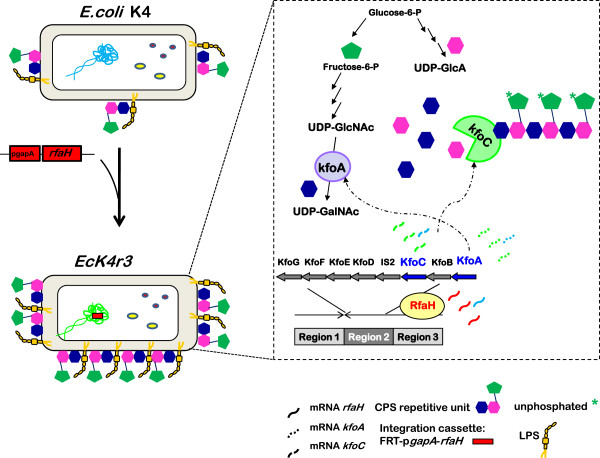
Quantitative real time PCR was used to analyse the level of expression of the rfaH gene in EcK4r3 grown on the medium containing glucose and yeast extract to investigate its expression levels and establish whether the presence of additional gene copies also affected RNA expression profiles of the kfoA and kfoC genes. Samples from shake flasks were collected during early exponential and mid to end stationary phases. Expression was normalised on the 16S rRNA. The overexpression folds are shown in Figure 1. The expression of all genes increased during growth in EcK4r3 compared to the wild type but, as expected, the increment of rfaH mRNA pools was higher compared to that of kfoC and kfoA transcripts.
Figure 1.
Analysis of rfaH, kfoC and kfoA gene expression in E.coli K4 and EcK4r3 in shake flask experiments. Histogram representing the fold overexpression of the genes under investigation in the recombinant strain EcK4r3 compared to the wild type E.coli K4.
Batch experiments in 2L bioreactors
Twenty-six hours batch growths were conducted to analyse the physiology of strain E. coli K4-pTrcrfaH, however, results were completely opposite from those expected after shake flask experiments. The amount of K4 CPS produced was on average 190 mg·L-1, almost 40% lower compared to that obtained from cultivating the reference strain in the same conditions. A growth defect and the loss of recombinant plasmid already after the first 8h of growth were observed (data not shown), explaining the low YK4.
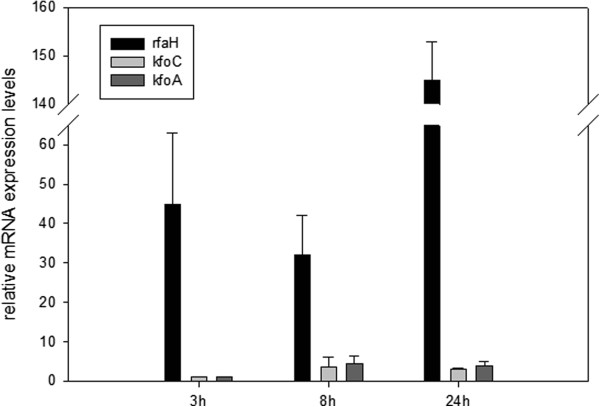
Batch experiments comparing strains EcK4r1 and EcK4r3 on the control medium are shown in Figure 2(a-b). The final titer of polysaccharide in the supernatant was higher by 34 and 55% compared to results obtained with E. coli K4 in the same growth conditions. EcK4r1 showed a lower growth, glycerol consumption and acid production rate (Figure 2b) compared to EcK4r3 and the concentration of total acids accumulated by the end of the process was higher. CPS production levels reached 402±20 and 466±25 mg·L-1,for EcK4r1 and EcK4r3, respectively.
Figure 2.
Time course of 24h batch fermentations on the control medium: comparison of strains EcK4r1 and EcK4r3. (a) Production of biomass and K4 CPS (b) Consumption of glycerol and production of total acids throughout the process.
The behaviour of EcK4r3 in the medium containing glucose and yeast extract was also analysed. Compared to the control medium only a slightly higher growth and carbon source consumption rate were observed, whereas the yield of K4 on biomass was unaffected. After 6h of growth the carbon source was exhausted, 4.2 gcdw·L-1 of biomass and 260 mg·L-1of K4 CPS were produced; the latter however doubled by the end of the process (24h) determining a final concentration of 532±30 mg·L-1of polysaccharide. The concentration of cell-bound and released LPS after 24h of growth was equal to 0.005 and 0.16 g·gcdw-1, respectively. Comparing the data to that obtained from the wild type strain a 1.58 and 1.55-fold increase were obtained for the two LPS fractions demonstrating the effect of rfaH overexpression on LPS production.
A hot methanol extraction method was used to determine the UDP-sugar precursors' content in fermentation samples by capillary electrophoresis. In particular biomass samples for the wt E. coli K4 (fermentation data not reported) and recombinant strain EcK4r3 after 5, 8 and 24h of growth were collected and analysed to identify eventual differences in UDP pools (Table 3). UDP-GlcA increased over time in both strains reaching after 24h a quite similar concentration of μmoles per gcdw; conversely, a 3.8 and 1.6-fold higher amount of sugar nucleotide is present in the wt after 5 and 8h of growth, respectively. The opposite trend is observed for UDP-GalNAc that decreases in the recombinant and wt strains; also in this case the precursors’s pool seems to be richer in the wt strain (2.3-fold higher) after 5 h of growth whereas more similar concentrations in the two strains are noticed after 8 and 24 h. Finally, E. coli K4 and EcK4r3 greatly differ for the content of intracellular UDP-Glucose (UDP-Glu) that shows opposite growing trends in the two strains, and quite diverse concentrations (Table 3).
Table 3.
Intracellular concentrations of UDP-Glu, UDP-GlcA and UDP-GalNAc in E.coli K4 (wt) and EcK4r3 during batch experiments
| |
|
E.coli K4 (wt) |
EcK4r3 |
|---|---|---|---|
| μmol∙gcdw-1 | μmol∙gcdw-1 | ||
| 5h |
UDP-Glu |
0.56 |
11.3 |
| |
UDP-GlcA |
0.42 |
0.11 |
| |
UDP-GalNAc |
3.77 |
1.63 |
| 8h |
UDP-Glu |
1.92 |
5.64 |
| |
UDP-GlcA |
1.33 |
0.82 |
| |
UDP-GalNAc |
0.61 |
0.51 |
| 24h |
UDP-Glu |
1.96 |
0.37 |
| |
UDP-GlcA |
1.85 |
1.93 |
| UDP-GalNAc | 0.07 | 0.05 |
Samples were collected after 5, 8 and 24h of growth. Experiments were performed in duplicate and the standard deviation was ≤ 15%.
DO-Stat fed-batch experiment
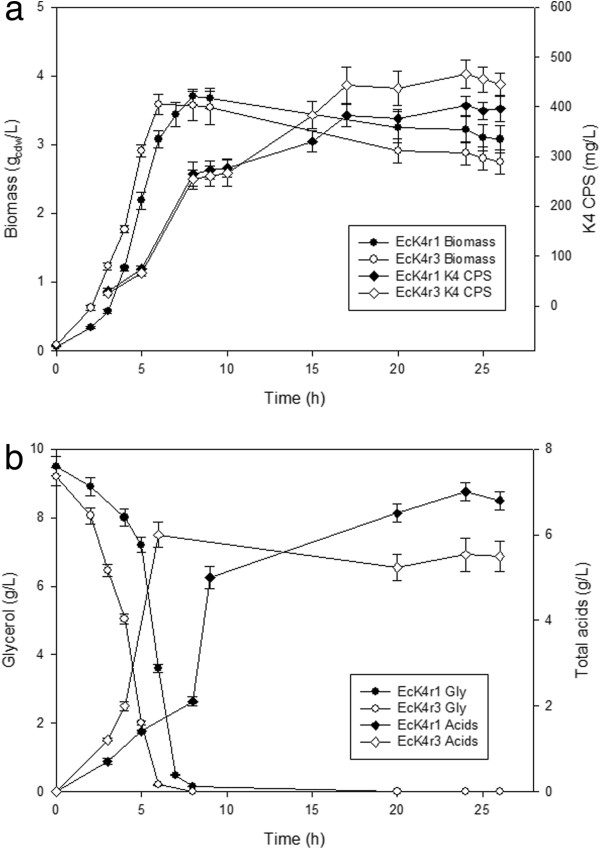
EcK4r3 was analysed in fed-batch fermentations using a DO-stat feeding strategy in order to meet the strain’s metabolic requests thus keeping overflow metabolism under control. On average within 49.5h of process the cell density reached 21.7±1.7 gcdw∙L-1 and 5.1±0.2 g∙L-1 of K4 CPS were attained (Figure 3). Compared to batch experiments on the same medium the YK4/X increased from 0.13±0.01 to 0.23±0.01 gCPS∙gcdw-1 whereas the YX/S decreased from 0.42±0.03 to 0.164±0.026 gcdw∙gS-1.
Figure 3.
Production of K4 CPS in 2L DO-Stat fed-batch experiments. Time course of biomass, polysaccharide production and residual glucose.
Discussion
Several microbial strains representing natural sources of GAG and GAG-similar molecules have so far been isolated and a great progress concerning the improvement of yields through the optimization of fermentation strategies was recently described [6,20]. Besides classical process optimization also genetic engineering of the strain is crucial for improving the efficiency of natural hosts. The so far reported metabolic re-tooling strategies mainly focused on increasing the supply of sugar precursors and of enzymes responsible for chain polymerization [6].
The transcription of region 2 of the CPS cluster of group II E. coli, that includes E. coli K4, is driven by a promoter located just downstream of region 3, consequently region 2 and 3 genes are transcribed as a single mRNA unit. Expression of group II K antigens requires the presence of the RfaH factor having the role of antiterminator thus providing transcription of promoter distal genes [21]. Previous work has indicated that loss of the RfaH protein in group II E. coli results in the decreased expression of region 2 genes [16] and K capsule [22]. It was therefore interesting to investigate whether, conversely, protein overexpression could lead to an increase in capsular polysaccharide production. At first the recombinant plasmid pTrc-rfaH containing the rfaH gene under the control of the Trc inducible promoter was introduced into E. coli K4 cells. Cells harbouring the above mentioned construct produced a protein of the expected molecular weight that increased over time in shake flask experiments and a significant increase in polysaccharide production was obtained after 8h and 24h of growth. The E. coli K4-pTrcrfaH recombinant strain only served as a proof of principle to determine whether the increased availability of RfaH would trigger CPS production. In fact, as already found previously for a similar expression construct [9], also in this case a severe loss of recombinant plasmid was observed in fermentation conditions thereby harshly affecting the productivity of the process. This finding therefore confirms that conventional plasmid driven expression constructs are suitable for investigating possible metabolic engineering targets in shake flasks but do not allow the scale up of the process with E. coli K4.
An expression cassette containing the rfaH gene under control of the P1 partial constitutive promoter of the glyceraldehyde-3-P dehydrogenase was designed and used to obtain integrative strains. As previously demonstrated E. coli K4 possesses an endogenous mobile element indicated as pK4EC05 that is stably inherited and shows homology to large conjugative plasmids [9]. pK4EC05 and the genome were used as integration targets for the construction of strains EcK4r1 and EcK4r3, respectively, to investigate whether the integration site could have an impact on improving CPS production. Results clearly show that growth of EcK4r1 and EcK4r3, and biomass formation were not affected by the introduction of recombinant DNA. Both strains demonstrated a significantly higher amount of polysaccharide in shake flask experiments on the control medium compared to the wt. A better performance was obtained from EcK4r3 that almost reached the titer produced from E. coli K4-pTrcrfaH. One major goal of this study was to investigate the stability of the newly constructed recombinant strains, a key feature in the attainment of master and working cell banks for biopharmaceutical applications. When shifting from shake flask to fermenter experiments the YK4/X for the wild type strain improves by 50%, specifically from 55 to 82mgCPS·gcdw-1[19]. The same YK4/X percentage increase was obtained when scaling up the batch process for EcK4r1 and EcK4r3. Also the YK4/X percentage increment for the two recombinant strains observed in shake flask experiments, in comparison to the wild type, was maintained in fermenter experiments (60% for strain EcK4r3 and 30% for strain EcK4r1). This conserved behaviour under the two different cultivation conditions is clearly demonstrating strain stability and overcomes the previously described problems. The development of biotech processes is often hindered by the use of hosts harbouring recombinant plasmids that can trigger stress responses that finally limit biomass and product yield, as observed for E. coli K4-pTrcrfaH. Similar restraints were not encountered for strain EcK4r1; as a matter of fact we observed that this endogenous element can host foreign DNA and keep it over generations and time, confirming its key importance for strain survival and it’s suitability for targeted gene integrations.
Fermentation experiments on the control medium in which the physiology of the integrative strains was studied confirm the superior performance of EcK4r3. Data highlighted a slightly faster growth for the latter that is reflected into the steeper substrate consumption and corresponding acid production rates. The significantly higher concentration of total acids (>25%) produced by EcK4r1 suggests a different carbon distribution which might influence the entire CPS biosynthesis process, resulting in a lower concentration of polymer. However, a key role might also be played by the number of gene copies integrated in the recombinant strains and by the integration site itself.
The use of glycerol as main carbon source in industrial microbiology is advantageous due to its low cost and handling ease. Nevertheless glucose is often preferable in large scale fed-batch fermentation processes. In this perspective an alternative medium containing glucose as main carbon source was evaluated. Also soy peptone, the main nitrogen source, was replaced by yeast extract that better supported growth and CPS production. A comparison of growth and CPS production for EcK4r3, the best performing integrative strain, and the wild type, on the two media under analysis is schematically represented in Figure 4. Considering all parameters studied best performances for both strains were obtained in the medium containing glucose and yeast extract. However, interestingly, the potential of the recombinant strain seems to be more expressed on the control medium since the improvement of all yields is higher, maybe indicating the occurrence of a certain type of limitation. Moreover the increment of biomass, K4 CPS, and relative yields observed for the wt strain following medium substitution, is lower for EcK4r3 further supporting the previous hypothesis. This may indicate that CPS production is subjected to various control levels one of which is RfaH-dependent, whereas the others controlled by medium components. Nevertheless, EcK4r3 demonstrated a yield of polysaccharide (mg·(gcdw·gsubS)-1) in batch experiments on this medium never described before, that is similar to that obtained with hyaluronic acid-producing Streptococcus strains [23,24] already used for industrial manufacturing processes. Compared to growth on the standard medium containing glycerol as the main carbon source we observed both for the wild type and for EcK4r3 an increase in the production of defructosylated polymer when glucose and yeast extract are present in the medium. Growth of both strains on the standard medium generates a final pH of about 6.4 whereas on the richer medium due to the higher amount of acids produces the pH is around 5. Chemical hydrolysis experiments of the K4 CPS in conditions simulating growth for 16h at pH 5 and 6 demonstrated a partial hydrolysis of the polysaccharide that is about 2-fold more pronounced at the lower pH. We therefore suggest that this might be one of the causes responsible for the presence of a polymer that is mostly defructosylated. However, probably several concurrent phenomena contribute to the production of mainly defructosylated polymer. For example since the growth and carbon consumption rates of both strains are higher on glucose another cause may be attributed to kinetic differences between polymerization and fructosylation and also to the channelling of fructose towards the synthesis of GalNAc that may also contribute to explaining the data observed.
Figure 4.
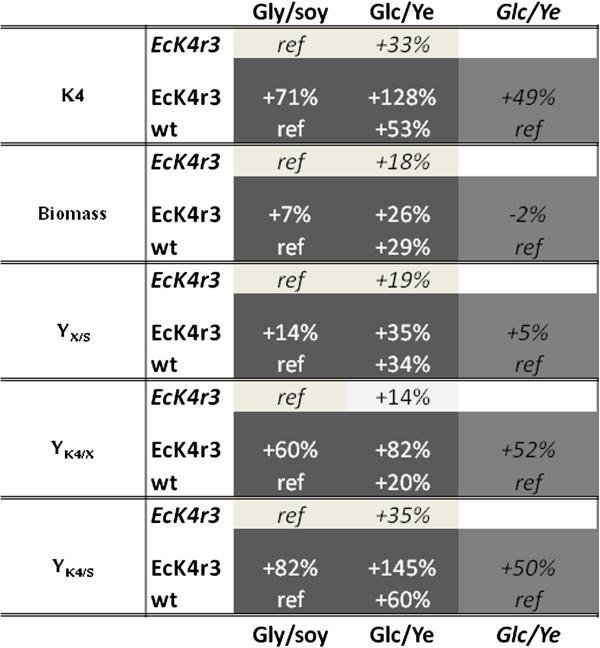
Relative changes of K4 CPS, biomass, and yields on different media for EcK4r3 and E.coli K4 (wt). Percentages in boxes with the same grey tone are referred to the combination of strain and medium indicated with ref (equal to 100%). Abbreviations: Glu (Glucose), Gly (Glycerol), Ye (Yeast extract).
To further improve the concentration of polysaccharide produced, a fed-batch process using a DO-stat feeding approach was developed. This technique allowed to satisfy the strain’s metabolic demand avoiding cell starvation or the accumulation of excess carbon thereby reaching the highest titer of K4 CPS produced through fermentation experiments up to date (5.3 g∙L-1). Compared to batch experiments on the same starting medium the amount of K4 CPS produced by EcK4r3 increased by 10-fold. Moreover growth conditions in fed-batch/DO-stat mode seemed to promote polysaccharide production to the detriment of biomass, since a 1.8-fold higher YK4/X was observed whereas the YX/S decreased by 2.6-fold compared to batch experiments.
Previous fermentation experiments on the wild type E. coli K4 performed by our group showed a significant improvement of the final K4 CPS concentration by using a membrane bioreactor combined with a fermentation strategy based on in situ product removal [25]. ECK4r3 would allow the use of well scalable fed-batch processes that do not require additional devices inside the fermentation vessel, which would be a step towards the actual production-scale of tons per year of chondroitin sulfate.
In order to investigate the increased availability of the antiterminator at the transcriptional level, the expression of rfaH in EcK4r3 was investigated through qPCR to analyse the impact of the additional pgapA regulated copy/ies on its expression. In particular the P1 site of the multipromoter system was used in this study [26]. As expected a significative increase of the rfaH mRNA pool was observed throughout growth with a peak after 24h. Stevens and coworkers [16] demonstrated that RfaH does not directly increase the initiation of region 2 transcripts whereas it supports their activity to obtain adequate levels of expression of the genes that are required to synthesize the K5 capsular polysaccharide. We analysed the expression of two genes belonging to region 2, namely kfoC and kfoA. Both genes were positively affected by rfaH overexpression without significative differences among the two transcripts. RfaH regulates bacterial operons engaged in the production of also other extracellular components involved in the virulence of pathogens such as the LPS, α-haemolysin toxin and F pilus. In fact, deletion of rfaH abolished the virulence of uropathogenic E. coli due to downregulation of several virulence factors [22]. To further deepen the study, production of cell-bound and released LPS in EcK4r3 was also investigated. The same CPS to LPS ratio observed for the wild type strain (2:1) is conserved in EcK4r3, although the latter demonstrates a 1.5/1.6-fold increase of both the cell-bound and released LPS portions, in accordance with the increment of K4 CPS production. These results clearly demonstrate the broader effect of rfaH overexpression.
A higher production of capsular polysaccharide, as in EcK4r3, does not necessarily imply a variation in the concentration of intracellular pools of precursor UDP-sugars; the former could in fact be due to a higher flux through the pool. However, it was interesting to find that rfaH overexpression affected both concentration values and time course profiles of UDP-Glu, UDP-GalNAc and UPD-GlcA, analysed in this work. The major difference, compared to the wt strain, regarded UDP-Glu. Wu and co-workers [27] cultivating Lactobacillus casei found an increase in the abundance of enzymes responsible for the biosynthesis of UDP-Glu and of precursor sugar-nucleotides for the synthesis of exopolysaccharides, in the early stationary phase. A similar trend was observed for the wt E. coli K4, whereas in EcK4r3 not only the intracellular concentration of UDP-Glu decreased towards the end of growth, but a 20-fold higher concentration compared to the wt, was observed when growth slowed down (5h). kfoF, belongs to region 2 and codes for a UDP-glucose dehydrogenase that converts UDP-Glu into UDP-GlcA. The induction of the gene, due to rfaH overexpression, might have increased the need for UDP-Glu that is an essential intermediate also for other biosynthesis processes (eg. Biosynthesis of cell wall components), so that the metabolism responded by increasing the available pool of this substrate. In both strains UDP-GlcA and UDP-GalNAc exhibited the same trends over time, however a lower intracellular availability of both sugar-nucleotides, that is most pronounced after 5h of growth, was observed in EcK4r3 compared to the wild type which might mirror the higher polysaccharide production rate of the recombinant strain. Overall rfaH overexpression seems to affect the intracellular concentration of UDP-sugar precursors further supporting an extensive metabolic response at least regarding the pathways involved in polysaccharide biosynthesis.
Conclusions
Overall in the present work a new engineered chondroitin production host was constructed by introducing additional copies of the rfaH gene in the unconventional strain E. coli K4. Homologous overexpression of the gene not only improved CPS production in shake flasks but also increased the final titer of polysaccharide in fermenter experiments in batch and fed-batch mode, thereby proving the possibility of achieving stable integrations in this peculiar capsulated strain. Furthermore the construct developed in this work does not require induction by IPTG or selection pressure (e.g. antibiotics) during the cultivation which makes the foreseen production process cheaper and reduces the impact on the environment. This work also shows that the know how acquired with studies carried on the wild type, and the fermentation processes developed, can be easily transferred to genetically engineered strains simplifying the approach towards the set up of large scale biotech production processes. A global overview of the results obtained in the present work is depicted in Figure 5.
Figure 5.
General overview on the effect of inserting additional copies of rfaH in E.coli K4. It shows that overexpression of rfaH leads to higher production of capsular polysaccharide that is probably due to an increase of expression of the genes belonging to region 2; in particular the mRNA levels of kfoA and kfoC were analysed in this study. The figure also indicates that rfaH overexpression increases lipopolysaccharide production.
Materials and Methods
Medium
The standard medium used for all shake flask and fermenter production studies consisted of a defined salts medium (KH2PO4 2 g∙L-1; K2HPO4 9.7 g∙L-1; Na3C6H5O7 0.5 g∙L-1; (NH4)2SO4 1 g∙L-1; MgCl2 0.1 g∙L-1) supplemented with glycerol or glucose (10 g∙L-1) as the main carbon source and neutralised soy peptone (1 g∙L-1) or yeast extract (2 g∙L-1), respectively, as additional nitrogen source (Oxoid). Luria-Bertani (LB) medium was used during transformation experiments. Transformed cells were grown on LB medium supplemented with ampicillin or kanamycin (50 μg∙mL-1). The wild type strain used in all experiments was E. coli K4 serotype O5:K4:H4 (CCUG 11307), purchased from the Culture Collection University of Göteborg.
Materials
Genomic DNA, plasmid DNA, and RNA were isolated using Qiagen DNeasy kit, Qiagen miniprep kit, Qiagen RNeasy kit (Qiagen, Valencia, CA) respectively according to the manufacturer’s instructions. Restriction endonuclease digestions, DNA ligations, SDS-PAGE and agarose gel electrophoresis were performed using standard techniques [28].
Construction of the rfaH overexpressing strains
Primers used for the construction of strains described in this study are reported in Table 4. Amplification of the rfaH gene and of the P1 gapA promoter for the construction of all strains were performed from E. coli K4 chromosomal DNA. Primers for the amplification of the gene and of the promoter were designed based on the sequence of rfaH and pgapA of E. coli K12 present in the database. Polymerase chain reaction (PCR) was carried out with Expand High fidelity PCR System (Roche, Monza, Italy) according to the manufacturer’s protocol. DNA fragments were recovered from agarose gels using the Qiaquick gel extraction kit (Qiagen, Valencia, CA). Restriction endonucleases were purchased from New England Biolabs and ligases were purchased from Invitrogen (Carlsband, CA). Nucleotide sequencing of all PCR fragments cloned was carried out at BMR Genomics (Padova, Italy) to verify that the sequences were correct.
Table 4.
Sequences of the primers used for constructing strains E.coli K4-pTrc rfaH , EcK4r1 and EcK4r3, and for real time PCR experiments
| Primer | Sequence |
|---|---|
|
rfaHpTrc1 |
5′-TAAAGGAGGTATACAAAAATGCAATCCTGGTATTTACTGTAC-3′ |
|
rfaHpTrc2 |
5′-TTAGAGTTTGCGGAACTCGGTAT-3′ |
|
16Sq_Up |
5′- GGTGTAGCGGTGAAATGCGTAGAG-3′ |
|
16Sq_Dw |
5′- CAAGGGCACAACCTCCAAGTC-3′ |
|
qkfoC_Up |
5′-TTTTCCCTGCCGCACGATCC-3′ |
|
qkfoC_Dw |
5′-TTCGGTCTGTTGAAGGAGCAATGG-3′ |
|
qkfoA_Up |
5′-GGATTTAGCGGAAGGGCATGTG-3′ |
|
qkfoA_Dw |
5′-TTCTGGTGATGACCAACTTTCAGC-3′ |
|
P1=pK4EC05_Up |
5′-CAGCACAGCAGAGCGAAGTGCATCATATCCTTCCAGATTTAAATTCTTCAAATTAACCCTCACTAAAGGGCG-3′ |
|
P2=pk4EC05_Dw |
5′-AAACTGTGATCGGGCGTAGGAACCCGCGTAGTCATCGTCGGCGCAGAAGTTTAGAGTTTGCGGAACTCGGTAT-3′ |
|
P3=LacZ_Up |
5′-CACCCTGGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGAAATTAACCCTCACTAAAGGGCGG-3′ |
|
P4=LacZ_Dw |
5′- AAAAGAATAAACCGAACATCCAAAAGTTTGTGTTTTTTAAATAGTACATAATGGATTTCCTTACGCGAAATACGGGCAGACATGGCCTGCCCGGTTATTTAGAGTTTGCGGAACTCGGTATTC-3′ |
|
P5=FRT_Dw |
5′-TAATACGACTCACTATAGGGCT-3′ |
|
P6=pGapA_Up |
5′-CCCTCGAGGGCCTTTAAAATTCGGGGCGCCGA-3′ |
|
P7=pGapA_Dw |
5′-CGGGATCCCGATATTCCACCAGCTATTTGTTAG-3′ |
| P8=rfaH_Up | 5′-CGGGATCCCGATGCAATCCTGGATTTTACTGTAC-3′ |
Strain E. coli K4-pTrcrfaH
rfaH was TA-cloned into pTrcHis (Invitrogen) not in frame with the N-terminal his tag contained in the vector. The forward primer contained a stop codon and a new ribosome binding site. Primers used for the amplification are rfaHpTrc1 and rfaHpTrc2 (Table 4).
Strain EcK4r1- EcK4r3
The recombinant strains EcK4r1 and EcK4r3 were constructed using the Gene deletion kit (Gene Bridges). The provided integration cassette was modified by the addition of the rfaH gene under the control of the gapA promoter just downstream of the kanamycin gene (Figure 6). E. coli K4 wt chromosomal DNA was used as template for the amplification of the glyceraldehyde-3-phosphate dehydrogenase promoter and the rfaH gene. All primers used for strain construction are reported in Table 4. A scheme of the resulting cassette is depicted in Figure 6. The pK4EC05_Up and pK4EC05_Dw oligos contain a 50bp region similar to the 5' and 3′portion of the PK4EC05 plasmid, respectively, to target the recombination event for the construction of strain EcK4r1.
Figure 6.
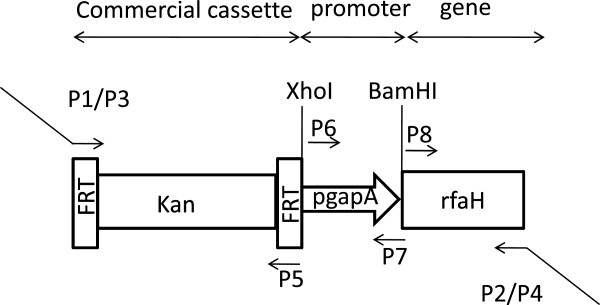
Schematic representation of the integration cassette used for the construction of EcK4r1 and EcK4r3.
The cassette used for the achievement of strain EcK4r3 was amplified with primers LacZ_Up and LacZ_Dw containing 100bp region similar to the 5' and 3′portion of the E. coli K4 genome, respectively, to target the recombination event.
Transformation
For the construction of all strains E. coli K4 electrocompetent cells were prepared according to the instrument instructions and transformed through electroporation using a Bio-Rad Gene Pulser (2mm cuvettes, 2.5 kV, 200 Ω, 25 μF). E. coli K4 was transformed with the vector pTrc-rfaH and plated on LB plates supplemented with 50 μg·mL-1 ampicillin to select for positive clones. Strains EcK4r1 and EcK4r3 were obtained following the instructions of the kit.
Strain characterization
The Minimum Inhibitory Concentration (MIC) of antibiotics, antifungal and antimycobacterial agents was established by performing the Etest® (BioMerieux Italia S.p.a.). Etest consists of a predefined gradient of antibiotic concentrations on a plastic strip and it was used to compare E. coli K4 and EcK4r3. The analysis was performed by the group of Prof. Donnarumma at the Department of Microbiology, Second University of Naples.
Gene expression
Quantitative real-time PCR was performed on E. coli K4 wild type and EcK4r3 recombinant strains to compare the mRNA levels of the genes of interest.
Three time points were chosen for analysis corresponding to 3, 8 and 24h of growth. RNA was treated with DNase using the DNA-free kit following the supplied protocol (Ambion Inc Austin, TX) and 500 ng were used for reverse transcription with the Reverse Transcription System (Promega, Madison, USA). PCR amplification was performed in an iQ5 instrument (Biorad California, USA). The 16S rRNA was used to normalise expression of rfaH, kfoC and kfoA genes. The primers used for the experiment are reported in Table 4.
Amplification was carried out in 25 μL containing 5 μL cDNA, 12.5 μL of iQ Syber Green Supermix (Biorad, California, USA) and 0.5 μL of each primer at a concentration of 0.4 μM. After incubation at 95°C for 3 min, amplification proceeded with 40 cycles of 95°C for 10 s and 62°C for 1 min. The efficiencies of the primer sets were measured using a dilution series of cDNA. The raw threshold cycle (Ct) values were converted to relative expression levels by the 2(-ΔΔCt) method [29]. Three biological samples each in triplicate were analysed for every timepoint in the wild type and recombinant strains.
Shake flask experiments
Shake flask experiments were performed in order to evaluate the effect of rfaH gene overexpression on the production of K4 polysaccharide. Before each experiment cells from 20% (w/v) glycerol stock preparations were streaked on agar plates and grown overnight (o/n) at 37°C. Single colonies were then used to inoculate pre-cultures that were incubated o/n at 37°C in shaking conditions.
The medium used for all shake flask production studies is described in the medium section. For strain E. coli K4-pTrcrfaH 100 μg∙L-1 ampicillin were supplemented before each experiment.
For all experiments 200 mL cultures of wild type and recombinant E. coli K4 were grown in 1 liter baffled flasks, keeping a 1:5 medium/air volume ratio, at 37°C and 200 rpm in a rotary shaker incubator (model Minitron, Infors, Bottmingen, Switzerland). For the strain E. coli K4-pTrcrfaH the cultures were grown until the OD600 reached 0.7 and at that point 0.4 or 1 mM β-isopropylthiogalactoside (IPTG) was added to the broth.
Strains EcK4r1 and EcK4r3 were grown for 24h without the addition of antibiotic and inducer. Samples were withdrawn during the course of the experiment to analyse polysaccharide production. Every shake flask experiment was repeated at least four times.
Fermentation experiments
Fermentation experiments were carried out as biological triplicates in a Biostat CT reactor (Sartorius Stedim; Melsungen, Germany), 2 L working volume. A constant pH of 7.5 was maintained via automated addition of 30% v/v NH4OH and 30% v/v H2SO4.
Seed cultures were prepared by inoculating a single colony in 200 mL of medium in 1L baffled shake flasks. The flasks were incubated overnight at 37°C and 200 rpm. For the duration of all cultivations 5 mL samples were withdrawn from the reactors at regular time intervals for the determination of substrates, extracellular metabolites and polysaccharide quantification.
For batch experiments the concentration of dissolved oxygen (DO) was maintained above 20% varying the air flow rate (1–1.5 L∙min-1) and stirring rate (200–800 rpm) according to oxygen demand. The media used for all experiments were that described in the medium section. For the strain E. coli K4-pTrcrfaH 100 μg∙L-1 ampicillin were added to the medium before each experiment and the cultures were grown until the OD600 reached 0.7 and at that point 0.4 or 1 mM β-isopropylthiogalactoside (IPTG) was added to the broth.
For fed-batch experiments cells were grown in the semidefined medium containing glucose and yeast extract. A DO-Stat controlled feeding strategy with a pO2 set point equal to 35% of air saturation was used. After the batch phase, a concentrated nutrient solution (450 g∙L-1 glucose and 90 g∙L-1 yeast extract, inorganic salts 20-fold concentrated) was fed to the culture in the following 42.5h of growth.
Extraction of the LPS
The biomass obtained from fermenter experiments was suspended in 90% v/v pre-warmed phenol. A volume of pre-warmed water was then added to the suspension and incubated for 30 minutes at 68°C in stirred conditions.
The solution was then centrifuged at 9000xg for 30 minutes at 4°C and the upper phase containing the LPS was extracted and preserved. This procedure was repeated for three times. The three fractions recovered were dialyzed against water (6000–8000 Da membranes) for two days. The upper phase was further treated for the quantification of the O-chain. Hydrolysis was performed adding 1% v/v acetic acid to the solution that was then incubated for 1.5 h at 100°C. The sample was then centrifuged at 9000xg, at 4°C for 30 minutes and the supernatant was analyzed through capillary electrophoresis as described in the following section (Quantification of CPS, LPS and UDP-sugars)
UDP-sugar extraction
UDP-sugars were extracted from biomass samples by slightly modifying the method reported by Maharjan and Ferenci [30]. Briefly, broth samples corresponding to about 0.8 mgcdw were cooled at 0°C in an ice-water bath and immediately centrifuged at 16000xg and 0°C for 5 minutes. Pellets were resuspended in 1mL of 50% v/v methanol and incubated at 70°C for 30 minutes. The supernatant was recovered by centrifugation and concentrated in a vacuum centrifuge to 70 μl for capillary electrophoresis analysis as described in the following section.
Hydrolisis of the K4 CPS
The K4 CPS produced in shake flask cultivations was used for an experiment of chemical hydrolysis. The polysaccharide (216 mg∙L-1) was incubated in the standard growth medium at pH = 5 and pH = 6 for 16 h. The temperature was kept constant at 37°C and agitation was set to 200 rpm. Samples were collected after 3, 6 and 16h of incubation and analysed by capillary electrophoresis.
Quantification of CPS, LPS and UDP-sugars
Broth samples were collected from shake flask and fermentation experiments and centrifuged at 1700xg. Supernatants were then ultrafiltered on 10 KDa centrifugal filter devices (YM-10 Centricon, Millipore, Bedford, MA, USA) at 5000xg and concentrated to 1/10th of their initial volume. The retentate was twice diafiltered and then analyzed for the content of K4 polysaccharide and LPS.
Capillary electrophoresis analysis were performed on a Beckman-Coulter HPCE instrument (P/ACE MDQ, Palo Alto, CA, USA) equipped with a diode array detector and a UV lamp, using an uncoated fused-silica tube (70 cm of total length, 60 cm of effective length, 50 μm I.D.) at 25°C. Separation and quantification of the K4 and K4 defructosylated polysaccharides and of the O-chain and LPS were performed according to the previously described method [31]. The same equipment was used for the determination of UDP-sugar concentrations by slightly modifying the method reported by Xu and colleagues [32]. In particular a buffer containing sodium phosphate 50mM, SDS 60mM and sodium borate 20mM, pH 9 was used for analysing the samples. Runs were performed at 18°C and 22 kV. UDP-Glu, UDP-GalNAc and UDP-GlcA were detected at λ = 200 nm. The concentration of standards used for the construction of calibration curves ranged between 0.013 to 1.7 mM for UDP-Glu and UDP-GalNAc, and from 0.026 to 1.7 mM for UDP-GlcA. Due to the complexity of the samples co-elutions with the pure standards were also performed to ascertain the identity of the different precursors.
Abbreviations
CPS: Capsular polysaccharide; DO: Dissolved oxygen; GAGs: Glycosaminoglycans; CS: Chondroitin sulfate; HA: Hyaluronic acid; wt: Wild type
Competing interests
The authors declare no competing interests.
Authors’ contribution
DC wrote the manuscript; DC, EC and AR performed experiments; DC, EC, MDR, AR and CS analysed the data; DC and CS conceived the work; all authors read and approved the manuscript.
Supplementary Material
Sequence of rfaH in E. coli K4.
Contributor Information
Donatella Cimini, Email: donatella.cimini@unina2.it.
Mario De Rosa, Email: mario.derosa@unina2.it.
Elisabetta Carlino, Email: elisabettacarlino85@yahoo.it.
Alessandro Ruggiero, Email: alessandro.ruggiero.79@gmail.com.
Chiara Schiraldi, Email: chiara.schiraldi@unina2.it.
Acknowledgements
The construction of strains EcK4r1 and EcK4r3 is described in the patent application PCT/EP2010/057129: Biotechnological production of chondroitin. We kindly thank Dr Odile F. Restaino and Dr Irene di Lauro for the help provided during analysis of polysaccharide samples and UDP-sugar determinations and Dr Loredana Carcarino for helpful discussions. We kindly thank Prof. Giovanna Donnarumma for providing data regarding the E-test.
References
- Volpi N. Anti-inflammatory activity of chondroitin sulphate: new functions from an old natural macromolecule. Inflammopharmacology. 2011;19(6):299–306. doi: 10.1007/s10787-011-0098-0. [DOI] [PubMed] [Google Scholar]
- Imada K, Oka H, Kawasaki D, Miura N, Sato T, Ito A. Anti-arthritic action mechanisms of natural chondroitin sulfate in humanarticular chondrocytes and synovial fibroblasts. Biol Pharm Bull. 2010;33(3):410–414. doi: 10.1248/bpb.33.410. [DOI] [PubMed] [Google Scholar]
- Achur RN, Kakizaki I, Goel S, Kojima K, Madhunapantula SV, Goyal A, Ohta M, Kumar S, Takagaki K, Gowda DC. Structural interactions in chondroitin 4-sulfate mediated adherence of Plasmodium falciparum infected erythrocytes in human placenta during pregnancy-associated Malaria. Biochemistry. 2008;47(47):12635–12643. doi: 10.1021/bi801643m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TW, Sun JS, Wu HC, Tsuang YH, Wang WH, Lin FH. The effect of gelatin-chondroitin sulfate-hyaluronic acid skin substitute on wound healing in SCID mice. Biomaterials. 2006;27(33):5689–5697. doi: 10.1016/j.biomaterials.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Pothacharoen P, Siriaunkgul S, Ong-Chai S, Supabandhu J, Kumja P, Wanaphirak C, Sugahara K, Hardingham T, Kongtawelert P. Raised serum chondroitin sulfate epitope level in ovarian epithelial cancer. J Biochem. 2006;140(4):517–524. doi: 10.1093/jb/mvj181. [DOI] [PubMed] [Google Scholar]
- Cimini D, De Rosa M, Schiraldi C. Production of glucuronic acid-based polysaccharides by microbial fermentation for biomedical applications. Biotechnol J. 2012;7(2):237–250. doi: 10.1002/biot.201100242. [DOI] [PubMed] [Google Scholar]
- Schiraldi C, Alfano A, Cimini D, De Rosa M, Panariello A, Restaino OF, De Rosa M. Application of a 22L scale membrane bioreactor and cross-flow ultrafiltration to obtain purified chondroitin. Biotechnol Prog. 2012;28(4):1012–1018. doi: 10.1002/btpr.1566. [DOI] [PubMed] [Google Scholar]
- Bedini E, De Castro C, De Rosa M, Di Nola A, Iadonisi A, Restaino OF, Schiraldi C, Parrilli M. A microbiological-chemical strategy to produce chondroitin sulfate A, C. Angew Chem Int Ed Engl. 2011;50(27):6160–6163. doi: 10.1002/anie.201101142. [DOI] [PubMed] [Google Scholar]
- Cimini D, De Rosa M, Viggiani A, Restaino OF, Carlino E, Schiraldi C. Improved fructosylated chondroitin production by kfoC overexpression in E. coli K4. J Biotechnol. 2010;150:324–331. doi: 10.1016/j.jbiotec.2010.09.954. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Roberts IS. Structure assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- Roberts IS, Mountford R, Hodge R, Jann KB, Boulnouis GJ. Common organization of gene clusters for production of different capsular polysaccharides (K antigens) in Escherichia coli. J Bacteriol. 1988;170:1305–1310. doi: 10.1128/jb.170.3.1305-1310.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AN, Boulnois GJ, Roberts IS. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990;4:1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- Trilli A, Busiello I, Daly S, Bagatin F. Biotechnological production of chondroitin. USP Application. 2011.
- Ninomiya T, Sugiura N, Tawada A, Sugimoto K, Watanabe H, Kimata K. Molecular cloning and characterization of chondroitin polymerase from Escherichia coli strain K4. J Biol Chem. 2002;277:21567–21575. doi: 10.1074/jbc.M201719200. [DOI] [PubMed] [Google Scholar]
- Suzuki K. Chondroitin producing bacterium and method of producing chondroitin. PCT WO 2008/13350 A1. 2008.
- Stevens MP, Clarke BR, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol Microbiol. 1997;24:1001–1012. doi: 10.1046/j.1365-2958.1997.4241780.x. [DOI] [PubMed] [Google Scholar]
- Marolda CL, Valvano MA. Promoter region of Escherichia coli O7 specific lipopolysaccharide gene cluster: structural and functional characterization of an upstream untranslated mRNA sequence. J Bacteriol. 1998;180:3070–3079. doi: 10.1128/jb.180.12.3070-3079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S, Hodson N, Griffiths G, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster: Evidence for the roles of H-NS, Bip-A and Integration host factor in regulation of group 2 capsule gene clusters in Pathogenic E. coli. J Bacteriol. 2000;182(10):2741–2745. doi: 10.1128/JB.182.10.2741-2745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Restaino OF, Catapano A, De Rosa M, Schiraldi C. Production of capsular polysaccharide from Escherichia coli K4 for biotechnological applications. Appl Microbiol Biotechnol. 2010;85(6):1779–1787. doi: 10.1007/s00253-009-2261-8. [DOI] [PubMed] [Google Scholar]
- De Angelis PL. Glycosaminoglycan polysaccharide biosynthesis and production: today and tomorrow. App Microbiol Biotechnol. 2012;94(2):295–305. doi: 10.1007/s00253-011-3801-6. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Hänfling P, Jann B, Jann K, Roberts IS. Regulation of Escherichia coli K5 capsular polysaccharide expression: evidence for involvement of RfaH in the expression of group II capsules. FEMS Microbiol Lett. 1994;124(1):93–98. doi: 10.1111/j.1574-6968.1994.tb07267.x. [DOI] [PubMed] [Google Scholar]
- Nagy G, Dobrindt U, Schneider G, Salam Khan A, Hacker J, Emodi L. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect Immun. 2002;70(8):4406–4413. doi: 10.1128/IAI.70.8.4406-4413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JH, Song JM, Kang JH, Kang DJ. Optimization of medium components for highmolecular-weight hyaluronic acid production by Streptococcus sp. ID9102 via a statistical approach. J Ind Microbiol Biotechnol. 2009;36:1337–1344. doi: 10.1007/s10295-009-0618-8. [DOI] [PubMed] [Google Scholar]
- Rangaswamy V, Jain D. An efficient process for production and purification of hyaluronic acid from Streptococcus equi subsp. zooepidemicus. Biotechnol Lett. 2008;30:493–496. doi: 10.1007/s10529-007-9562-8. [DOI] [PubMed] [Google Scholar]
- Restaino OF, Cimini D, De Rosa M, Catapano A, De Rosa M, Schiraldi C. High cell density cultivation of Escherichia coli K4 in a microfiltration bioreactor: a step towards improvement of chondroitin precursor production. Microb Cell Fact. 2011;16:1–10. doi: 10.1186/1475-2859-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B, Branlant C. The Escherichia coli gapA gene is transcribed by the vegetative RNA polymerase holoenzyme E sigma 70 and by the heat shock RNA polymerase E sigma 32. Bacteriol. 1994;176(3):830–839. doi: 10.1128/jb.176.3.830-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wang W, Yu D, Zhang W, Li Y, Sun Z, Wu J, Meng H, Zhang H. Proteomics analysis of Lactobacillus casei Zhang, a new probiotic bacterium isolated from traditional home-made koumiss in inner Mongolia of China. Mol Cell Proteomics. 2009;8(10):2321–2338. doi: 10.1074/mcp.M800483-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning. A Laboratory Manual. New York: Cold Spring Harbour Laboratory Press; 2001. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maharjan RM, Ferenci T. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem. 2003;313:145–154. doi: 10.1016/S0003-2697(02)00536-5. [DOI] [PubMed] [Google Scholar]
- Restaino OF, Cimini D, De Rosa M, De Castro C, Parrilli M, Schiraldi C. High-performance CE of Escherichia coli K4 surface polysaccharides. Electrophoresis. 2009;30(22):3877–3883. doi: 10.1002/elps.200900279. [DOI] [PubMed] [Google Scholar]
- Xu G, Lehmann R, Schleicher E, Häring HU, Liebich H. Advantages in the analysis of UDP-sugars by capillary electrophoresis-comparison of the conventional HPLC method with two new capillary electrophoretic micro-procedures. Biomed Chromatogr. 1998;12(3):113–115. doi: 10.1002/(SICI)1099-0801(199805/06)12:3<113::AID-BMC772>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of rfaH in E. coli K4.