Abstract
Rice white tip nematode, Aphelenchoides besseyi, is a kind of plant parasitic nematodes that cause serious losses in rice and many other crops. Fatty acid and retinoid binding protein (FAR) is a specific protein in nematodes and is related to development, reproduction, infection to the host, and disruption of plant defense reactions, so the inhibition of FAR function is the potential approach to control A. besseyi. The full-length of Ab-far-1 cDNA is 805 bp, including 546 bp of ORF that encodes 181 amino acids. Software analysis revealed that the Ab-FAR-1 was rich in α-helix structure, contained a predicted consensus casein kinase II phosphorylation site and a hydrophobic secretory signal peptide, but did not have glycosylation sites. The Ab-FAR-1 had 52% homology to Gp-FAR-1 protein. The Ab-FAR-1 and Gp-FAR-1 were grouped in the same branch according to the phylogenetic tree. Fluorescence-based ligand binding analysis confirmed that the recombinant Ab-FAR-1 (rAb-FAR-1) bound with the fluorescent analogues 11-((5-dimethylaminonaphthalene-1-sulfonyl) amino) undecannoic acid (DAUDA), cis-parinaric acid and retinol, but the oleic acid would compete with the binding site. Quantitative PCR (qPCR) was used to assess the expression level of Ab-far-1 at different development stages of A. besseyi, the highest expression was found in the females, followed by eggs, juveniles and males. Using in situ hybridization technique, Ab-far-1 mRNA was present in the hypodermis of juveniles and adults, the ovaries of females and the testes of males. When A. besseyi was treated with Ab-far-1 dsRNA for 48 h, the silencing efficiency of Ab-far-1 was the best and the number of nematodes on the carrot was the least. Thus FAR plays important roles in the development and reproduction of nematodes. This study is useful and helpful to figure out a new way to control the plant parasitic nematodes.
Introduction
Rice white tip nematode, Aphelenchoides besseyi, is a kind of foliar nematodes that feed ecto- or endoparasitically above-ground parts of plant, it can parasite in more than 200 kinds of plants in 35 genera. Rice (Oryza sativa) and strawberry (Fragaria ananasa) are the most common hosts [1], [2]. A. besseyi is widely distributed and occurs in most rice growing areas of the world, and rice yield is reduced by 10%–71% in the occurred paddies [3]. At present, chemical seed treatment and soil application are the main approaches to control A. besseyi. But there are some risks of damaging the germination after treatment. On the other hand, chemical nematicide has not been recommended due to its high toxicity.
With the development of molecular biology, the genetic engineering technology has been applied in the genome research of plant parasitic nematodes. The molecular methods are widely used to study the effective and safe way to prevent plant parasitic nematodes. Therefore, the studies on the genes that are related to the life activity and infection mechanism of plant parasitic nematodes are very important.
In the processes of development and infection to the host, plant parasitic nematodes require fatty acids and retinol for lipid biosynthesis and assembly of macromolecular structures, but they are unable to synthesize fatty acids and retinol by themselves. To sustain their life activities, plant parasitic nematodes have to obtain these metabolites from their hosts and the environment through the lipid binding proteins (LBPs) [4], [5]. Nematodes have been found to produce a series of unusual proteins that exhibit high affinity binding to lipid, and these proteins can be divided into two different classes according to their molecular weight and structure features: polyprotein allergens/antigens (NPAs) and fatty acid and retinoid binding proteins (FARs) [6], [7]. FAR proteins which are rich in helix structure have different structure from proteins of similar biochemical function from other organism groups. FARs promote the absorption, transportation and specific localization of fatty acid and retinoid [4], [8]. Retinol plays important roles in gene activation, cell signaling, and tissue differentiation and repairation [5], [7], [8]. Thus the secreted FARs can help nematodes not only to obtain the lipid nutrition from the host, but also to infect the host and inhibit the host defense mechanism. The first identified FAR protein was the Ov-FAR-1 (formerly known as Ov20) from Onchocerca volvulus [9]. Ov-FAR-1 can promote the infection by reducing the defense of host, and cause worse damage to the host [9], [10]. Eight FARs have been identified in the model nematode Caenorhabditis elegans, named Ce-far-1 to 8 [7]. FARs have also been found in animal parasitic nematodes, including Ancylostoma caninum, Brugia malayi, Brugia pahangi, Ancylostoma ceylanicum, Acanthocheilonema viteae, Ascaris suum, Heligmosomoides polygyrus, Onchocerca gutturosa, Onchocerca dukei, Onchocerca ochengi, Loa loa, Wuchereria bancrofti, and Litomosoides sigmodontis [5], [8], [11], [12], [13], [14]. In plant parasitic nematodes, however, only one FAR from Globodera pallida (Gp-FAR-1) has been identified, which is present in the hypodermis of second-stage juvenile and is related to the defense system of plant host at the initial infection stage [15].
FAR plays a critical role in the development and infection processes of plant parasitic nematodes [15]. As an effective target, FAR has been attracted much attention to control plant parasitic nematodes. According to the expressed sequence tag (EST) of FAR gene from cDNA library of A. besseyi, we amplified the full-length cDNA of FAR gene from A. besseyi (Ab-far-1) and analyzed the structure and feature of Ab-far-1 gene with the bioinformatic method. We used fluorescence-based binding assays to investigate the binding activity of bacterial recombinant Ab-FAR-1 to fatty acid and retinol. We localized the Ab-far-1 mRNA at different development stages by in situ hybridization and detected the expression level of Ab-far-1 by quantitative PCR (qPCR). In addition, the function of Ab-far-1 gene was investigated by RNA interference (RNAi) approach. This is the first study to identify and analyze the Ab-far-1 gene, and to investigate its function using RNAi technique.
Materials and Methods
Ethics statement
We collected the nematodes in areas where rice white tip nematodes occurred and no specific permit was required. The field for nematodes collection was neither privately owned nor protected, and did not involve endangered or protected species.
Nematodes
A. besseyi used in this study was collected from the leaves of infected O. sativa in Nanjing City, Jiangsu Province, China where A. besseyi occurred and identified by laboratory of plant nematology, South China Agricultural University. A. besseyi was preserved and cultured on excised carrot (Daucus carota) disks in Petri dishes at 25°C in dark incubator [3], [16].
Nematode extraction
The carrot callus inoculated with A. besseyi for 30 days was mashed with a blender. The mashed solution was filtered through the combine sieves with aperture size of 0.147 mm and 0.026 mm. Nematodes were collected from 0.026 mm aperture sieve in a beaker and Petri dishes.
Cloning of full-length FAR gene from A. besseyi
Total RNA of 20,000 mixed stages nematodes was extracted with TRIzol reagent (Invitrogen, Carlbard, CA, USA), then treated with RQ1 RNase-Free DNase (Promega, Madison, WI, USA) at 37°C for 15 min. The cDNA sequences were amplified by a SMART RACE cDNA Amplification kit (Clontech, Japan). According to the EST sequences of A. besseyi FAR gene that was screened from the cDNA library of A. besseyi in our lab (unpublished, Fig. S1), 5' RACE primers (FAR-R1 and FAR-R2) and 3' RACE primers (FAR-F1and FAR-F2) (Table 1) were designed to amplify the cDNA sequence. The amplification products were purified and ligated into the pMD 18-T vector (Takara, Japan), and then transformed into Escherichia coli JM109 competent cells. The positive clones were sent to BGI Company for sequencing. Based on the sequencing results of the 5' and 3' RACE products, the specific primers of QCFF and QCFR (Table 1) were designed and used to amplify the full-length cDNA of A. besseyi FAR gene.
Table 1. Primers used in this study.
| Primer | Sequence | Source |
| FAR-F1 | 5′-GAGACTTTCCCGCAAATCACC-3′ | |
| FAR-F2 | 5′- ATCGACACCTACAAGAAATTGGCTG -3′ | |
| FAR-R1 | 5′- GCAGATGATTGTGACCGTTTAGTTC -3′ | |
| FAR-R2 | 5′- GATTGAAGCAGATGATTGTGACCGT -3′ | |
| QCFF | 5′- CGTTGTTTCAGTAACTCACATCTTT -3′ | |
| QCFR | 5′- GAACATACGAATACAACTAAAAGGA -3′ | |
| FARf-BamHI | 5′-GAGAGAGGATCCAGCCTTCGTACCTTCGTTGT-3′ | |
| FARr-XhoI | 5′-GAGAGACTCGAG GCAGATGATTGTGACCGTTTA-3′ | |
| 18sF | 5′- CTCGTGGTGGCTGGTATGCTG -3′ | |
| 18sR | 5′- GTTTCCCGTGTTGAGTCAAATTAAG -3′ | |
| qPCR-F | 5′- TCGCTCTTCTGTCTTGCCATG -3′ | |
| qPCR-R | 5′- GATGGATTTGTCTTCATCGGTA -3′ | |
| FAR-IN-T7S1 | 5′-TAATACGACTCACTATAGGGGCCTTCGTACCTTCGTTGTCTTG -3′ | |
| FAR-IN-A1 | 5′- GTTCTGGCACTTGTTGAATGCTC -3′ | |
| FAR-IN-T7A1 | 5′-TAATACGACTCACTATAGGGGTTCTGGCACTTGTTGAATGCTC-3′ | |
| FAR-IN-S1 | 5′- GCCTTCGTACCTTCGTTGTCTTG -3′ | |
| FARRiF | 5′- TACGCCGAACTTACCGATGAA -3′ | |
| FARRiT7R | 5′-TAATACGACTCACTATAGGGCAAGTCTGGCTTCTCTTCACCC -3′ | |
| FARRiT7F | 5′-TAATACGACTCACTATAGGGTACGCCGAACTTACCGATGAA -3′ | |
| FARRiR | 5′- CAAGTCTGGCTTCTCTTCACCC -3′ | |
| G-T7S | 5′-GGATCCTAATACGACTCACTATAGGGCACAAGTTCAGCGTGTCCGGCG-3 -3′ | Zhang et al., 2012 |
| G-A | 5′- CGATGCGGTTCACCAGGGTGTCG -3′ | Zhang et al., 2012 |
| G-T7A | 5′-GGATCCTAATACGACTCACTATAGGG CGATGCGGTTCACCAGGGTGTCG -3′ | Zhang et al., 2012 |
| G-S | 5′- CACAAGTTCAGCGTGTCCGGCG -3′ | Zhang et al., 2012 |
Sequence analysis, alignment and phylogenetic studies
Sequence homology comparisons were conducted using BLASTX and BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The protein transmembrane regions, theoretical isoelectric point, molecular weight, hydrophobicity and glycosylation sites were predicted by the Protein Machine software available at the Expasy site (http://www.expasy.ch/tools/). Predictions of a signal peptide for secretion and the cleavage site were performed at http://www.cbs.dtu.dk/services/SignalP/. The prediction of protein localization site was performed at http://psort.hgc.jp/form2.html. A theoretical 3D structure of Ab-FAR-1 was constructed with the automated mode of SWISS-MODEL. The amino acid sequences of FAR protein from A. besseyi and other six FAR proteins from four species of nematodes [17] were aligned using DNAMAN software (Lynnon Biosoft, Canada). Based on the amino acid sequences of Ab-FAR-1 and other 25 FAR proteins from 16 species of nematodes, a phylogenetic tree was constructed by the neighbor-joining method [18] with MEGA (Molecular Evolutionary Genetics Analysis, USA).
Expression and purification of recombinant Ab-FAR-1
To obtain purified Ab-FAR-1 protein,the full-length Ab-far-1 was amplified from the plasmid with primers FARf-BamHI and FARr-XhoI (Table 1), then cloned into prokaryotic expression vector pET-32a (Novagen, Madison, WI, USA) after digestion with BamHI and XhoI. The plasmid was introduced into E. coli DH5α for sequence confirmation. Recombinant plasmid DNA was introduced into E. coli BL21(DE3)for expression. Expression of the recombinant protein was examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining after treatment with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG). The recombinant fusion Ab-FAR-1 protein with His-tag at the N-terminus was purified by affinity chromatography using Ni Sepharose High Performanc (GE Healthcare, Sweden) according to the manufacturer's instructions. The purity of purified recombinant protein was confirmed by SDS-PAGE.
Ligand-binding experiments
The binding activity of purified Ab-FAR-1 protein to fatty acid was measured using the fluorescent analogue 11-((5-dimethylaminonaphthalene-1-sulfonyl) amino) undecannoic acid (DAUDA) (Sigma, USA), the naturally fluorescent cis-parinaric acid (Molecular Probes) (Cayman, USA), retinol (Sigma, USA) and oleic acid (Sigma, USA). Fluorescence measurements were performed at 20°C with FluoroMax 4 (HORIBA Jobin Yvon, France) in a total volume of 3 ml per well as described previously [15], [19]. The excitation wavelengths used for DAUDA, retinol and cis-parinaric acid were 345, 350 and 319 nm, respectively. DAUDA, cis-parinaric acid and oleic acid were prepared at 10 mM stock solution in ethanol. DAUDA and oleic acid were used at 1∶1000 dilutions in PBS; while cis-parinaric acid was diluted at 1∶2000 in PBS. Free retinol was freshly prepared at 10 mM in ethanol and further diluted at 1∶1000 in ethanol and added directly to the protein solutions.
Dissociation constants were estimated in fluorescence titration experiments as described previously [5], [15], [19]. The dissociation constant (Kd) for rAb-FAR-1 binding to DAUDA and cis-parinaric acid were estimated by adding increasing concentrations of rAb-FAR-1 to 10 µM DAUDA in PBS and 10 µM cis-parinaric acid in PBS (total volume of 3 ml), respectively. To determine the Kd for rAb-FAR-1 binding to retinol, increasing concentrations of fluorescent ligand were added to 10 µM Ab-FAR-1 solution in Tris-HCl buffer. Fluorescence data were corrected for dilution and fitted by standard nonlinear regression techniques (using Microcal ORIGIN software) to a single noncompetitive binding model to give estimation of the dissociation constant (Kd) and maximal fluorescence intensity (Fmax).
Expression of Ab-far-1 mRNA at different development stages of A. besseyi
qPCR was used to assess the expression levels of Ab-far-1 at different development stages of A. besseyi: females, males, juveniles and eggs. RNA samples were prepared from 100 nematodes at each developmental stage using the MicroElute total RNA kit (OMEGA) according to the manufacturer's instructions. Total RNA was treated with RQ1 RNase-Free DNase (Promega) as described above. The RNA was quantified by a Nano-drop spectrophotometer and stored at -80°C for further analysis. All RNAs used for qPCR were prepared in triplicate.
cDNA was synthesized by a ReverTra Ace qPCR RT kit (TOYOBO) with random primers according to the manufacturer's instructions. The specific primers qPCR-F and qPCR-R (Table 1) for Ab-far-1 were designed to detect Ab-far-1 expression level. The 140 bp of 18S rRNA (AY508035) was amplified as a reference gene using the primers 18sF and 18sR (Table 1). The qPCR was performed on CFX-96 (Bio-Rad) qPCR machine using SYBR Green Real-time PCR Master Mix-plus kit (TOYOBO) according to the manufacture's protocol. All assays were performed in triplicate. Initial data analysis was carried out to create Ct values using Bio-Rad CFX-96 manager software and to determine the relative expression level by the melt curves. 18S rRNA was used as a positive control in all experiments. All experiments were performed in triplicate.
Localisation of Ab-far-1 mRNA by using in situ hybridization
In situ hybridization was performed as described by De Boer et al. [20] with some modifications. 10,000 mixed stages nematodes, including females, males and juveniles separated from carrot callus were concentrated to a 30–50 µl pellet and fixed in 3% paraformaldehyde at 4°C for 16 h. The specific sense (FAR-IN-T7S1, FAR-IN-A1) and antisense (FAR-IN-T7A1, FAR-IN-S1) (Table 1) primers were designed to amplify DIG-labelled sense and antisense RNA probes (Roche, Germany) from full-length cDNA of Ab-far-1 gene. DIG-labelled sense or antisense RNA probe was added to the hybridization solution containing the nematode sections, and then rotated at 47°C for 12 h. After hybridization, the nematodes were examined and photographs were taken with differential interference microscopy.
Synthesis of Ab-far-1 dsRNA
The fragment of Ab-far-1 ORF was cloned into the vector pMD18-T (Takara, Japan), and confirmed by sequencing. Two primer pairs of FARRiF/FARRiT7R and FARRiT7F/FARRiR (Table 1) were designed to amplify the sense and antisense single-stranded RNA (ssRNA) products. The sense and antisense ssRNAs were transcribed from Ab-far-1 plasmid using the ScriptMAX Thermo T7 Transcription KIT (TOYOBO, Japan) according to the manufacturer's instructions, and mixed together in equal proportions and heated at 94°C for 10 min, then cooled to room temperature to allow the annealing of complementary strands. The dsRNA was purified with 1/10 amount of KAc (3 mol/l) and 2-fold amount of 95% ethanol overnight, then washed with 75% ethanol for 3 times and dissolved in deionized water. The quantity of dsRNA was measured by a Nano-drop spectrophotometer and analyzed by 1.2% agarose gel electrophoresis, and finally stored at −80°C for later use. Non-endogenous control dsRNA (125 bp) (green fouorescent protein gene, gfp) was generated with specific primers (G-T7S, G-A, G-T7A, and G-S) (Table 1) [21].
Knockdown of Ab-far-1 using dsRNA
500 mixed stage nematodes separated from carrot callus were collected in a DEPC-treated Eppendorf tube and washed twice with DEPC water, and then soaked in 50 µl Ab-far-1 dsRNA solution (2 µg/μl) and lightly shook in a rotary incubation (200 rpm) for 12 h, 24 h, 36 h and 48 h in the dark at 25°C. Non-endogenous gfp dsRNA solution 50 µl (2 µg/μl) was used as a control. After soaking in dsRNA, the nematodes were washed three times with DEPC water and the total RNA was then extracted by the MicroElute total RNA kit (OMEGA) as described above. qPCR was used to analyze the transcript suppression of Ab-far-1 in A. besseyi after dsRNA treatment. All assays were performed in triplicate.
To test the reproductive capacity of Ab-far-1 silenced nematodes, thirty female nematodes separated from carrot callus were treated with dsRNA for 12 h, 24 h, 36 h and 48 h respectively. Then all nematodes were inoculated onto carrot callus and maintained in a dark incubator at 25°C for 35 days. Isolating and calculating the number of total nematodes in the carrot callus at last. Each treatment was carried out in 5 duplicates. All data in this study were subjected to analysis of variance (ANOVA) and multiple comparisons of means were conducted by Duncan's Multiple Range Test at p = 0.01 using SAS (Release 8.01).
Results
Cloning of full-length FAR gene from A. besseyi
According to the EST sequences (486 bp) of FAR gene from cDNA library of A. besseyi, the specific primers for FAR gene were designed to amplify the 805 bp full-length cDNA sequence and cloned into pMD 18-T vector. The plasmid was named Ab-far-1, which included a 546 bp of open reading frame (ORF) for encoding a deduced 181 amino acids (GenBank accession number JQ686690) (Fig. S2). The ORF began with an ATG initiation codon at nucleotide 93 and terminated with TAA at nucleotide 638 (Fig. S2).
Sequence analysis of the Ab-FAR-1 protein
The Ab-FAR-1 protein encoded 181 amino acids with theoretical molecular mass of 20.5217 kDa, the molecular formula was C923H1514N238O280S3 with a theoretical isoelectric point of 6.66. The location site of Ab-FAR-1 protein was predicted in extracellular compartment (including cell wall). Ab-FAR-1 sequences had the highest similarity with the Gp-FAR-1 protein from G. pallida (GenBank accession number CAA70477.2, 52% identity and 73% similarity, E-value 1e-51) and the FAR from O. ochengi (GenBank accession number ACB70198.1, 51% identity and 73% similarity, E-value 1e-54). Ab-FAR-1 also showed higher similarities with the Ov-FAR-1 from O. volvulus (GenBank accession number Q25619, 50% identity and 73% similarity, E-value 8e-56) and Og-FAR-1 from O. gutturosa (GenBank accession number Q8WT59, 50% identity and 73% similarity, E-value 1e-55).
Like other six FARs from four species of nematodes [17], Ab-FAR-1 protein contained a hydrophobic secretory signal peptide and was rich in α-helix but no extended β structure (Fig. S3). Moreover, the Ab-FAR-1 also contained a conserved casein kinase II phosphorylation site, but did not have the glycosylation sites like Ce-FAR-1 and Ce-FAR-2.
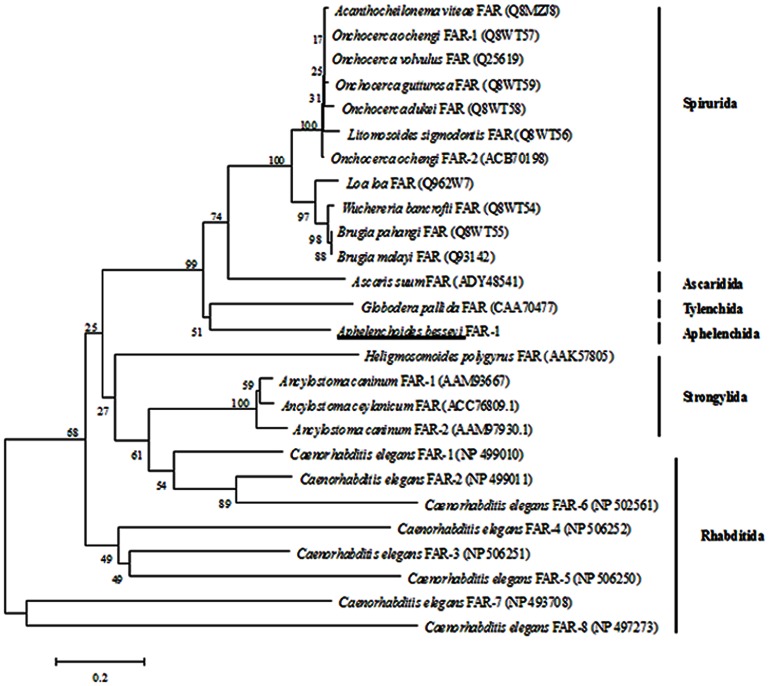
In the phylogenetic tree (Fig. 1) constructed by the Ab-FAR-1 amino acid sequences and other 25 FAR proteins from 16 species of nematodes, Ab-FAR-1 of Aphelenchida nematode A. besseyi and Gp-FAR-1 of Tylenchida nematode G. pallida were present in the same branch of the phylogenetic tree, which suggests that they have a closer genetic relationship. In addition, all these 26 FAR proteins from 17 species of nematodes were divided into 6 groups: Spirurida, Ascaridida, Tylenchida, Aphelenchida, Strongylida, and Rhabditida.
Figure 1. Phylogenetic relationships among the amino acid sequences of fatty acid and retinoid binding proteins in nematodes.
Phylogram was constructed according to amino acid sequences depicting the evolutionary relationships among fatty acid and retinoid binding proteins of 26 different nematode species, Aphelenchoides besseyi FAR-1 highlighted by the underline. Accession number of the sequences was in brackets.
Ligand binding
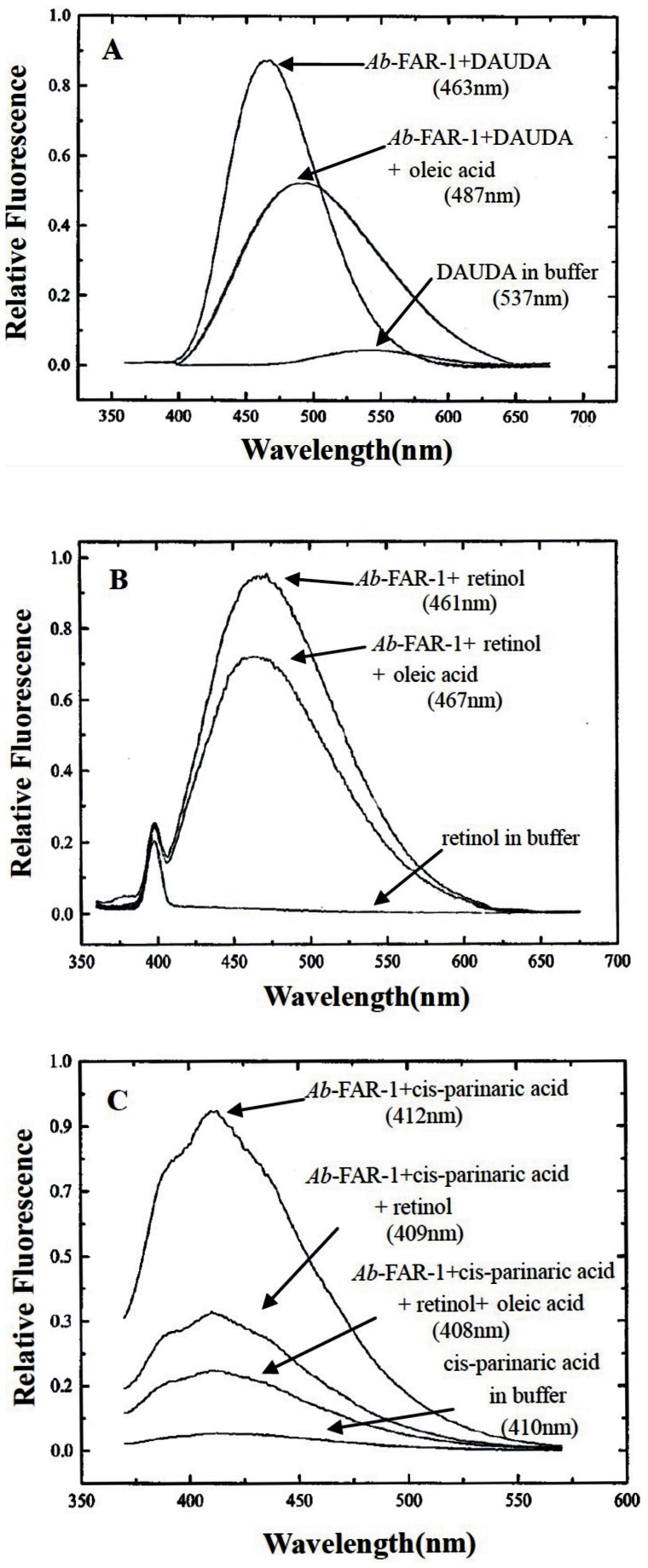
SDS-PAGE showed that the recombinant Ab-FAR-1 was well purified, with only one band of approximately 19-20 KDa, which was consistent with theoretical molecular mass (20 KDa) of Ab-FAR-1 (Fig. S4). The purified rAb-FAR-1 was found to bind the dansylated fatty acid DAUDA because a significant wavelength shift of peak fluorescence emission was observed (Fig. 2A). In buffer alone, the peak emission of DAUDA occurred at 537 nm, but moved to 463 nm upon addition of rAb-FAR-1 (Fig. 2A). The wavelength shift indicates that rAb-FAR-1 has a highly apolar binding site, and the degree of shift is unusually large for lipid transporter proteins but typical for FAR protein. These characteristics have been demonstrated by previous studies [13], [15], [17], [22], [23], [24], [25], [26]. After the addition of oleic acid to DAUDA, protein complex produced a pronounced drop in the fluorescence intensity (487 nm) (Fig. 2A), indicating the oleic acid can displace DAUDA from the binding site. The retinol binding activity of purified Ab-FAR-1 test showed that the fluorescence emission of retinol was minimal in buffer alone, but was substantially increased to 461 nm when the retinol was added to the rAb-FAR-1 solution (Fig.2B). After the addition of oleic acid to retinol, the protein complex produced an obvious drop in the fluorescence intensity (467 nm) (Fig.2B). rAb-FAR-1 was also found to bind the intrinsically fluorescent cis-parinaric acid. In control buffer, the emission peak of cis-parinaric acid appeared at 410 nm, but moved to 412 nm upon addition of rAb-FAR-1 (Fig.2C). The effects of subsequent addition of retinol to the rAb-FAR-1+cis-parinaric acid and addition of oleic acid solution to the rAb-FAR-1+cis-parinaric acid+ retinol were different (Fig.2C). These results indicate that the retinol and fatty acid binding sites are congruent, overlapping, or interactive, and the addition of oleic acid can competitively displace cis-parinaric acid.
Figure 2. Ligand binding by rAb-FAR-1.
(A) Ab-FAR-1. The reverse change of DAUDA emission was observed after the addition of oleic acid to the rAb-FAR-1+DAUDA complex. The wavelengths of peak emission by DAUDA were different. (B) Fluorescence emission spectra (excitation at 350 nm) of retinol in ethanol alone or after the addition of rAb-FAR-1. The competitive effect of oleic acid was also found. (C) Fluorescence emission spectra (excitation at 319 nm) of cis-parinaric acid alone or after the addition of rAb-FAR-1.The effects of the subsequent addition of retinol to the rAb-FAR-1+cis-parinaric acid and addition of oleic acid solution to the rAb-FAR-1+cis-parinaric acid+ retinol were examined.
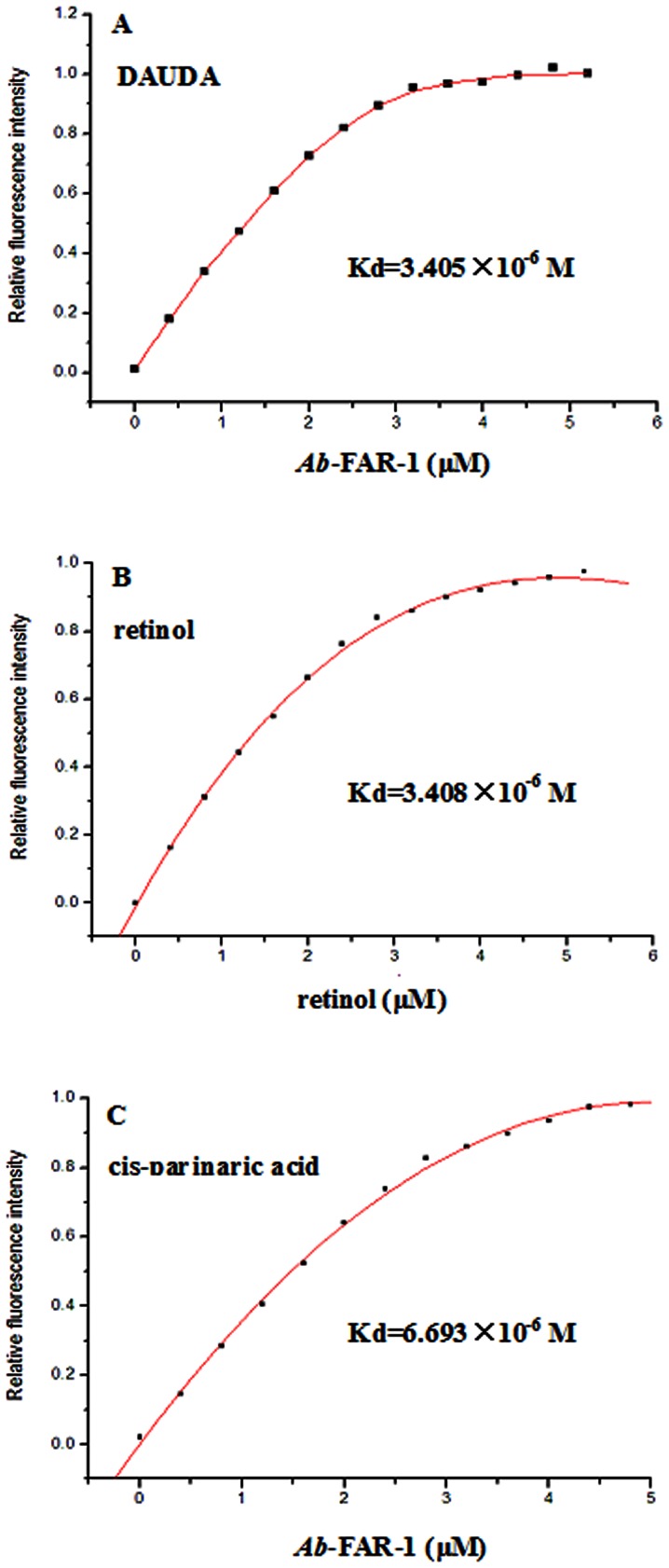
Fluorescence titration experiments were performed to determine the binding affinity of DAUDA, retinol and cis-parinaric acid. Figure 3 showed typical saturation binding curves for each of the three fluorescent ligands. The titration curve (corrected for the background fluorescence of DAUDA) in Figure 3A predicted a Kd of 3.405×10−6 M for the Ab-FAR-1: DAUDA interaction. We also demonstrated binding of Ab-FAR-1 to retinol (Kd of 3.408×10−6 M) (Fig. 3B) and cis-parinaric acid (Kd of 6.693×10−6 M) (Fig. 3C). These values were within the general range of dissociation constants reported for other FAR proteins [5], [7], [8], [15], [19]. The results indicate that Ab-FAR-1 is a functional protein and has binding activities to fatty acid and retinol in vitro.
Figure 3. Fluorescent titration lipid binding analysis of rAb-FAR-1.
(A) Change in relative fluorescence intensity (excitation at 345 nm) of DAUDA (10 µM) in the presence of increasing concentrations of rAb-FAR-1. The best fit curve was used to determine the dissociation constant (Kd) for the DAUDA: rAb-FAR-1 interaction. (B) Change in relative fluorescence intensity (excitation at 350 nm) of 10 µM rAb-FAR-1 in the presence of increasing concentrations of retinol. The curve was used to derive the equilibrium dissociation constant (Kd) for the retinol: rAb-FAR-1 interaction. (C) Change in relative fluorescence intensity (excitation at 319 nm) of cis-parinaric acid (10 µM) in the presence of increasing concentrations of rAb-FAR-1. The best fit curve was used to determine the dissociation constant (Kd) for the cis-parinaric acid: rAb-FAR-1 interaction.
Expression and localization of Ab-far-1 mRNA
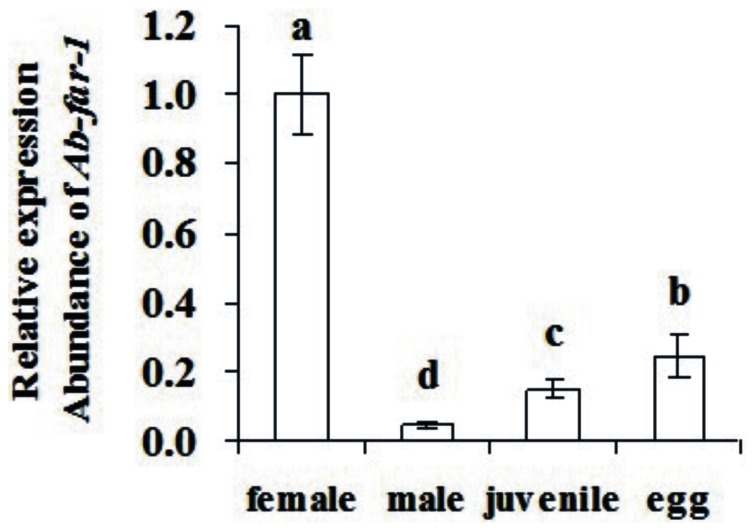
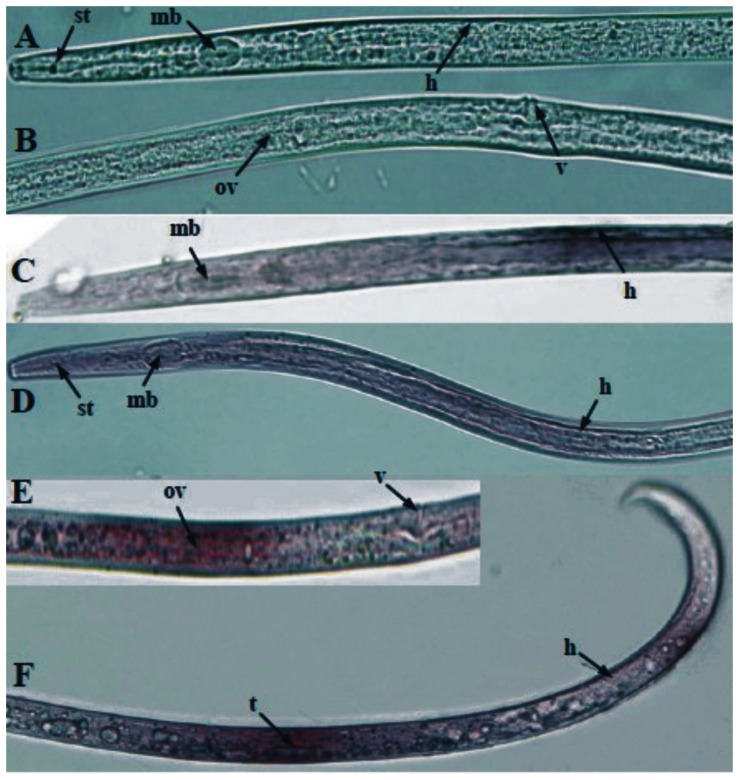
The qPCR results showed that Ab-far-1 mRNA transcript was present in all developmental stages, the highest transcript levels noted in females, while its expression in eggs, juveniles and males accounted for 25%, 14.9%, and 4.7% of the expression level in females, respectively (Fig. 4). The results of in situ hybridization showed that Ab-far-1 mRNA was present in the hypodermis of juveniles (Fig. 5C), the hypodermis and ovaries of females (Fig. 5 D, E), and in the hypodermis and testes of males (Fig. 5F). No hybridization signal was detected in the nematodes after incubation with the control sense probe (Fig. 5A, B).
Figure 4. Expression of the Ab-far-1 in 100 eggs, females, juveniles and males, respectively of Aphelenchoides besseyi.
Bars indicate standard errors of mean data (n = 3) and different letters indicate significant differences (p<0.01) between treatments.
Figure 5. Tissue localisation of Ab-far-1 mRNA in juveniles, females and males of Aphelenchoides besseyi using in situ hybridization.
(A and B) No signal in A. besseyi section was hybridized with sense Ab-far-1 DIG- Labeled RNA probe. (C) Ab-far-1 was present in the hypodermis of juveniles. (D) Ab-far-1 was present in the hypodermis of females. (E) Ab-far-1 was present in the ovaries of females. (F) Ab-far-1 was present in the hypodermis and testes of males. st: stylet, mb: medium bulb, h: hypodermis, ov: ovaries, v: vulva, t: testis.
The effect of RNAi
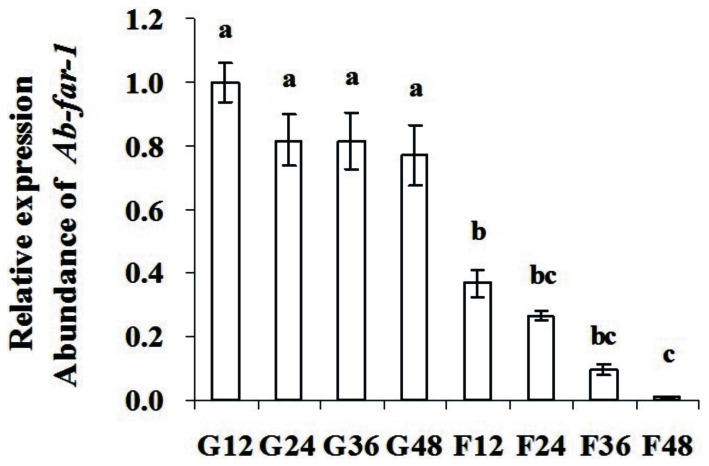
After treatment with Ab-far-1 dsRNA, the expression of Ab-far-1 mRNA in A. besseyi was detected by the qPCR. Compared with the relative expression level of Ab-far-1 mRNA in the corresponding control nematodes (gfp dsRNA treatment), the relative expression of Ab-far-1 mRNA in nematodes treated with Ab-far-1 dsRNA was reduced by 63.2% at 12 h, 55.1% at 24 h, 71.8% at 36 h, and 76% at 48 h (Fig. 6). The expression level of Ab-far-1 mRNA was reduced with the increasing incubation time with dsRNA, while the non-endogenous gfp dsRNA control had no effect on the expression of Ab-far-1 mRNA at different time points.
Figure 6. Expression of the Ab-far-1 mRNA in Aphelenchoides besseyi treated with Ab-far-1 double-stranded (ds) RNA.
G12, G24, G36 and G48 indicates expression of Ab-far-1 mRNA in control nematodes soaked by non-endogenous gfp dsRNA solution for 12 h, 24 h, 36 h and 48 h, respectively. F12, F24, F36 and F48 indicates expression of Ab-far-1 mRNA in nematodes soaked by Ab-far-1 dsRNA for 12 h, 24 h, 36 h and 48 h, respectively. Bars indicate standard errors of mean data (n = 3) and different letters indicate significant differences (p<0.01) between treatments.
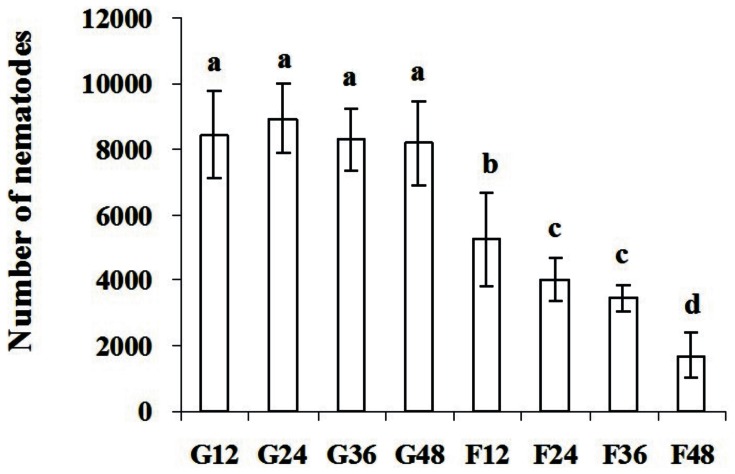
After being treated with dsRNA for 12 h, 24 h, 36 h and 48 h and inoculated onto carrot callus for 35 days, A. besseyi treated with Ab-far-1 dsRNA had significantly (p<0.01) lower reproduction than those treated with gfp dsRNA at all time points. The reproduction of nematodes was significantly (p<0.01) decreased with the increasing exposure time to Ab-far-1 dsRNA. The reproduction of nematodes treated with gfp dsRNA did not show difference at different treatment time points (Fig. 7).
Figure 7. Number of Aphelenchoides besseyi on carrot callus after inoculation of 30 females 35 days later, respectively.
G12, G24, G36 and G48 indicates number of A. besseyi after inoculating 30 females treated by non-endogenous gfp dsRNA solution for 12 h, 24 h, 36 h and 48 h, respectively. F12, F24, F36 and F48 indicates number of A. besseyi after inoculating 30 females treated by Ab-far-1 dsRNA for 12 h, 24 h, 36 h and 48 h, respectively. Bars indicate standard errors of mean data (n = 5) and different letters indicate significant differences (p<0.01) among treatments.
Discussion
FAR protein is a specific protein only in the nematodes, which plays a critical role in the development and reproduction in nematodes, and in the processes of infecting host [7], [15], [27]. In this study, we obtained the full-length sequences of Ab-far-1 in A. besseyi, which is the first report for the FAR gene in Aphelenchida nematodes. The secondary structure of Ab-FAR-1 was rich in α-helix but without β extended structure, which is an important factor that sets FAR apart from the similar lipid-binding proteins of vertebrates [17], [19], [28], [29], [30], [31]. There are four conserved Proline residues in Ab-FAR-1 and other six FAR proteins amino acid sequences in sequence alignment. Proline is an inclined interrupt helical structure and cannot generate the hydrogen bond to sustain the conformation of α-helix and is an amino acid which requires less free energy to fold [32]. But there is Glutamic or Alanine residue behind the two Proline residues in the middle of first helix. The Glutamic and Alanine are the strongest nucleate α-helices [33], which may influence the secondary structure of FAR. It has been reported that FAR proteins are secreted from the cell to the extracellular environment where they presumably play roles in the sequestration and transport of fatty acids and retinoids to maintain the nematodes normal life activity during the infection to the host [9], [13], [15], [17]. A hydrophobic secretory signal peptide was identified in the Ab-FAR-1 sequences, which indicates Ab-FAR-1 may be secreted from the cell to the extracellular environment. Furthermore, the Ab-FAR-1 also contained a conserved casein kinase II phosphorylation site, which is appeared in all known nematodes FAR proteins [13], [15], [17], [19]. Phosphorylation is known to regulate the biological activities of many proteins, including gene regulation, homodimerisation control and the stability of α-helices [34], [35], [36], [37], [38], [39]. Thus we assume that the phosphorylation sites may play an important role in the process of the infection of A. besseyi to the hosts.
It is believed that FAR protein is necessary for the nematodes not only to bind lipids like linoleic and linolenic acids to maintain the metabolism, destroy the plant defense systems and complete the infection, but also to bind retinoid in collagen synthesis, embryonic development and reproduction [8], [15]. In this study, the expression of Ab-far-1 mRNA at different nematode stages showed the expression was highest in females and lowest in males, higher in eggs than in juveniles, which conformed to their individual biological functions. The female of A. besseyi plays key roles in infection and reproduction, while the number and infection ability of male is lower than female and the male is not necessary for the reproduction of the nematode as it is capable of parthenogenesis. The egg and juvenile are responsible for reproduction and infection, respectively, which indicates that this protein may play an important role in the formation of the embryo and cell differentiation. It was also found that the FAR gene expression at different developmental stages was associated the biological functions in animal parasitic nematodes. The Hc-FAR-1 gene expression of Haemonchus contortus was higher in adult than in larvae [27] and AceFAR-1 mRNA expression was lowest in male than other developmental stages of A. ceylanicu [5]. In plant parasitic nematodes G. pallida, Prior [15] reported the gp-far-1 mRNA was present in the second-stage juveniles, virgin females and adult female, but did not analyze the differences of its expression level.
In situ hybridization results revealed that Ab-far-1 mRNA was present in the hypodermis of A. besseyi. The cuticle layer, hypodermis and internal muscle layer form the nematode body wall, and the hypodermis is one of the most active areas in the nematodes. The expression of Ab-far-1 in the hypodermis may help nematodes to utilize the fatty acid and retinoid from the hosts and the environment quickly to maintain the autologous metabolism. Meanwhile, Ab-far-1 in the hypodermis may help nematodes to neutralize plant defense and to complete the infection. The gp-far-1 mRNA was localized in the hypodermis of the infectious second-stage juveniles of G. pallida [15]. In addition, Ab-far-1 mRNA was also present in the ovaries of females and the testes of males, but signals appeared diffuse, whether is related to the internal structure of ovaries and testes is not clear because the nematodes were cut off, fixed and hybridized in situ hybridization, it needs further study to be confirmed.
In this paper, RNAi was employed to further verify the Ab-far-1 function. To our knowledge, it has not been reported in other nematodes FAR gene study. After the treatment with Ab-far-1 dsRNA, the expression of Ab-far-1 and the reproduction of nematodes were decreased with the increasing exposure time. After treatment with dsRNA for 48 h, the silencing efficiency of Ab-far-1 was the maximum and the reproduction of nematodes was the least. This may be because the FAR protein can bind with fatty acids and retinol. Since the retinol is thought to be essential for collagen synthesis and embryonic development, down-regulation of Ab-far-1 by RNAi leads to less binding with retinol, and thus causes the reduced reproduction in A. besseyi. This evidence further proves the roles of Ab-FAR-1 in the development and reproduction of A. besseyi.
The transgenic plant with the expression of specific dsRNA to the target gene in nematodes would result in the inhibition of target gene in nematodes after feeding [40]. Thus, the expression of specific dsRNAs for nematode development-related and parasitic genes in transgenic plants is a potentially way to enhance the resistance to nematodes and to control plant parasitic nematodes. FAR protein is not only a kind of protein related to development, reproduction, infection and interfered the plant defense system, but also a specific protein only existed in the nematodes [7], [27]. Moreover, the Ab-FAR-1 gene has not been found in mammal and other animals, so this transgenic plant containing Ab-FAR-1 dsRNA will not cause any safety issue to the environment and other organisms. Therefore, it is imperative to consider the generation of transgenic plant containing Ab-FAR-1 dsRNA, which will be an effective method to control A. besseyi.
Supporting Information
EST sequence of Aphelenchoides besseyi fatty acid and retinoid binding protein gene.
(TIF)
Ab-far-1 cDNA sequence and its deduced amino acid sequence. atg, Translation starting signal. taa, Translation termination signal. Nucleotide sequence are denoted in lowercase and deduced amino acid sequence are marked in dark gray.
(TIF)
The structural predictions of Aphelenchoides besseyi Ab -FAR-1 and its amino acid sequence alignments with other nematode FAR proteins. Ov-FAR-1, Onchocerca volvulus FAR (Q25619); Bm-FAR-1, Brugia malayi FAR (Q93142); Gp-FAR-1, Globodera pallida FAR (CAA70477); Ce-FAR-1, Caenorhabditis elegans FAR-1 (NP 499010); Ce-FAR-2, C. elegans FAR-2 (NP 499011); Ce-FAR-6, C. elegans FAR-6 (NP 502561); Ab-FAR-1, A. besseyi FAR-1; Lowercase, putative secretory signal peptides; shaded boxes, positions where amino acids are conserved in all the sequences; underscored, consensus N-linked glycosylation sites; boldface, conserved casein kinase II phosphorylation site; In Consensus line, uppercase amino acids which are conserved at that position in all of the sequences, lowercase amino acid which are conserved at that position in more than half of the sequences, # indicates any of NDQEBZ. The Jpred line shows the secondary strucure prediction from submission of the multiple alignment to the Jpred secondary structure prediction programme. H Prediction for α-helix; gaps regions for which no structural prediction emerged. No β-structure was predicted by Jpred or any other secondary structure prediction programmes.
(TIF)
Sodium dodecyl sulphate polyacrylamide gel of purified His- Ab -FAR-1 and r Ab -FAR-1. Lane 1, purified His-Ab-FAR-1; Lane 2, purified rAb-FAR-1; Lane M, PageRuler Prestained protein Ladder (Sofar Technology, China).
(TIF)
Funding Statement
This work was funded by National Natural Science Foundation of China (Nos. 30871629 and 31071665). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Franklin MT, Siddigi MR (1972) Aphelenchoides besseyi. CIH. Descriptions of plant-parasitic nematodes. UK: Commonwealth Agricultural Bureaux Press. 21–23p.
- 2.Bridge J, Luc M, Plowright RA (1990) Nematode parasites of rice. In: Luc M, Sikora RA, Bridge J, editors. Plant parasitic nematodes in subtropical and tropical agriculture. Oxford: CABI Publishing. pp. 69–76. [Google Scholar]
- 3. Pei YY, Cheng X, Xu CL, Yang ZF, Xie H (2012) Virulence of part populations of Aphelenchoides besseyi on rice in China. Chinese Journal of Rice Science 26: 218–226. [Google Scholar]
- 4. Kennedy MW (2000) The polyprotein lipid binding proteins of nematodes. Biochim Biophys Acta 1476: 149–164. [DOI] [PubMed] [Google Scholar]
- 5. Fairfax KC, Vermeire JJ, Harrison LM, Bungiro RD, Grant W, et al. (2009) Characterisation of a fatty acid and retinol binding protein orthologue from the hookworm Ancylostoma ceylanicum . Int J Parasitol 39: 1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McDermott L, Cooper A, Kennedy MW (1999) Novel classes of fatty acid and retinol binding protein from nematodes. Mol Cell Biochem 192: 69–75. [PubMed] [Google Scholar]
- 7. Garofalo A, Rowlinson MC, Amambua NA, Hughes JM, Kelly SM, et al. (2003) The FAR protein family of the nematode Caenorhabditis elegans. Differential lipid binding properties, structural characteristics, and developmental regulation. J Biol Chem 278: 8065–8074. [DOI] [PubMed] [Google Scholar]
- 8. Basavaraju S, Zhan B, Kennedy MW, Liu YY, Hawdon J, et al. (2003) Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: gene transcription pattern, ligand binding properties and structural characterisation. Mol Biochem Parasitol 126: 63–71. [DOI] [PubMed] [Google Scholar]
- 9. Tree TIM, Gillespie AJ, Shepley KJ, Blaxter ML, Tuan RS, et al. (1995) Characterisation of an immunodominant glycoprotein antigen of Onchocerca volvuluswith homologues in other filarial nematodes and Caenorhabditis elegans . Mol Biochem Parasit 69: 185–195. [DOI] [PubMed] [Google Scholar]
- 10. Bradley JE, Nirmalan NJ, Klager SL, Faulkner H, Kennedy MW (2001) River blindness: a role for parasite retinoid-binding proteins in the generation of pathology?. Trends Parasitol 17: 471–475. [DOI] [PubMed] [Google Scholar]
- 11. Mei B, Kennedy MW, Beauchamp J, Komuniecki PR, Komuniecki R (1997) Secretion of a novel, developmentally regulated fatty acid-binding protein into the perivitelline fluid of the parasitic nematode, Ascaris suum . J Biol Chem 272: 9933–9941. [DOI] [PubMed] [Google Scholar]
- 12. Nirmalan N, Cordeiro NJ, Klager SL, Bradley JE, Allen JE (1999) Comparative analysis of glycosylated and non-glycosylated filarial homo-logues of the 20-kilodalton retinol binding protein from Onchocerca volvulus (Ov20). Infect Immun 67: 6329–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garofalo A, Klager SL, Rowlinson M-C, Nirmalan N, Klion A, et al. (2002) The FAR proteins of filarial nematodes: secretion, glycosylation and lipid binding characteristics. Mol Biochem Parasitol 122: 161–170. [DOI] [PubMed] [Google Scholar]
- 14. Bath JL, Robinson M, Kennedy MW, Agbasi C, Linz L, et al. (2009) Identification of a Secreted Fatty Acid and Retinol-Binding Protein (Hp-FAR-1) from Heligmosomoides polygyrus . J Nematol 41: 228–233. [PMC free article] [PubMed] [Google Scholar]
- 15. Prior A, Jones JT, Blok VC, Beauchamp J, McDermott L, et al. (2001) A surface-associated retinol-and fatty acid-binding protein (Gp-FAR-1) from the potato cyst nematode Globodera pallida: lipid binding activities, structural analysis and expression pattern. Biochem J 356: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reise RW, Huettel RN, Sayre RM (1987) Carrot Callus Tissue for Culture of Endoparasitic Nematodes. J Nematol 19: 387–389. [PMC free article] [PubMed] [Google Scholar]
- 17. Garofalo A, Kennedy MW, Bradley JE (2003) The FAR proteins of parasitic nematodes: their possible involvement in the pathogenesis of infection and the use of Caenorhabditis elegans as a model system to evaluate their function. Med Microbiol Immunol 192: 47–52. [DOI] [PubMed] [Google Scholar]
- 18. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 19. Kennedy MW, Garside LH, Goodrick LE, McDermott L, Brass A, et al. (1997) The Ov20 protein of the parasitic nematode Onchocerca volvulus. A structurally novel class of small helix-rich retinol-binding proteins. J Biol Chem 272: 29442–29448. [DOI] [PubMed] [Google Scholar]
- 20. De Boer JM, Yan Y, Smant G, Davis EL, Baum TJ (1998) In-situ hybridization to messenger RNA in Heterodera glycines . J Nematol 30: 309–312. [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang C, Xie H, Xu CL, Cheng X, Li KM, et al. (2012) Differential expression of Rs-eng-1b in two populationsof Radopholus similis (Tylenchida: Pratylecnchidae) and its relationship to pathogenicity. Eur J Plant Pathol 133: 899–910. [Google Scholar]
- 22. Macgregor RB, Weber G (1986) Estimation of the polarity of the protein interior by optical spectroscopy. Nature 319: 70–73. [DOI] [PubMed] [Google Scholar]
- 23. Wilkinson TC, Wilton DC (1986) Studies on fatty acid-binding proteins. The detection and quantification of the protein from rat liver by using a fluorescent fatty acid analogue. Biochem J 238: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kennedy MW, Brass A, McCruden AB, Price NC, Kelly SM, et al. (1995) The ABA-1 allergen of the parasitic nematode Ascaris suum: fatty acid and retinoid binding function and structural characterization. Biochemistry 34: 6700–6710. [DOI] [PubMed] [Google Scholar]
- 25. Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ (1999) Binding studies of tear lipocalin: the role of the conserved tryptophan in maintaining structure, stability and ligand affinity. Biochim Biophys Acta 1433: 307–320. [DOI] [PubMed] [Google Scholar]
- 26. Suire S, Stewart F, Beauchamp J, Kennedy MW (2001) Uterocalin, a lipocalin provisioning the preattachment equine conceptus: fatty acid and retinol binding properties, and structural characterization. Biochem J 356: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuang L, Colgrave ML, Bagnall NH, Knox MR, Qian M, et al. (2009) The complexity of the secreted NPA and FAR lipid-binding protein families of Haemonchus contortus revealed by an iterative proteomics–bioinformatics approach. Mol Biochem Parasitol 168: 84–94. [DOI] [PubMed] [Google Scholar]
- 28. Cowan SW, Newcomer ME, Jones TA (1993) Crystallographic studies on a family of cellular lipophilic transport proteins. Refinement of P2 myelin protein and the structure determination and refinement of cellular retinol-binding protein in complex with all-trans-retinol. J Mol Biol 230: 1225–1246. [DOI] [PubMed] [Google Scholar]
- 29. Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, et al. (1994) Lipid-binding proteins: a family of fatty acid and retinoid transport proteins. Adv Protein Chem 45: 89–151. [DOI] [PubMed] [Google Scholar]
- 30. Flower DR (1996) The lipocalin protein family: structure and function. Biochem J 318: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noy N (2000) Retinoid-binding proteins: mediators of retinoid action. Biochem J 348: 481–495. [PMC free article] [PubMed] [Google Scholar]
- 32. Matthews BW (1987) Genetic and structural analysis of the protein stability problem. Biochemistry 26: 6885–6888. [DOI] [PubMed] [Google Scholar]
- 33. Singh R, Deol SK, Sandhu PS (2010) Chou-Fasman Method for Protein Structure Prediction using Cluster Analysis. World Academy of Science, Engineering and Technology 72: 980–985. [Google Scholar]
- 34. Buelt MK, Xu Z, Banaszak LJ, Bernlohr DA (1992) Structural and functional characterization of the phosphorylated adipocyte lipid-binding protein (pp15). Biochemistry 31: 3493–3499. [DOI] [PubMed] [Google Scholar]
- 35. Ganjeizadeh M, Zolotarjova N, Huang WH, Askari A (1995) Interactions of Phosphorylation and Dimerizing Domains of the α-Subunits of Na+/K+-ATPase. J Biol Chem 270: 15707–15710. [DOI] [PubMed] [Google Scholar]
- 36. Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, et al. (1996) Dimerization Is Required for the Activity of the Protein Histidine Kinase CheA That Mediates Signal Transduction in Bacterial Chemotaxis. J Biol Chem 271: 939–945. [DOI] [PubMed] [Google Scholar]
- 37. Brownlie P, Ceska TA, Lamers M, Romier C, Stier G, et al. (1997) The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure 5: 509–520. [DOI] [PubMed] [Google Scholar]
- 38. Olson JK, Bishop GA, Grose C (1997) Varicella-zoster virus Fc receptor gE glycoprotein: serine/threonine and tyrosine phosphorylation of monomeric and dimeric forms. J Virol 71: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szilak L, Moitra J, Krylov D, Vinson C (1997) Phosphorylation destabilizes alphahelices. Nat Struct Biol 4: 112–114. [DOI] [PubMed] [Google Scholar]
- 40. Urwin PE, Lilley CJ, Atkinson HJ (2002) Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol Plant Microbe Interact 15: 747–752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EST sequence of Aphelenchoides besseyi fatty acid and retinoid binding protein gene.
(TIF)
Ab-far-1 cDNA sequence and its deduced amino acid sequence. atg, Translation starting signal. taa, Translation termination signal. Nucleotide sequence are denoted in lowercase and deduced amino acid sequence are marked in dark gray.
(TIF)
The structural predictions of Aphelenchoides besseyi Ab -FAR-1 and its amino acid sequence alignments with other nematode FAR proteins. Ov-FAR-1, Onchocerca volvulus FAR (Q25619); Bm-FAR-1, Brugia malayi FAR (Q93142); Gp-FAR-1, Globodera pallida FAR (CAA70477); Ce-FAR-1, Caenorhabditis elegans FAR-1 (NP 499010); Ce-FAR-2, C. elegans FAR-2 (NP 499011); Ce-FAR-6, C. elegans FAR-6 (NP 502561); Ab-FAR-1, A. besseyi FAR-1; Lowercase, putative secretory signal peptides; shaded boxes, positions where amino acids are conserved in all the sequences; underscored, consensus N-linked glycosylation sites; boldface, conserved casein kinase II phosphorylation site; In Consensus line, uppercase amino acids which are conserved at that position in all of the sequences, lowercase amino acid which are conserved at that position in more than half of the sequences, # indicates any of NDQEBZ. The Jpred line shows the secondary strucure prediction from submission of the multiple alignment to the Jpred secondary structure prediction programme. H Prediction for α-helix; gaps regions for which no structural prediction emerged. No β-structure was predicted by Jpred or any other secondary structure prediction programmes.
(TIF)
Sodium dodecyl sulphate polyacrylamide gel of purified His- Ab -FAR-1 and r Ab -FAR-1. Lane 1, purified His-Ab-FAR-1; Lane 2, purified rAb-FAR-1; Lane M, PageRuler Prestained protein Ladder (Sofar Technology, China).
(TIF)