Abstract
Myogenin (myog) encodes a highly conserved myogenic regulatory factor that is involved in terminal muscle differentiation. It has been shown in mammals that methylation of cytosines within the myog promoter plays a major role in regulating its transcription. In the present study, the Senegalese sole (Solea senegalensis) myog putative proximal promoter was identified and found to be highly conserved among teleosts. Therefore, it is plausible that it plays a similar role in controlling myog expression. Cytosine methylation of the myog promoter in skeletal muscle of Senegalese sole larvae undergoing metamorphosis was influenced by rearing temperature. A lower temperature (15°C) significantly increased myog promoter methylation in skeletal muscle, particularly at specific CpG sites, relatively to higher rearing temperatures (18 and 21°C). Myog transcription was downregulated at 15°C, whereas expression of dnmt1 and dnmt3b was upregulated, consistently with the higher myog methylation observed at this temperature. Rearing temperature also affected growth and fast muscle cellularity, producing larger fibers at 21°C. Taken together, our data provide the first evidence of an epigenetic mechanism that may be underlying the temperature-induced phenotypic plasticity of muscle growth in teleosts.
Keywords: Solea senegalensis, thermal plasticity, myogenin, methylation, epigenetic regulation, myogenesis
Introduction
The development of vertebrates is largely regulated by epigenetic events such as DNA methylation of specific cytosine residues in the genome, which is stably inherited through cell division.1 DNA cytosine methylation is performed by a group of DNA methyltransferases, known as Dnmts. Dnmt1 is involved in maintaining existing methylation patterns and has a direct role in histone methylation,2 whereas Dnmt3a and 3b are two functionally related proteins that are essential for de novo methylation.1,3,4
Myogenin (myog) belongs to a family of four myogenic regulatory factors (MRFs), which are critical regulators of myogenesis and highly conserved among vertebrates. It encodes a transcription factor of the basic-helix-loop-helix (bHLH) protein family and plays an essential role in the specification and differentiation of myoblasts.5 In zebrafish embryos, muscle-specific expression of myog was shown to be controlled by MEF2 and MEF3 binding sites in the promoter and by two non-canonical Enhancer Boxes (E-box), which are the MRF protein binding sites conferring muscle-specificity.6 Also in striped bass (Morone saxatilis), a 0.6 kb sequence of the myog promoter containing regulatory elements was enough to drive muscle-specific myog expression.7 In the mouse embryo myog promoter, the Myocyte Enhancer Factor-2 (MEF2) (which is bound by elements of the MEF2 family of transcription factors) and MEF3 binding sites were shown to be critical for the correct temporal and spatial expression of myog.8,9 Moreover, the E-box (E1) present between the TATA box and the transcription start site (TSS) was shown to be the binding site for myogenic bHLH protein complexes, and mutation of this E-box was shown to block myog expression in the myotome during development.10 The myog promoter has a relatively low density of CpG residues11 but methylation of cytosine nucleotides within this region plays a role in the negative regulation of transcription,11,12 During mouse early development, myog promoter was shown to be initially methylated, but it becomes demethylated as development proceeds.12 Furthermore, in murine mesenchymal progenitor cell cultures treated with a DNA methylation inhibitor, myog was upregulated at the myoblast stage and myogenesis was promoted.13
Myog expression is known to vary with temperature in some teleost species. For example, in tiger pufferfish (Takifugu rubripes) embryos, the peak myog expression occurred later with respect to developmental stage at a higher incubation temperature.14 In early larvae of rainbow trout (Oncorhynchus mykiss) and seabass (Dicentrarchus labrax), myog expression increased toward the temperature to which these species are naturally exposed in the wild.15 Nevertheless, in spite of the vital importance of myog in muscle development and growth, virtually nothing is known about the molecular mechanisms underlying the epigenetic regulation of myog expression by temperature. In natural conditions, the marine flatfish Senegalese sole (Solea senegalensis) faces temperature fluctuations between 13 and 28°C,16 and large thermal variation have also been observed in aquaculture conditions.17 Incubation temperature has been reported to influence muscle cellularity up to 30 d post-hatching (dph), as, for example, larvae initially incubated at 18 or 21°C had 11 and 9% more muscle fibers than those incubated at 15°C, respectively.18 An increase in muscle growth was observed particularly during and after metamorphosis. Rearing temperature was also found to highly influence protein metabolism in Senegalese sole larvae and post-larvae (Campos C, Castanheira M.F, Engrola S., Valente L.M.P., Fernandes J.M.O and Conceição L.E.C., unpublished). In the present study, we hypothesized that rearing temperature post-hatching could influence the muscle phenotype of Senegalese sole larvae and the methylation status of the myog promoter in skeletal muscle.
Results
Growth
Rearing temperature significantly affected dry weight (DW) and total length (LT) of Senegalese sole larvae at stage 2 of metamorphosis (Table 1). DW was 0.7 ± 0.2, 0.5 ± 0.2 and 0.3 ± 0.2 mg at 21°C, 18°C and 15°C, respectively (p < 0.001).
Table 1. Age, dry weight and total length of Senegalese sole larvae reared at 15, 18 or 21°C.
| Metamorphosis | 21°C | 18°C | 15°C |
|---|---|---|---|
|
Days post hatching |
12 |
15 |
23 |
|
DW (mg) (Mean ± SD) |
0.7 ± 0.2a |
0.5 ± 0.2b |
0.3 ± 0.2c |
| LT (mm) (Mean ± SD) | 6.9 ± 0.8a | 6.7 ± 0.8a | 5.5 ± 0.8b |
Different superscript letters indicate statistically significant differences (p < 0.05) between treatments.
Characterization of the putative promoter and 5′ UTR of the myog gene in Senegalese sole
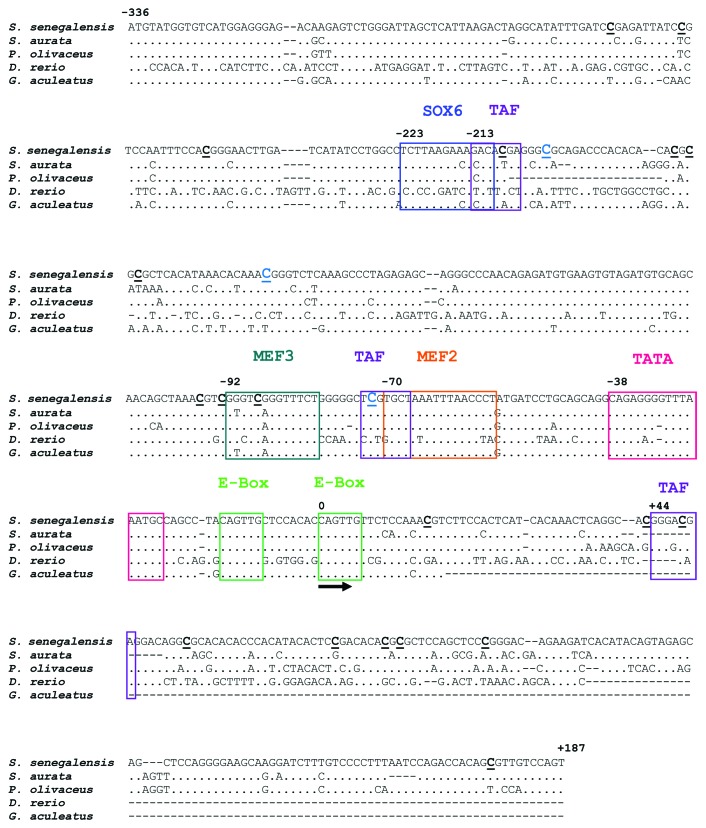
A sequence of 523 bp comprising 187 bp of the putative myog 5′ untranslated region (UTR) and 336 bp of its proximal promoter has been identified in Senegalese sole (GenBank accession number KC404969). Binding sites for MEF2, MEF3, TATA, Sox6 and TAF (putative TATA box binding protein associated factor) were present in this sequence and two putative non-canonical, muscle-specific E-box binding sites (5′-CAGTTG-3′) were found separated by 8 bp (Fig. 1). The most proximal elements to the translation initiation codon (ATG) are the two E-boxes in the 5′ UTR. Putative binding sequences for MEF3 and MEF2 were located at positions -92 and at -70 from the transcription starting site (TSS), respectively, and three putative TATA box binding protein associated factors (TAF) were found at positions -213, -73 and +44 bp from the TSS.
Figure 1. Comparison of the 5′-flanking UTR and putative promoter sequence of myog genes from Senegalese sole and other fish sequences. SOX6, TAF, MEF3, MEF2, TATA binding sites and the two proximal E-boxes are indicated. The sequence and localization the E-boxes, MEF2, MEF3 and TATA are particularly conserved among myog promoters from all five fish species. The predicted transcription starting site is indicated (arrow). The location of the 22 CpG sites is indicated by bold, underlined Cs. GenBank accession numbers for the sequences are: KC404969 (Solea senegalensis), EF462192 (Sparus aurata), EF144128 (Paralichthys olivaceus) and AY124482 (Danio rerio). The Ensembl accession number for Gasterosteus aculeatus is ENSGACG00000000349.
The organization of putative E-boxes, MEF2 and MEF3-B binding sites in the Senegalese sole myog promoter was conserved among different fish species (Fig. 1). Identity at the nucleotide level across Acanthopterygii was 100%, 92% and 100% for the E-boxes, MEF3 and MEF2, respectively.
In muscle of Senegalese sole larvae, DNA methylation of the myog promoter is affected by temperature
To avoid any possible bias of the sequencing, the CpG site located in the reverse primer was not included in the analysis. Therefore, 21 CpG nucleotides up to 523 bp upstream of the ATG start codon were analyzed for global methylation patterns and total number of methylated cytosines (Fig. 1). A rearing temperature of 15°C significantly increased overall methylation levels of the myog putative promoter (21.8 ± 4.9) in muscle of larvae undergoing metamorphosis relatively to the 21°C (17.8 ± 6.0) (p < 0.05) (Fig. 2A). The methylation patterns proportions, expressed as percentage of the 144 bisulfite patterns in all temperature groups (CG, CHG and CHH, where H = A, C or T) were similar among temperatures: around 15% for CGN (CpG methylation), 20% for CHG (non-CpG methylation) and 65% for CHH (non-CpG methylation).
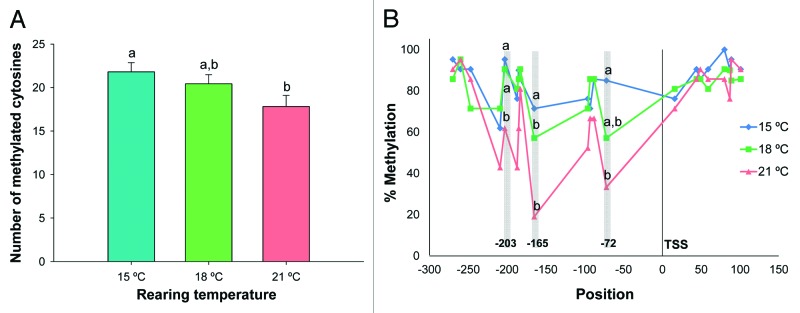
Figure 2. (A) Total number of methylated cytosines in the Senegalese sole myog proximal promoter in muscle of larvae reared at 15, 18 and 21°C. Error bars indicate the standard error of the means for each treatment (n = 21). Different letters (and no common letters) indicate statistically significant differences between temperatures (p < 0.05) as determined by ANOVA. (B) Percentage of cytosine methylation at 21 CpG sites in Senegalese sole myog proximal promoter in muscle of larvae reared at different temperatures (15, 18 and 21°C). Different letters indicate significant differences between temperatures at each CpG position (p < 0.05), as determined by AMOVA. TSS represents the transcription starting site.
At several CpG sites, the cytosines were differentially methylated between temperature groups (Fig. 2B). Significant differences were observed at position -203 (located 4 bp downstream of a TAF binding site) (Fst = 0.122, p = 0.015), position -165 (Fst = 0.221, p = 0.000) and position -72 from the TSS (located in a TAF binding site and separated by 1 bp from a MEF2 site) (Fst = 0.148, p = 0.013). CpG sites were also identified in the MEF3 (-88 bp) and in the TAF binding sites (-209 bp), even if no significant differences were observed between temperatures. In fact, the CpG located in this latter TAF motif was one of the less methylated across all temperatures. Analysis of myog methylation in a non-muscle tissue (gut, n = 6) revealed that all CpG sites located in critical binding sites for muscle myog expression, such as MEF2 or MEF3, were 100% methylated at all temperatures (Fig. S1).
Expression of myog and dnmts homologs in Senegalese sole larvae changes with temperature
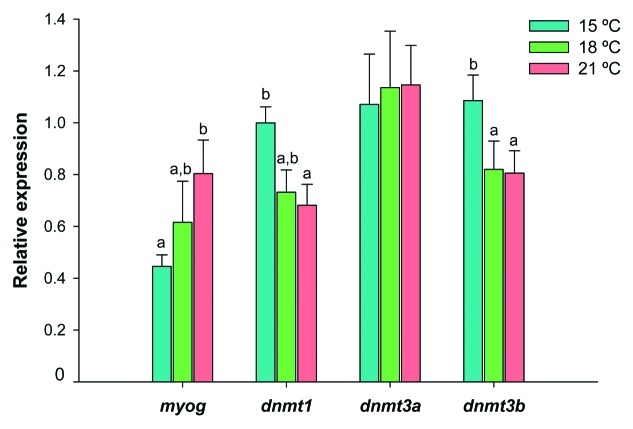
During metamorphosis, myog expression in Senegalese sole larvae was affected by rearing temperature (Fig. 3) and 1.8-fold higher at 21°C than at 15°C (p < 0.05). In contrast, dnmt1 expression was higher in the 15°C group than in the 21°C one (p < 0.05) and dnmt3b transcript levels were higher at 15°C than at either 18 or 21°C (p < 0.05). The dnmt3a paralogue had a uniform expression profile across temperatures (Fig. 3).
Figure 3. Relative expression of myog, dnmt1, dnmt3a and dnmt3b in Senegalese sole larvae reared at 15°C (blue bars), 18°C (green bars) and 21°C (red bars) until metamorphosis stage 2 (see main text for details). Transcript levels were determined by qPCR and normalized within each developmental stage, using eef1a1, rps4 and ubq as endogenous reference genes. Error bars indicate the standard error of the means for each treatment (n = 6). For each gene, significant differences between temperatures are indicated by different letters (without letters in common, p < 0.05).
Phenotypic plasticity of muscle growth in Senegalese sole larvae
Rearing temperature significantly influenced Senegalese sole fast skeletal muscle growth (Fig. 4). Larvae reared at 21°C showed a 1.6-fold increase in fiber diameter (p < 0.001) (Fig. 4A) (6.0 ± 2.9, 6.1 ± 2.6 and 9.5 ± 3.8 µm at 15°C, 18°C and 21°C, respectively) and a 3.1-fold increase in total cross sectional area (A) (p < 0.05) (Fig. 4B), relatively to fish from the 15°C group. The mean number of fast fibers (N) was not significantly different between temperatures, despite the moderately higher number of fibers of the 21°C treatment.
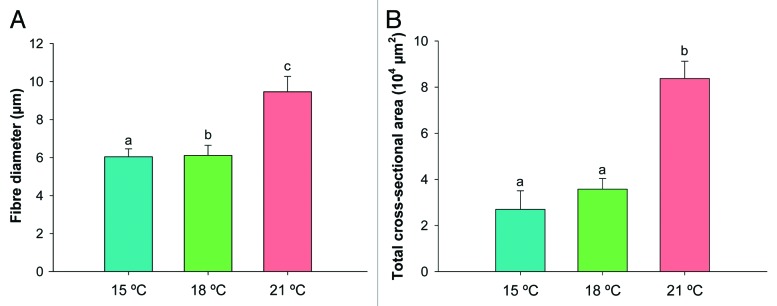
Figure 4. (A) Fiber diameter (µm) and (B) total cross-sectional muscle area A (µm2) of fast muscle in Senegalese sole larvae reared at 15°C (blue bars), 18°C (green bars) and 21°C (red bars). Error bars indicate the standard error of the means for each treatment (n = 6) and different letters indicate significant differences between temperatures (p < 0.05).
Discussion
The putative proximal promoter of Senegalese sole myog is highly conserved and its methylation levels in skeletal muscle increase with lower rearing temperatures
A higher methylation level of the myog promoter in skeletal muscle of Senegalese sole larvae was observed at 15°C both in the global methylation of the promoter and specifically in CpG sites (Fig. 2A and B). Non-CpG methylation has been shown to play an important role in mammals. For example, non-CpG hypermethylation of the peroxisome proliferator-activated receptor-γ co-activator promoter in skeletal muscle was found to decrease the mitochondrial density in diabetics type 2, contributing to impaired fat oxidation and excess lipid storage.19 In the Senegalese sole myog promoter, several CpC sites were also found dispersed throughout the sequence, raising the hypothesis that their differential methylation may influence myog transcription, alongside with methylation at specific CpG sites. It has been suggested that CpG and non-CpG methylation are in a dynamic equilibrium controlling myog transcription, and that active non-CpG demethylation during muscle differentiation occurs faster than CpG demethylation.11
The Senegalese sole myog promoter putative regulatory sequences were conserved relatively to other vertebrate species and it is likely that they play a similar role in controlling transcriptional activity. This region of the myog promoter contains several conserved DNA binding elements, such as the E-boxes, MEF2 and MEF3 binding sites and the TATA box. In the present study, three CpG sites were significantly hypermethylated at 15°C compared with 21°C, and most of the others CpGs also had higher methylation levels at 15°C, albeit not statistically significant (Fig. 2B). One of the 15°C-hypermethylated CpGs was located in a TAF (putative TATA box binding protein associated factor) binding site, which is involved in establishing the transcription initiation factor TFIID multimeric protein complex and plays a central role in mediating promoter responses to activation and repression.20 Another 15°C-hypermethylated CpG was also located 4 bp downstream of a putative TAF motif (-209 bp), which is relevant for the regulation of myog transcription. The putative TATA binding site was found to be highly conserved across species.
The MEF2 site, which is bound by elements of the MEF2 family of transcription factors (such as MEF2A, MEF2C or MEF2D), seems to be required for the activation of myog in some developmental contexts.21 In seabream (Sparus aurata) and zebrafish, deletion of the MEF2 binding site significantly reduced muscle-specific expression of myog.6,22 Given the known importance of the MEF2 regulatory motif, the status of cytosine methylation at this site is likely to interfere with the transcriptional activity of the myog promoter in Senegalese sole. The closest CpG site to the E-boxes significantly affected by temperature was the one located in a TAF and very close to the MEF2 motifs. It is plausible that the methylation status of regions flanking E-boxes in the of Senegalese sole myog promoter affect the binding of bHLH complexes, since it has been reported that flanking sequences, methylation status and interaction with adjacent regulatory elements contribute to selection of particular protein complexes by the E-boxes.23,24
Myog and dnmt expression are affected by temperature in Senegalese sole larvae
In teleosts, myog transcripts are present during early development from somitogenesis onwards14 and in post-embryonic stages its transcription is related to muscle growth by hyperplasia and hypertrophy.25 For example, in brown trout (Salmo trutta) larvae, myog expression persisted in zones of intense muscle hyperplasia at the dorsal and ventral apices of the myotome and next to the horizontal septum.26
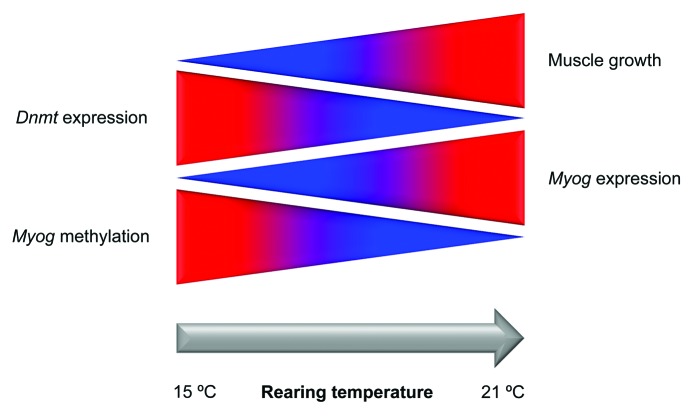
In a previous study with Senegalese sole juveniles, myog expression in fast skeletal muscle was influenced by diet and positively correlated with growth parameters such as daily growth index or protein gain, which is indicative of myog importance in growth and protein accretion processes.27 We have found that expression of myog in Senegalese sole larvae undergoing metamorphosis was lower at 15°C compared with 21°C. This is consistent with the cytosine methylation pattern of the myog proximal promoter observed in muscle at different rearing temperatures, supporting the hypothesis that methylation of CpG sites and/or non-CpG methylation may influence myog expression in Senegalese sole larvae (Fig. 5).
Figure 5. Simplified model of temperature-induced phenotypic plasticity of muscle growth in Senegalese sole. A higher rearing temperature (21°C vs. 15°C) lead to lower methylation levels of the myog proximal promoter in fast muscle, which correlated with a decrease in dnmt1 and dnmt3b transcripts and an increase in myog expression and muscle growth. Blue and red indicate lower and higher levels of methylation, gene expression and muscle growth, respectively.
Expression of dnmt1 and dnmt3b was significantly higher at 15°C during metamorphosis, whereas lower mRNA levels were observed in larvae from the 18 and 21°C° groups. This corroborates the methylation results discussed above, since Dnmts are directly involved in DNA methylation of cytosines. In mammals, DNA methylation patterns are somatically heritable through the action of Dnmt1, which is the maintenance methyltransferase.28 The Senegalese sole Dnmt1 putative partial protein was shown to be highly conserved (94% and 83% identity with zebrafish and human counterparts, respectively) and it is likely that it plays similar functions across different vertebrate taxa. Few studies on dnmt1 expression have been performed in fish but Rai et al.2 reported that zebrafish dnmt1 morphants exhibited a dramatic reduction of genomic cytosine methylation. Interestingly, expression patterns of dnmt3a and dnmt3b paralogues during zebrafish embryonic development were strikingly different. Moreover, temperature clearly influenced expression of these two genes in a different manner, suggesting that dnmt3 paralogues are diverging and that dnmt3a and dnmt3b may play different roles in thermal epigenetic regulation of gene expression during early development in zebrafish.29 Given the higher similarity of zebrafish dnmt3a across vertebrate taxa and low non-synonymous/synonymous substitution ratios in the sequence, dnmt3a may have a more conserved function in vertebrate physiology than dnmt3b.29 It is not known if a specific Dnmt targets specific cytosines according to its location in a CpG or non-CpG context in the myog promoter. Nevertheless, such possibility should not be discarded given the distinct expression of dnmt3 genes in Senegalese sole larvae among temperatures.
Muscle cellularity and growth are affected by temperature in Senegalese sole larvae
The ultimate size of a fish is shaped by the balance between recruitment and enlargement of muscle fibers and water temperature is one of the main constraints affecting muscle phenotype but the molecular basis of such phenotypic plasticity is still poorly understood.25 However, temperature impact on muscle growth depends largely on the species and on the temperatures to which they are exposed in their natural environment. For example, raising seabass larvae at 15°C induces a higher muscle growth than higher or lower temperatures, but in rainbow trout, optimal growth is observed at 4°C compared with 8°C.15
Senegalese sole is a species of relatively temperate waters and natural spawning of broodstock in captivity has been observed between 16°C and 22°C (for a review see ref. 17). In the present study, fast muscle growth and cellularity in this species were largely affected by rearing temperature. To the best of our knowledge, this is the first study indicating that an epigenetic mechanism such as myog methylation in muscle of Senegalese sole larvae reared at different temperatures can affect gene expression and muscle growth (Fig. 5). It is plausible that developmental time difference, an indirect effect of temperature, also affected DNA methylation and myog expression. It remains to be seen if the epigenetic events herein reported are conserved in other temperate-water fish species, with its implications for the aquaculture industry.
Material and Methods
Fish husbandry and sampling
The Senegalese sole incubation experiment took place at the LEOA facility, University of Algarve, Portugal. Embryos were incubated in a 200L cylindro-conical tank at 20.2 ± 0.5°C. Newly hatched larvae were then transferred to 9 fiberglass conical tanks (100 L) tanks in a closed recirculation system with an initial density of 60 larvae L−1. Water temperature, salinity, O2, pH and nitrogenous compounds were monitored regularly and larvae were exposed to an artificial photoperiod of 12h light: 12h dark. They were reared at three temperatures: 15.2 ± 0.5, 18.3 ± 0.6 or 21.1 ± 0.4°C, in triplicate groups until complete metamorphosis. At mouth opening, larvae were fed rotifers (Brachionus sp) enriched with DHA Selco® (Inve) for 6 h prior to harvesting. Artemia AF Strain nauplii (na) (Inve) were introduced between 5 dph and 8 dph and metanauplii between 9 and 16 dph, according to rearing temperature. Artemia enrichment was done at 250,000 nauplii/metanauplii L−1, with 0.4 g L−1 supplementation of Easy DHA Selco® (Inve) and Micronised Fishmeal® (Ewos) added in two doses of a 1: 1 mixture (weight basis).
Metamorphic larvae (Met) at stage 2, according to the eye-translocation stage,30 were sampled at 12 dph at 21°C, 15 dph at 18°C and 23 dph at 15°C. Larvae were killed by over-anesthesia with MS-222 (400 mg·L−1). Samples for nucleic acid extraction were then snap-frozen in liquid nitrogen and stored at -80°C. Dry weight (DW, mg) was measured on -80°C freeze-dried sole larvae (n = 3 in pools of 25 larvae, ± 0.001 mg) and total length (LT, mm) was measured on 20–30 larvae per tank using an analytical scales.
Animal handling protocols were conducted according to the directive of November 24th, 1986 (86/609/EEC) from the European Economic Community concerning animal experimentation guidelines.
RNA and genomic DNA extraction
Total RNA was extracted from 6 pools of 15 larvae per temperature group (2 pools per tank) using Qiazol (Qiagen). Assessment of RNA quality was performed by agarose gel electrophoresis and its quantity determined with a Nanodrop spectrophotometer (Nanodrop Technologies/Saven Werner). Absorbance ratios (260/280 nm) were greater than 1.9, indicating high purity RNA. Genomic DNA was isolated from larvae skeletal muscle (6 larvae per temperature) using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer’s instructions. Quality and quantity of genomic DNA were determined as indicated above for RNA. Genomic DNA was also extracted from Senegalese sole gut (n = 6), to serve as a control of methylation levels (see below).
Cloning and sequencing the putative 5`UTR and proximal promoter of the myog gene
Senegalese sole myog putative 5`UTR and promoter were isolated by PCR as two overlapping DNA fragments from sole DNA libraries using the Genome Walker™ Universal Kit (Clontech). Briefly, Senegalese sole genomic DNA was completely digested with restriction enzymes (DraI, EcoRV, PvuII and StuI) to produce blunt-ended DNA fragments. The digested DNA was then ligated with a DNA adaptor (Clontech) and the resulting DNA fragments were used as templates for two rounds of primary and nested PCR amplifications using two adaptor-specific primers together with two myog specific primers (GSPs) in the first exon (GenBank accession number EU934044). The remaining sequence of the first exon, the 5` flanking region and part of the promoter was isolated using the GSP1 5`-CGCCTCCAGACAGACTCGCACACAAG and GSP2 5`-CTGGTCAGGGAAGAAATAGGGGTTGGTC; the remaining part of the promoter was isolated using the GSP1 5`- GGACGGATAATCTCGGATCAAATATGCC, GSP2 5`-TCTTGTCTCCCTCCATGACACCATACAT. Cloning and sequencing were essentially performed as previously reported.27
DNA bisulfite treatment and sequencing
Bisulfite modification of muscle genomic DNA extracted from stage 2 larvae was performed using the EpiTect Bisulfite Kit (Qiagen), according to the manufacturers’ instructions. Bisulfite treated DNA from each temperature group (15, 18 and 21°C) were pooled, and then amplified by PCR using the following primers: forward, 5`-ATGTATGGTGTTATGGAGGGAGA; reverse, 5`-ACTAAACAACGCTATAATCTAAA TTA). A 523 bp fragment of the putative 5`UTR and promoter of the Senegalese sole myog gene were obtained. PCR products were gel-purified using the QIAquick gel extraction kit (Qiagen) and cloned onto a pCR4-TOPO® plasmid vector (Invitrogen), as reported.27 Twenty one individual colonies per temperature were picked and lysed in H20 at 99°C for 5 min. Following centrifugation at 16 x g for 3 min, 1 µl of the supernatant was used for PCR amplification with M13 primers and an annealing temperature of 55°C. PCR products were visualized on agarose gel, purified with the ExoSAP-IT® PCR Product Cleanup (Affymetrix) and sequenced as described elsewhere.27
The efficiency of the bisulfite conversion step and methylation levels at each CpG site were evaluated with the Bisulfite Sequencing data Presentation and Compilation software (http://biochem.jacobsuniversity.de/BDPC/) and the BiQ Analyzer (http://biq-analyzer.bioinf.mpi-inf.mpg.de/). The mean percentage of converted cytosines was 96.2, 96.4 and 96.6% for the 15, 18 and 21°C group, respectively. The total number of methylated cytosines (CpG sites + non-CpG sites) was calculated as the average of total number of methylated cytosines across the 21 clones, for each temperature.
Dnmt cloning and sequencing
BLAST similarity searches against the nr database (http://blast.ncbi.nlm.nih.gov) and Ensembl (http://www.ensembl.org/) were performed to identify orthologs of dnmt1, dnmt3a and dnmt3b in other teleost species. Degenerate primers were designed against the most conserved regions of the sequences. Cloning and sequencing were essentially performed as previously reported.27 DNA sequences were analyzed with the CodonCode Aligner v.3.7.1 software (CodonCode Corporation) and their identity determined by BLASTN similarity searches against the NCBI and Ensembl databases. Partial coding sequences of 259 bp, 750 bp and 469 bp were obtained for Senegalese sole dnmt1, dnmt3a and dnmt3b, respectively (See Table 2 for GenBank Accession numbers).
Table 2. Primers used in qPCR.
| Gene | Forward sequence (5′→3′) | Reverse sequence (5′→3′) | Accession (GenBank) | Size (bp) | E (%) |
|---|---|---|---|---|---|
|
myog |
GTCACAGGAACAGAGGACAAAG |
TGGTCACTGTCTTCCTTTTGC |
EU934044 |
118 |
92 |
|
dnmt1 |
GATCCCAGTGAGGAGTACGG |
AAGAAGGTCCTCATAAGTAGCGTC |
KC129104 |
117 |
93 |
|
dnmt3a |
AACTGCTGTAGGTGTTTCTGTGTG |
CGCCGCAGTAACCCGTAG |
KC129105 |
134 |
90 |
|
dnmt3b |
ATCAAGCGATGTGGCGAGC |
CGATGCGGTGAAAGTCAGTCC |
KC129106 |
91 |
97 |
|
eef1a1 |
ATTGGCGGCATTGGAACA |
CATCTCCACAGACTTGACCTC |
AB326302 |
117 |
91 |
|
rps4 |
CTGCTGGATTCATGGATGTG |
GGCAGTGATGCGGTGGAC |
AB291557 |
101 |
90 |
| ubq | TCTGCGTGGTGGTCTCATC | TGACCACACTTCTTCTTGCG | AB291588 | 135 | 92 |
For each gene, its GenBank accession number, amplicon size (bp) and amplification efficiency (E) are indicated. The annealing temperature of all primer pairs is 60°C.
Quantitative real-time PCR (qPCR)
One microgram of total RNA from each pool of larvae during metamorphosis was used to synthesize cDNA with the QuantiTect reverse transcription kit (Qiagen), as reported elsewhere.31 Specific qPCR primers were designed for the sole myog and dnmts sequences. Whenever possible, primers were designed to span at least one intron/exon border to avoid amplification of potential contaminating genomic DNA, and then analyzed with Netprimer (http://www.premierbiosoft.com/), as previously described.31 Elongation factor 1α (eef1a1), ribosomal protein 4 (rps4) and ubiquitin (ubq) were used as endogenous reference genes to normalize target gene expression. Primer sequences, amplicon sizes and qPCR amplification efficiencies are shown in Table 2. Quantification of gene expression was performed by qPCR with SYBR Green chemistry (Roche) on a LightCycler® 480 (Roche), as detailed elsewhere.29,31
Muscle morphometry
Two fish at stage 2 metamorphosis were collected per tank and fixed in 4% paraformaldehyde (Sigma-Aldrich) in phosphate buffered saline (PBS tablets, Sigma-Aldrich) for 6–12 h, and washed in PBS. Samples were then dehydrated in a graded ethanol (AGA) series, cleared in xylol (Prolabo, VWR International LLC) and finally included in paraffin Histosec® (Merck). Larvae were sectioned (10 µm) transversely to the body axis just posterior to the anus. Sections were then mounted on slides coated with aminopropyltriethoxysilane (APES) (Sigma-Aldrich), to improve adhesion and then stained with hematoxylin–eosin (Merck). Total cross-sectional muscle area [A (mm2) (muscle)], the total number of fiber [N (fibers)] and the fiber cross-sectional area [ā (µm2) (muscle fiber)] were measured. The total cross-sectional muscle area [A (fast muscle)] was computed after tracing the physical limits of interest of the section on the monitor, at a 200 × magnification. The outlines of muscle fiber were traced using a 400 × magnification. The fiber diameter (µm) was estimated indirectly, as the diameter of a circle having the same area of a fiber in a perfect cross-section. This study was performed using an Olympus BX51 microscope (Olympus Europa GmbH) with the Cell^B Basic imaging software.
Statistical analysis
The effects of the temperature on fast muscle fibers were evaluated using a covariance analysis (ANCOVA), in which temperature was set as the independent variable while the total length was set as a covariate.
Evaluation of expression stability for the three reference genes was performed using the statistical application geNorm (http://medgen.ugent.be/), as previously reported.31 Expression of target genes was evaluated by the relative quantification method.31 Differences in the expression of target genes with temperature were examined by a one-way ANOVA with Holm-Sidak post-hoc tests using the SigmaPlot 11.0 statistical package (Systat software). When the data did not meet the normality or equal variance requirements, a Kruskal-Wallis one-way ANOVA on ranks with a median test was performed instead. Significance levels were set at p < 0.05.
To check for differences in methylation levels at specific CpG positions of the myog putative promoter, an analysis of molecular variance (AMOVA) was performed using the Arlequin 3.5.1.2 software (http://cmpg.unibe.ch/software/arlequin3/). Each CpG site was trimmed from the original sequence (7–26 nucleotides in length), with two possible variants for each cytosine nucleotide: C if methylated and T if unmethylated. Then, all sequences from the same temperature were considered as one group, with size equivalent to the number of sequences analyzed for each temperature (n = 21). Significance level was set at p < 0.05.
Supplementary Material
Acknowledgments
The authors acknowledge J. Sendão, M.F. Castanheira, H. Teixeira, F. Rocha and A. Santos (CCMAR, Portugal), for their invaluable help during the experimental setup and sampling of Senegalese sole larvae. We are grateful to A. Sundaram (University of Nordland, Norway) for his assistance during the laboratory experiments. This study was supported by Project EPISOLE - PTDC/MAR/110547/2009, granted by Fundação para a Ciência e a Tecnologia and Programa Operacional Temático Factores de Competitividade (COMPETE), FEDER, with additional support from the GrowCod project (ref. 190350/S40) funded by the Research Council of Norway. C. Campos and S. Engrola acknowledge the financial support by Fundação para a Ciência e Tecnologia, Portugal, through grants SFRH/BD/43633/2008 and SFRH/BPD/49051/2008, respectively.
Disclosure of Potential Conflicts of Interest
There is no conflict of interest that I should disclose. This work represents unbiased, independent research funded by public agencies from Portugal and Norway.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/24178
References
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, et al. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–85. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–59. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 2001;21:2404–12. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du SJ, Gao J, Anyangwe V. Muscle-specific expression of myogenin in zebrafish embryos is controlled by multiple regulatory elements in the promoter. Comp Biochem Physiol B Biochem Mol Biol. 2003;134:123–34. doi: 10.1016/S1096-4959(02)00194-X. [DOI] [PubMed] [Google Scholar]
- 7.Tan X, Hoang L, Du S. Characterization of muscle-regulatory genes, Myf5 and myogenin, from striped bass and promoter analysis of muscle-specific expression. Mar Biotechnol (NY) 2002;4:537–45. doi: 10.1007/s10126-002-0013-1. [DOI] [PubMed] [Google Scholar]
- 8.Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, et al. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci U S A. 1998;95:14220–5. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng TC, Wallace MC, Merlie JP, Olson EN. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science. 1993;261:215–8. doi: 10.1126/science.8392225. [DOI] [PubMed] [Google Scholar]
- 10.Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7(7A):1277–89. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- 11.Fuso A, Ferraguti G, Grandoni F, Ruggeri R, Scarpa S, Strom R, et al. Early demethylation of non-CpG, CpC-rich, elements in the myogenin 5′-flanking region: a priming effect on the spreading of active demethylation. Cell Cycle. 2010;9:3965–76. doi: 10.4161/cc.9.19.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palacios D, Summerbell D, Rigby PWJ, Boyes J. Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Mol Cell Biol. 2010;30:3805–15. doi: 10.1128/MCB.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hupkes M, Jonsson MK, Scheenen WJ, van Rotterdam W, Sotoca AM, van Someren EP, et al. Epigenetics: DNA demethylation promotes skeletal myotube maturation. FASEB J. 2011;25:3861–72. doi: 10.1096/fj.11-186122. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes JM, Mackenzie MG, Wright PA, Steele SL, Suzuki Y, Kinghorn JR, et al. Myogenin in model pufferfish species: Comparative genomic analysis and thermal plasticity of expression during early development. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:35–45. doi: 10.1016/j.cbd.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Wilkes D, Xie SQ, Stickland NC, Alami-Durante H, Kentouri M, Sterioti A, et al. Temperature and myogenic factor transcript levels during early development determines muscle growth potential in rainbow trout (Oncorhynchus mykiss) and sea bass (Dicentrarchus labrax) J Exp Biol. 2001;204:2763–71. doi: 10.1242/jeb.204.16.2763. [DOI] [PubMed] [Google Scholar]
- 16.Vinagre C, Fonseca V, Cabral H, Costa MJ. Habitat suitability index models for the juvenile soles, Solea solea and Solea senegalensis, in the Tagus estuary: Defining variables for species management. Fish Res. 2006;82:140–9. doi: 10.1016/j.fishres.2006.07.011. [DOI] [Google Scholar]
- 17.Imsland AK, Foss A, Conceição LEC, Dinis MT, Delbare D, Schram E, et al. A review of the culture potential of Solea solea and S. senegalensis. Rev Fish Biol Fish. 2003;13:379–407. doi: 10.1007/s11160-004-1632-6. [DOI] [Google Scholar]
- 18.Campos C, Valente LMP, Conceição LE, Engrola S, Sousa V, Rocha E, et al. Incubation temperature induces changes in muscle cellularity and gene expression in Senegalese sole (Solea senegalensis) Gene. 2013;516:209–17. doi: 10.1016/j.gene.2012.12.074. [DOI] [PubMed] [Google Scholar]
- 19.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–98. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–42. doi: 10.1016/0968-0004(96)10044-X. [DOI] [PubMed] [Google Scholar]
- 21.Faralli H, Dilworth FJ. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp Funct Genomics. 2012;2012:836374. doi: 10.1155/2012/836374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Codina M, Bian YH, Gutiérrez J, Du SJ. Cloning and characterization of myogenin from seabream (Sparus aurata) and analysis of promoter muscle specificity. Comp Biochem Physiol Part D Genomics Proteomics. 2008;3:128–39. doi: 10.1016/j.cbd.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Salero E, Giménez C, Zafra F. Identification of a non-canonical E-box motif as a regulatory element in the proximal promoter region of the apolipoprotein E gene. Biochem J. 2003;370:979–86. doi: 10.1042/BJ20021142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lüscher B, Larsson LG. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene. 1999;18:2955–66. doi: 10.1038/sj.onc.1202750. [DOI] [PubMed] [Google Scholar]
- 25.Johnston IA. Environment and plasticity of myogenesis in teleost fish. J Exp Biol. 2006;209:2249–64. doi: 10.1242/jeb.02153. [DOI] [PubMed] [Google Scholar]
- 26.Steinbacher P, Haslett JR, Obermayer A, Marschallinger J, Bauer HC, Sänger AM, et al. MyoD and Myogenin expression during myogenic phases in brown trout: a precocious onset of mosaic hyperplasia is a prerequisite for fast somatic growth. Dev Dyn. 2007;236:1106–14. doi: 10.1002/dvdy.21103. [DOI] [PubMed] [Google Scholar]
- 27.Campos C, Valente LM, Borges P, Bizuayehu T, Fernandes JM. Dietary lipid levels have a remarkable impact on the expression of growth-related genes in Senegalese sole (Solea senegalensis Kaup) J Exp Biol. 2010;213:200–9. doi: 10.1242/jeb.033126. [DOI] [PubMed] [Google Scholar]
- 28.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 29.Campos C, Valente LM, Fernandes JM. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene. 2012;500:93–100. doi: 10.1016/j.gene.2012.03.041. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez-Diaz C, Yufera M, Canavate JP, Moyano FJ, Alarcon FJ, Diaz M. Growth and physiological changes during metamorphosis of Senegal sole reared in the laboratory. J Fish Biol. 2001;58:1086–97. doi: 10.1111/j.1095-8649.2001.tb00557.x. [DOI] [Google Scholar]
- 31.Fernandes JMO, Mommens M, Hagen O, Babiak I, Solberg C. Selection of suitable reference genes for real-time PCR studies of Atlantic halibut development. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:23–32. doi: 10.1016/j.cbpb.2008.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.