The hexameric Mcm2–7 complex, which is composed of the evolutionarily related Mcm2 to Mcm7 subunits, forms the core of the eukaryotic replicative helicase (Fig. 1). In the late M to G1 phase, Mcm2–7 is recruited to replication origins with the assistance of at least three essential replication factors, ORC, Cdc6 and Cdt1, to form the pre-replicative complex (pre-RC). In the S phase, Cdc45 and GINS associate with Mcm2–7 via the action of two kinases, DDK and S-CDK, and other replication factors to assemble the active Cdc45-Mcm-GINS helicase, which translocates away from origins to unwind double-stranded DNA at replication forks.1 Mcm2–7 appears to be the key factor for the control of DNA replication, as most known regulatory mechanisms are dedicated to Mcm2–7, rather than to DNA polymerases.
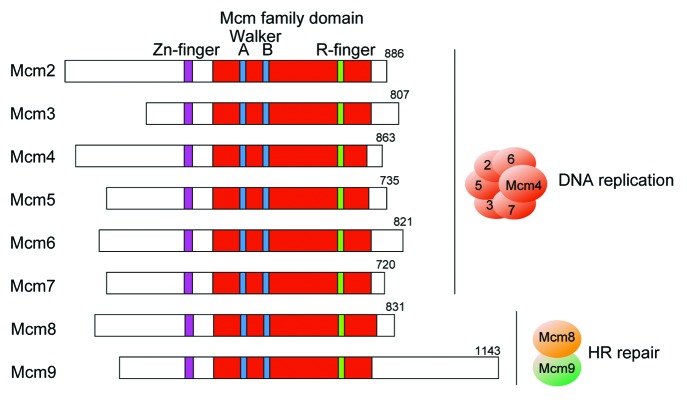
Figure 1. Schematic illustration of the Mcm family proteins of Xenopus laevis. The Walker A and B motifs within the Mcm family domain, and the zinc (Zn)- and arginine (R)-finger motifs are indicated. Mcm2 to Mcm7 form a hexameric complex that is essential for DNA replication. Mcm8 and Mcm9 form a dimer that is likely to be involved in HR repair.
Mcm8 and Mcm9 are evolutionarily related to the other Mcm proteins, all of which contain the Walker A and B motifs for ATP hydrolysis within the MCM family domain, as well as the zinc- and arginine-finger motifs (Fig. 1). A phylogenetic analysis has suggested that the last common ancestor of eukaryotes had the MCM8 and MCM9 genes.2 These two genes are conserved in many eukaryotic species but seem to have been lost together in yeast and C. elegans, suggesting the co-evolution of Mcm8 and Mcm9. Drosophila species are an exception, as they have only the MCM8 homolog, termed REC.2 As can be imagined, based on the role of Mcm2–7 in DNA replication, early studies suggested independent and controversial roles for Mcm8 and Mcm9 in DNA replication. In Xenopus egg extracts, Mcm8 was shown to be involved in elongation of the replication fork.3 Conversely, in the same system, Mcm9 was reported to interact with Cdt1 for the loading of Mcm2–7, i.e., pre-RC formation.4 Recently, knockout mice and chicken DT40 cells lacking the MCM8 or MCM9 gene were successfully generated, clearly indicating that the two proteins are not essential for DNA replication in these organisms.5-7 Moreover, consistent with the notion that the MCM8 and MCM9 genes have co-evolved in most eukaryotes, Mcm8 and Mcm9 form a complex and are required for tolerance to DNA damage, suggesting that the Mcm8–9 complex plays a role in DNA repair.6,7
In the April 15, 2013 issue of Cell Cycle, Gambus and Blow re-examined the role of Mcm8 and Mcm9 in DNA replication in Xenopus egg extracts to address the discrepancies observed in previous reports. As is the case with humans, mice and chickens,6,7 the authors demonstrated that Xenopus Mcm8 and Mcm9 form a stable complex; however, neither of them associates with Cdt1 and Mcm2–7. Using glycerol gradient and size-exclusion experiments, the Mcm8–9 complex was found to be a dimer, unlike the previous suggestion of the formation of a hexamer in DT40 cells.7 During the replication of sperm chromatin DNA, Mcm8 and Mcm9 associate with chromatin from the late S to G2 phase, suggesting that they are not essential for DNA replication. In fact, depletion of Mcm9 from egg extracts, which eventually co-depletes Mcm8, has a marginal effect on normal DNA replication. Finally, the authors showed that various types of DNA damage induce association of Mcm8–9 to chromatin, which is consistent with the observation that Mcm8 and Mcm9 are required for resistance to DNA damage.6,7
The most important future question regarding this issue is: “What is the role of the Mcm8–9 complex in resistance to DNA damage?” Taking into account the fact that the knockout mice and DT40 cells lacking MCM8 or MCM9 exhibit a severe defect in gametogenesis and homologous recombination (HR), respectively,5-7 it is likely that the Mcm8–9 complex has a conserved function in HR repair. Another question is: “Does the Mcm8–9 complex function as a helicase?” If so, is the dimeric Mcm8–9 complex converted to a hexameric complex similar to Mcm2–7, and are there any accessory factors, such as Cdc45 and GINS in the case of Mcm2–7, that are required for its helicase activity? The characterization of Mcm8 and Mcm9 might shed light on a new mechanism of HR repair. Future analyses are warranted.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24670
References
- 1.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Richards TA, Aves SJ. Ancient diversification of eukaryotic MCM DNA replication proteins. BMC Evol Biol. 2009;9:60. doi: 10.1186/1471-2148-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maiorano D, Cuvier O, Danis E, Méchali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–28. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Lutzmann M, Méchali M. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol Cell. 2008;31:190–200. doi: 10.1016/j.molcel.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Hartford SA, Luo Y, Southard TL, Min IM, Lis JT, Schimenti JC. Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression. Proc Natl Acad Sci USA. 2011;108:17702–7. doi: 10.1073/pnas.1113524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, et al. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell. 2012;47:523–34. doi: 10.1016/j.molcel.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura K, Ishiai M, Horikawa K, Fukagawa T, Takata M, Takisawa H, et al. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol Cell. 2012;47:511–22. doi: 10.1016/j.molcel.2012.05.047. [DOI] [PubMed] [Google Scholar]


