Abstract
The transcription factor p63 is critical for many biological processes, including development and maintenance of epidermal tissues and tumorigenesis. Here, we report that the TAp63 isoforms regulate cell metabolism through the induction of the mitochondrial glutaminase 2 (GLS2) gene both in primary cells and tumor cell lines. By ChIP analysis and luciferase assay, we confirmed that TAp63 binds directly to the p53/p63 consensus DNA binding sequence within the GLS2 promoter region. Given the critical role of p63 in epidermal differentiation, we have investigated the regulation of GLS2 expression during this process. GLS2 and TAp63 expression increases during the in vitro differentiation of primary human keratinocytes, and depletion of GLS2 inhibits skin differentiation both at molecular and cellular levels. We found that GLS2 and TAp63 expression are concomitantly induced in cancer cells exposed to oxidative stresses. siRNA-mediated depletion of GLS2 sensitizes cells to ROS-induced apoptosis, suggesting that the TAp63/GLS2 axis can be functionally important as a cellular antioxidant pathway in the absence of p53. Accordingly, we found that GLS2 is upregulated in colon adenocarcinoma. Altogether, our findings demonstrate that GLS2 is a bona fide TAp63 target gene, and that the TAp63-dependent regulation of GLS2 is important for both physiological and pathological processes.
Keywords: glutaminolysis, p63, skin differentiation, colon carcinoma
Introduction
p63 is a transcription factor belonging to the p53 family.1 Like other members of the p53 family,2 p63 is expressed as multiple isoforms arising by alternative promoter usage or differential splicing events at the C terminus.3,4 In particular, TAp63 isoforms contain a complete N-terminal transcriptional domain and are capable of efficiently transactivating different p53-responsive genes, thus largely mimicking p53 tumor-suppressive activities.5 Accordingly, ectopic expression of TAp63 can induce cell cycle arrest and apoptosis in different tumor cell lines, and different cellular stresses induce TAp63 expression by both transcriptional and post-translational mechanisms.6-9 ΔNp63, which lacks the N-terminal transcriptional domain, mainly acts as a dominant negative of p53 and TAp63 isoforms, even though it posses two transactivation domains,10-12 which endows it with a specific transcriptional activity, mainly towards epithelial specific genes.13-15 Accordingly, genetic studies in mice have indicated that p63 is crucially involved in the maintenance of epithelial stem cells,16-18 and mice genetically depleted of p63 display a complete lack of skin as well as defects in limb development and other epithelial-derived tissues.16,19,20 The role of p63 in skin formation has been further supported by ex vivo and in vitro experiments indicating a prevalent expression of ΔNp63 in the basal layer of the epidermis.21,22
Although the p63 gene is rarely mutated in human tumors, numerous studies have highlighted the importance of p63 in tumor development, and recent reports have demonstrated that TAp63 acts as a putative tumor suppressor gene.23-25 Loss or impaired expression of TAp63 has been associated with tumor progression and poor prognosis in some invasive human cancers,26 and TAp63-knockout mice develop metastatic tumors, suggesting that the deregulated expression of TAp63 isoforms could be important in regulating tumor progression and metastasis.27 Although TAp63 activity has been recently linked to the regulation of lipid and glucose metabolism,28 its role in the regulation of energy metabolism and cellular antioxidant defenses is not completely characterized.
Enhanced mitochondrial glutamine catabolism and increased rate of glycolysis are the two major metabolic alterations exhibited by many cancer cells in order to support their rapid proliferation.29-36 Glutaminolysis involves the initial deamination of glutamine by glutaminase, yielding glutamate and ammonia. In mammals, two different genes located on distinct chromosomes encode for glutaminase enzymes, the kidney-type isozymes (GLS1) and the liver-type isozymes GLS2.37,38 Although GLS1 and GLS2 share a considerable degree of sequence similarity, the two enzymes have different kinetic, immunological and molecular features. GLS1 is activated by high phosphate levels and is inhibited by the end-product glutamate, while GLS2 is activated by low phosphate levels and is not inhibited by glutamate.39 Although GLS2 has been initially reported to be expressed specifically in postnatal liver, recent evidence indicate that this enzyme is also expressed in other tissues and in some cancer cells.40,41
On the basis of their ability to convert glutamine into glutamate, an important intermediate for energy production and antioxidant cellular defense, it is not surprising that glutaminase expression is often deregulated in cancer cells. Loss of GLS2 expression has been reported in hepatocellular carcinomas, in highly malignant glioblastoma and anaplastic astrocytomas.40,41 Moreover, the ectopic expression of GLS2 reduces cell colony formation in hepatocarcinoma cell lines, suggesting that GLS2 might negatively regulate tumorigenesis.42
Although it is currently well established that the tumor-suppressive activity of p53 is intimately linked to the transcriptional regulation of a subset of genes involved in cell metabolism, energy production and intracellular redox control,43 our knowledge on the potential link between TAp63 activity and cell metabolism is limited. Here we report that TAp63 isoforms are able to induce the expression of GLS2, and that the TAp63-dependent regulation of GLS2 is an important mechanism regulating cell differentiation and ROS-dependent apoptosis.
Results
GLS2 is a p63 target gene
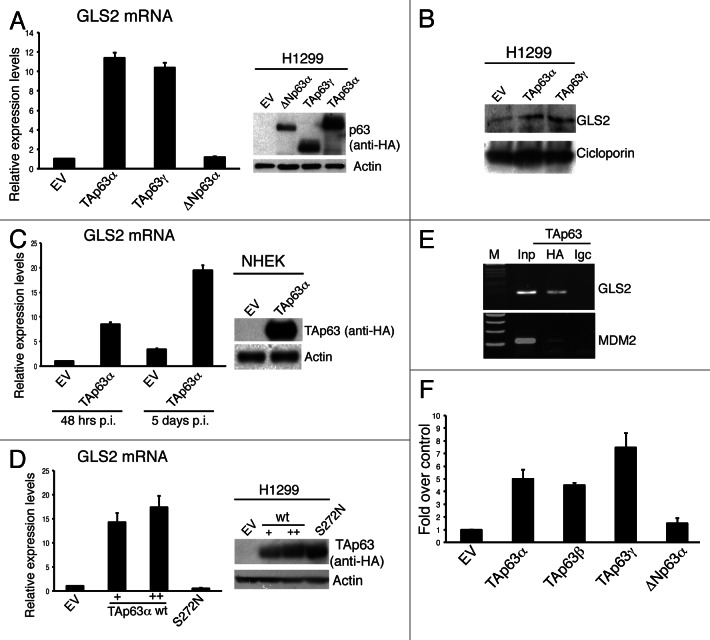
In a previous screening for epithelial transcriptional targets of p63,17,21 we observed a potential regulation of metabolic genes. Among these, we decided to further validate GLS2 as p63 target gene. To this end, we firstly verified whether p63 is able to regulate GLS2 expression. We overexpressed different p63 isoforms in the p53-null H1299 lung adenocarcinoma cell line and analyzed GLS2 mRNA levels by qRT-PCR. As shown in Figure 1A, ectopic expression of TAp63α and TAp63γ, but not ΔNp63α, isoforms markedly increased GLS2 expression. In order to validate this data at the protein level, we extracted mitochondrial proteins from transfected cells and analyzed GLS2 protein levels. As shown in Figure 1B, GLS2 protein levels were also upregulated upon TAp63 expression. Furtermore, retrovirally expressed TAp63α was able to induce GLS2 expression in normal human epidermal keratinocytes (NHEK) (Fig. 1C).
Figure 1. GLS2 expression is regulated by TAp63. (A) H1299 cells were transfected with the indicated HA-tagged p63 constructs and, 24 h after transfection, total RNA was extracted and utilized for reverse transcription and quantitative real-time PCR (qRT-PCR) (left panel) using specific primers for human GLS2 and β actin (for quantity normalization). Results are shown as mean ± SD of three independent experiments. Concomitantly, whole-cell extracts were utilized for western blot analysis using the antibodies to the indicated proteins (right panel). (B) H1299 cells were treated as in (A), and mitochondrial proteins were extracted (see Materials and Methods) and analyzed by IB using antibodies to the indicated proteins. (C) NHEK cells were retrovirally infected with HA-tagged TAp63α expressing virus and 48 h or 5 d after infection GLS2 mRNA levels were measured by q-RT-PCR (left panel). TAP63 protein levels were quantified by immunoblotting (IB) using anti-HA antibody (right panel). (D) H1299 cells were transfected, as indicated, with an empty vector (EV), increasing amount of HA-tagged wild-type TAp63α, or HA-tagged TAp63α (S272N) mutant. Transfected cells were either subjected to RNA isolation for the quantification of GLS2 mRNA by qRT-PCR (left panel) or to immunoblotting for the analysis of the expression levels of wild-type and mutant TAp63α proteins using antibodies to the indicated proteins. (E) Chromatin immunoprecipitation analysis of the human GLS2 promoter was carried out by purifying chromatin from Tet-On/HA-TAp63α-SaOs 2 inducible cell line treated with doxycycline for 24 h and then immunoprecipitating it using HA-specific antibody or IgC-unspecific antibody (see also Materials and Methods). Binding of TAp63α to the MDM2 promoter was used as a control. (F) HEK293E cells were transfected with pGL2 luciferase gene construct holding human GLS2 promoter fragment with either an empty vector (EV) or with the indicated HA-tagged p63 constructs. Co-transfection of a renilla luciferase control plasmid was used to normalize the transfection efficiency. Luciferase assay was performed 24 h after transfection. For all panels, data are shown as the mean ± SD of three replicates.
TAp63 acts as a transcription factor able to bind the DNA promoter region of its target genes.5,44 In order to verify whether the TAp63-dependent regulation of GLS2 depends on the ability of TAp63 to bind DNA, we transfected H1299 cells with vectors encoding wild-type TAp63α or its S272N ectodermal dysplasia-derived mutant, which is unable to bind DNA.45 In contrast to wild-type, TAp63α S272N mutant did not induce GLS2 expression (Fig. 1D). Analysis of the human GLS2 promoter region revealed a p53/p63 consensus DNA binding element located at −787 with respect to the transcription start. To verify direct binding of TAp63α to the putative responsive element within the GLS2 promoter in vivo, we performed a chromatin immunoprecipitation assay (ChIP) using SAOS-2 cells in which the expression of HA-tagged TAp63α can be induced by doxicyclin. As shown in Figure 1E, we found that TAp63 binds to a DNA fragment of the GLS2 promoter containing the p53/p63-responsive element. To confirm that this DNA binding element confers TAp63-dependent transcriptional activity, we performed luciferase assays using the pGL2 firefly luciferase reporter vector in which one copy of this p53/p63 binding element was cloned upstream of the firefly reporter genes. The expression of TAp63 isoforms greatly enhanced luciferase activity, while ΔNp63α had no effect (Fig. 1F). All these results demonstrate the TAp63 is able to regulate GLS2 expression by recognizing the p53/p63 binding element located in the GLS2 promoter.
GLS2 regulates skin differentiation
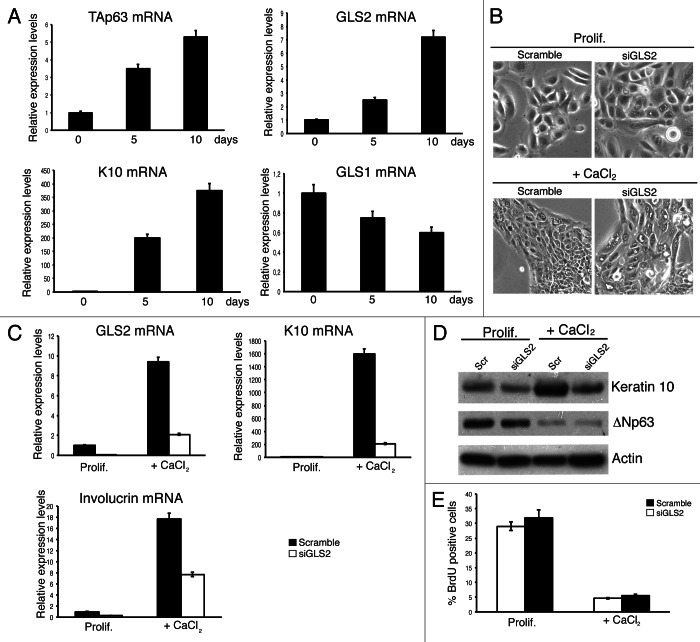
It is has been proposed that ΔNp63 is essential for the maintenance of the progenitor population in the basal layer compartment, while TAp63 would be required to allow complete suprabasal differentiation.46 Since GLS2 expression has been associated with cell differentiation, and its expression is not restricted to liver tissue,40 we analyzed the expression of GLS2 by qRT-PCR during the in vitro differentiation of normal human epidermal keratinocytes (NHEK). We found that GLS2 and TAp63 expression are upregulated in parallel during skin differentiation (Fig. 2A). In contrast, GLS1 expression showed a small, non-significant decrease during the in vitro differentiation. To assess the importance of GLS2 in skin differentiation, we used an siRNA approach to downregulate GLS2 in human keratinocytes. As shown in Figure 2B, GLS2-depleted keratinocytes showed a massive cell vacuolization, which is more evident under differentiation conditions. In some cells, especially at later times, several vacuoles seemed to have fused together. These morphological changes were accompanied by dysregulated expression of some skin differentiation markers. Specifically, GLS2 depletion reduced the upregulation of keratin 10 and involucrin both at the mRNA (Fig. 2C) and protein level (Fig. 2D). However, GLS2 depletion did not influence proliferation, as assessed by BrdU incorporation (Fig. 2E), suggesting that the defects of keratinocyte differentiation are not due to deregulation of cell cycle. These results suggest that GLS2 is important in keratinocyte differentiation.
Figure 2.
GLS2 regulates keratinocytes differentiation. Normal human epidermal keratinocytes (NHEKs) were cultured in KBM medium with KGM-2 growth supplements. Cells were induced to differentiate by adding 1.2 mM CaCl2 to the culture medium. Cells were collected at the indicated time points, and total RNA was isolated and utilized to quantify by qRT-PCR the mRNA levels of TAp63, GLS2, keratin 10 (K10) and GLS1. K10 was used as control of keratinocytes differentiation. (B) Morphology of NHEKs after GLS2 depletion. NHEKs were transfected twice with siRNA oligos targeting a non-relevant mRNA (scramble) or GLS2 mRNA. After 24 h of the second transfection, cells were induced to differentiate by adding 1.2 mM CaCl2 to the culture medium. Cell images, by phase-contrast microscopy, were taken both in proliferating condition and after 3 d in differentiation medium. (C) GLS2, K10 and involucrin mRNA levels were quantified by qRT-PCR in NHEKs transfected as in (B). (D) NHEKs transfected as in (B) were subjected to IB analysis using the antibodies to the indicated proteins. (E) NHEKs treated as in (B) were fixed at indicated time points and stained with anti-bromodeoxiuridine antibody. The percentage of BrdU-positive nuclei was counted by confocal microscopy. Data are shown as the mean ± SD of three replicates.
Endogenous TAp63 induces GLS2 expression under stress conditions
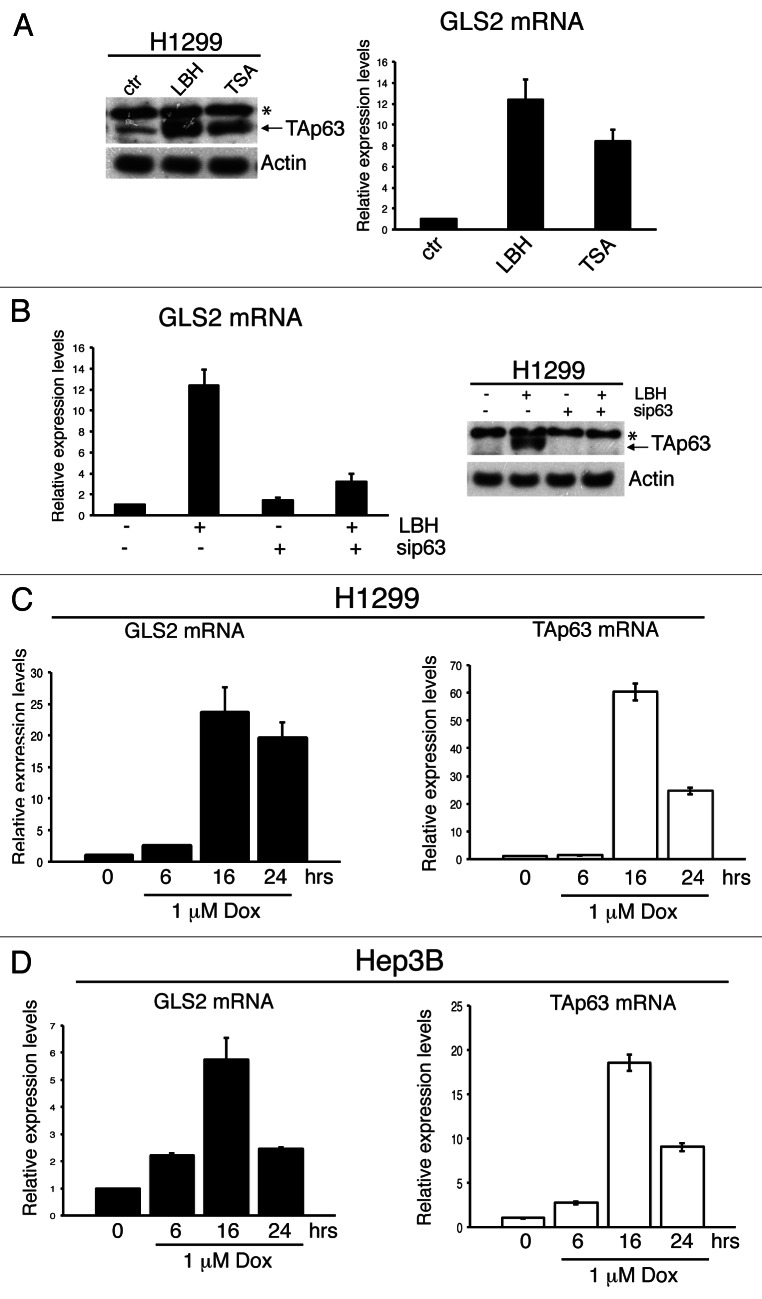
Besides its role in controlling physiological processes such as aging and maintenance of epidermal and dermal precursors, TAp63 activity has also been implicated in pathological processes, such as cancer development.27,47 TAp63 expression is deregulated in human cancers, and treatment of cancer cells with chemotherapeutic drugs or with histone deacetylase (HDAC) inhibitors results in induction of TAp63 expression.6,7,48,49 To verify whether GLS2 expression can be modulated following stress stimuli that activate TAp63, we treated H1299 cells with two chemical HDAC inhibitors, namely Trichostatin A (TSA) and LBH589, and analyzed the expression of both TAp63 and GLS2. Both TSA and LBH589 increased the expression of GLS2 in parallel with a marked induction of TAp63 (Fig. 3A). Similar results were obtained in HCT116 cells (Fig. S1). To verify whether GLS2 induction upon HDAC inhibition was specifically mediated by TAp63, we performed siRNA-mediated silencing of TAp63 in LBH589-treated cells. As shown in Figure 3B, downregulation of TAp63 abrogated the LBH589-induced increase of GLS2 expression, indicating that GLS2 upregulation is not a pleitropic effect resulting from HDAC inhibition. To evaluate also the effect of chemotherapeutic drugs on GLS2 expression, we treated p53-null cell lines with doxorubicin. Following drug administration, GLS2 expression was augmented in H1299 cells (Fig. 3C) and in the hepatocarcinoma cell line Hep3B (Fig. 3D) in parallel with TAp63 upregulation. These results indicate that activation of endogenous TAp63 under stress conditions positively regulates GLS2 expression in the absence of functional p53.
Figure 3. HDAC inhibitors and DNA damage agents regulate GLS2 expression through TAp63 induction. (A) H1299 cells were treated with TSA (1 μM) or LBH589 (2 μM) for 18 h, and whole-cell extracts were analyzed by IB using antibodies to the indicated proteins (left panels). In parallel total RNA was extracted, and GLS2 expression was analyzed by qRTR-PCR (right panels). (B) H1299 cells were transfected twice with siRNA oligos targeting a non-relevant mRNA or p63 mRNA. After 48 h of each siRNA oligo transfection, cells were treated with HDAC inhibitors as described in (A). Cells were used either for quantification of GLS2 mRNA by qRT-PCR (right panel) or subjected to IB analysis utilizing antibodies to the indicated proteins (left panel). H1299 (C) and Hep3B (D) cells were treated with 1 μM doxorubicin for the indicated time points. Total RNA was extracted and used to measure GLS2 and TAp63 mRNA levels by qRT-PCR. Data shown are the mean ± SD of three replicates.
GLS2 protects cells from ROS-dependent apoptosis and is upregulated in colon adenocarcinoma
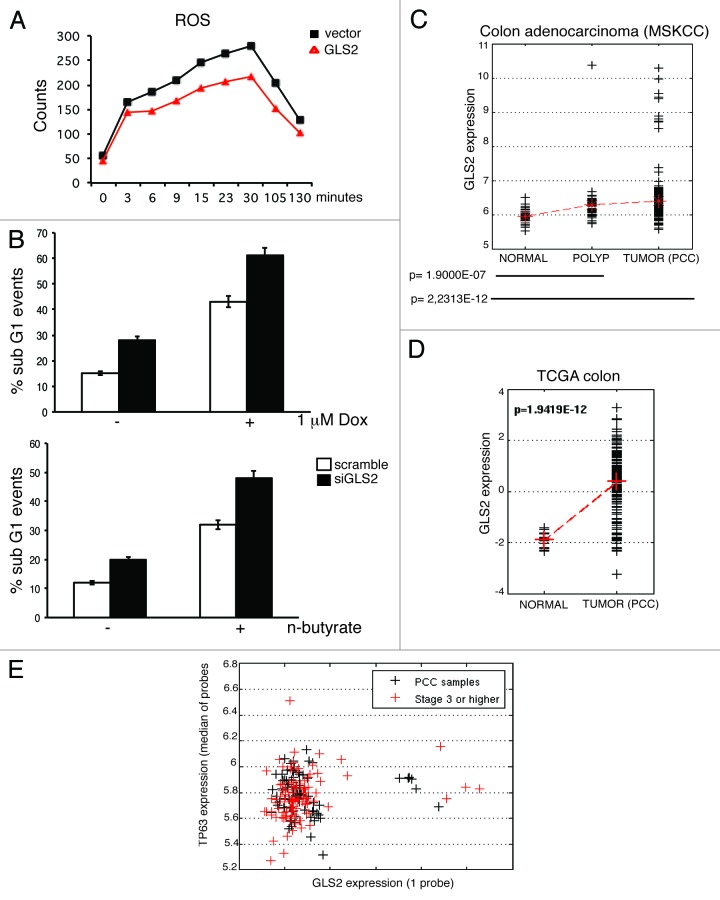
GLS2 catalyzes the conversion of glutamine into glutamate, which can either be converted to α-ketoglutarate, thus stimulating the production of ATP via the TCA cycle, or support the production of two major intracellular scavengers of reactive oxygen species (ROS) glutathione and NADPH.39,50 Consistent with its enzymatic function, we found that GLS2 overexpression increased the intracellular levels of glutamate, α-ketoglutarate and ATP (Fig. S2). To verify whether these metabolic changes were also associated with an increase in cellular defense against oxidative stresses, we measured the amount of intracellular ROS in GLS2-overexpressing cells after exposure to H2O2. As shown in Figure 4A, GLS2-overexpressing cells showed a decrease in the level of ROS produced upon oxidative stress, suggesting that GLS2 can act as a cellular defense against oxidative stress. To further investigate this, we tested whether GLS2 depletion by siRNA could sensitize cells to oxidative stress-induced apoptosis. We transfected H1299 cells with siRNA oligos directed against GLS2 mRNA, and then evaluated the rate of cell death in response to doxorubicin or n-butyrate, two known inducers of ROS-dependent apoptosis.51 Depletion of GSL2 increases the percentage of apoptotic cells (Fig. 4B), thus confirming the role of GLS2 in the regulation of the apoptotic response mediated by oxidative stresses. Since the intracellular redox balance has been suggested to be an important factor in determining human tumors’ response to chemotherapic agents, we analyzed the expression levels of GLS2 in different datasets of epithelial tumors. As shown in Figure 4C, GLS2 expression is significantly increased in a cohort of 186 colon carcinoma tumor samples with respect to benign polyps (p value = 1.9 E-07) or normal colon tissue (p value = 2.23E-12). We also observed the same trend in a TCGA colorectal dataset, which contains 101 colon carcinomas (p value = 1.9419E-12) (Fig. 4D), in nasopharyngeal carcinomas (GSE12452) and in two lung cancer datasets (GSE19188 and GSE11969) (Fig. S3). We also analyzed the correlation between p63 and GLS2 expression in colon carcinoma, and found that p63 correlates with GLS2 expression in tumor samples (Fig. 4E) but not in normal tissues (coefficient 0.0951, p = 0.4940, Spearman correlation).
Figure 4. GLS2 protects tumor cells from ROS-mediated apoptosis and is upregulated in epithelial tumors. (A) H1299 cells were transfected with an empty vector (vector) or Flag-tagged GLS2 (GLS2) and 24 h after transfection cells were treated with 200 μM H202. Intracellular ROS levels were measured by DCF staining. (B) H1299 cells were transfected twice with siRNA oligos targeting a non-relevant mRNA (scramble) or human GLS2 mRNA. After 48 hours of each siRNA oligo transfection, cells were treated with either 1 μM doxorubicin (left panel) or n-butyrate for 18 h. The percentage of sub-G1 cells was measured by FACS analysis. Representative results are shown (mean ± s.d. n=3). (C and D) Box and whisker plots of GLS2 expression in a cohort of 186 colon carcinoma tumor samples respect to polyps or normal colon tissue (C) and in TCGA colorectal dataset (D). (E) Spearman correlation between GLS2 and p63 expression in colon carcinoma. See “Materials and Methods” for details of data analysis.
Together, these data indicate that colon carcinoma, lung and HNSCC tumors displayed markedly increased levels of GLS2. Notably, the upregulation of GLS2 expression in these tumors is accompanied by a concomitant upregulation of p63 levels, suggesting that the connection between p63 and GLS2 in human tumors could be functionally important to drive and maintain glutamine catabolism necessary to maintain the energy supply and antioxidant cellular defense.
Discussion
The activity of tumor suppressor genes is intimately linked to the regulation of cell metabolism, energy production and levels of ROS. Specifically, p53 has been demonstrated to play a critical role in the regulation of cell metabolism, and different p53 target genes involved in this pathway have been identified so far.43,52-54 For example, p53 decreases the glycolytic rate by inhibiting the expression of glucose transporters55 and phosphoglycerate mutase,56 while it increases the expression of TIGAR, which reduces fructose-2 6-biphosphate levels.57 On the other hand p53 is also able to promote some steps in glycolysis,58 suggesting that the regulation of cell metabolism and energy production exerted by p53 could generate opposite outcomes, likely depending on the rate of p53 activation, cell context and levels of cellular stresses. A similar scenario has also been described for the p53-dependent regulation of ROS levels. In fact, among transcriptional targets of p53, there are both several genes whose products potentially generate ROS and genes acting as antioxidants, including GLS2.59-61 In addition, p73, another family protein, has been recently involved in the regulation of cell metabolism through the transcriptional control of Cox4A.62 Although p63 shares some biological functions with p53, our knowledge on whether and how p63 regulates cell metabolism and energy production is limited. Recently, the analysis of the phenotype of the TAp63-knockout mice has highlighted the importance of TAp63 in controlling glucose metabolism.28 TAp63−/− mice are characterized by obesity, glucose intolerance and insulin resistance. Following caloric restriction, TAp63 is able to transcriptionally activate Sirt1, which, in turn, positively regulates the expression of GLS2.28 Here we showed that TAp63 can also directly regulate the expression of GLS2. These two TAp63-dependent transcriptional mechanisms (direct or indirect via Sirt1), although not mutually exclusive, could act together or independently to assure the energy supply necessary to the cells to survive in a caloric restriction period. GLS2 indeed catalyzes the conversion of glutamine into glutamate, which, in turn, can be converted to α-ketoglutarate, thus stimulating the production of ATP via the TCA cycle.
In addition to being important for supplying ATP, glutamate also represents an important precursor of glutathione, which is the main ROS intracellular scavenger. Accordingly we found that the upregulation of GLS2 expression decreases the cellular levels of ROS, while its depletion sensitizes lung cancer cells to ROS-dependent apoptosis. Thus, these results suggest that TAp63 might regulate the cellular antioxidant defense mechanisms under stress conditions. In addition, TAp63 is also able to regulate pro-oxidant genes such as REDD1.63 REDD1 is a stress protein that can be induced following stress conditions, including glucose deprivation and hypoxia through both p53 and p63-dependent mechanisms. Overexpression of REDD1 sensitizes cell to ROS-induced apoptosis, while its downmodulation decreases intracellular ROS production. Therefore we can speculate that TAp63, similarly to what has been already postulated for p53, might regulate both pro-oxidant genes, like REDD1, and antioxidant genes, like GLS2, depending on the levels of stress stimuli and/or cellular context. Moreover, the N-terminal truncated isoform of p63, ΔNp63, is able to regulate the expression of genes, which counteracts ROS production, like glutathione peroxidase GPX2.64 Therefore, the p63-dependent regulation of ROS cellular balance might also be dependent on the relative levels of the expression of its isoforms.
Different data have suggested that GLS2 function is associated to cell resting, quiescent cell states and cell differentiation.40 Accordingly, downregulation of GLS2 expression has been reported in some human tumors, such as hepatocellular carcinomas and highly malignant glioblastoma.42,65,66 In contrast to these data, we found that in a cohort of 186 human primary colon carcinoma GLS2 expression is significantly (p value= 2.2313 E-12) upregulated with respect to normal colon tissue. Moreover, in colon carcinoma and in head and neck squamous cell carcinoma p63 correlates with GLS2 expression among tumor samples, but not among normal samples, suggesting that in some human tumors p63/GLS2 axis can be functionally important to protect tumor cells against oxidative stresses. Accordingly, glutaminase activity is positively correlated with malignancy in tumors.50,67 It is thus possible that GLS2 expression might be under positive selection in order to fully support glutaminolysis, which is required for both energy supply and antioxidant cellular defense. The fact that GLS2 expression is downmodulated in some tumors (hepatocellular carcinomas and highly malignant glioblastoma) while upregulated in others (colon carcinoma, lung and HNSCC) suggests that different tumors might have differential requirements of glutaminolysis, reflecting the tumor-specific relationship between ROS levels, glutamine catabolism and ROS-dependent apoptosis.
Besides analyzing the role of GLS2 in pathological processes, we have also evaluated the impact of GLS2-dependent glutaminolysis in physiological processes. Specifically, we found that GLS2 and TAp63 mRNA levels are concomitantly upregulated during in vitro differentiation of human primary keratinocytes. Conversely, GLS1 expression, which is not regulated by TAp63 or p53, does not vary during this process, strengthening the functional relationship between TAp63 and GLS2. Notably, we found that GLS2-depleted keratynocites showed defects in their capacity to differentiate both at morphological and molecular levels. GLS2 depletion indeed decreases the expression of skin differentiation markers, such as keratin 10 and involucrin. At the morphological level GLS2-depleted cells showed cytoplasmic vacuolization, which is more severe upon differentiation stimuli. Such severe cytoplasmic vacuolization may result by dysfunction of different processes, such us mytochondrial dysfunction or endoplasmatic reticulum (ER) stress.68 Although we observed a dilation of the ER cisternae indicating that the vacuoles observed upon GLS2 depletion were formed by the ER, the expression of markers of ER stress were not affected by GLS2 depletion (data no shown). Although we do not yet know the specific GLS2-dependent metabolic pathway accounting for its role during keratinocytes differentiation, we can hypothesize that deregulation of oxidative balance might play role in GLS2-mediated inhibition of skin differentiation. Different reports have demonstrated that ROS signaling is not only important for mediating and activating cell death, but also regulates intracellular signaling involved in differentiation through multiple tyrosine growth factor receptors, including the epidermal growth factor (EGF-R).69,70 Regulation of EGF signaling by ROS may therefore contribute to the effect of GLS2 on keratinocytes differentiation in vitro. In conclusion, here we described a TAp63-dependent regulation of the glutamine turnover through the transcriptional activation of the GLS2 gene. Furthermore, we showed that the TAp63/GLS2 axis might be an important pathway regulating ROS-dependent apoptosis in the absence of functional p53.
Materials and Methods
Cell culture, transfection, plasmids and drug treatment
Human lung carcinoma H1299 and human colon carcinoma HCT116 cell lines were cultured in Dulbecco’s modified Eagle’s and McCoy’s medium (Gibco, Invitrogen), respectively. Human hepatocarcinoma Hep3B cell line was grown in Minimal Essential Medium (Gibco, Invitrogen). All media were supplemented with 10% fetal bovine serum (FBS), 100 µg/ml penicillin and 100 µg/ml streptomycin (all Gibco, Invitrogen) and cultured at 37 C° with 5% CO2. Cryopreserved normal human epidermal keratinocytes (NHEK Lonza, Basel, Switzerland) were cultured in dishes coated with calf skin collagen type III (Sigma) in keratinocyte growth medium (Epilife, Cascades Biologics, Gibco) supplemented with human keratinocyte growth supplements (Cascades Biologics, Gibco). To induce keratinocyte differentiation, CaCl2 (Sigma) was added to the medium to a final concentration of 1.2 mM. For H1299 transfection experiments, cells were seeded in 10 cm well plate at 80% of cell confluency and, after 24 h, were transfected with Lipofectamine Plus (Invitrogen) following manufacturer’s instructions. Doxorubicin and n-butyrate were added to the cell medium to a final concentration of 1 μM and 5 mM, respectively. For histone deacetylase inhibition cells were treated for 18 h with LBH589 and Trichostatin A (TSA) to a final concentration of 2 μM and 5 μM, respectively.
Plasmids
The pcDNA3.1 expression vector for HA-tagged p63 constructs were previously described.45 Human GLS2 expressing vector was kindly provided by Dr. Zhaohui Feng (Cancer Institute of New Jersey). For retrovirus production, wild-type HA-tagged p63 isoforms were subcloned into the retroviral vector LZRSpBMN by PCR. All cDNAs were sequenced.
siRNA transfection, retroviral infection and detection of apoptosis
siRNA oligos transfection was previously described.71 Briefly, cells were seeded at a density of 1.4 x 105 cells/well in a 6-well plate and transfected with oligos twice (at 24 and 48 hours after plating) using RNAimax (Invitrogen) according to manufacturer’s instructions. Smart pool siRNA oligos direct against GLS2 mRNA, p63 mRNA and non-relevant gene (scramble) were purchased by Dharmacon. Cells were collected after 48 h, and lysates were subjected to immunoblotting. Retroviral particles encoding for HA-TAp63α and HA-TAp63γ were produced by co-transfection of packaging GP-293 cells (Clontech) with LZRSpBMN empty vector, LZRSpBMN-HA-TAp63α or LZRSpBMN-HA-TAp63γ together with VSVG expressing vector by FuGENE (Roche). Forty-eight hours after transfection, the virus-containing medium was collected and supplemented with 8 µg/ml polybrene (Sigma). Cells were then infected by replacing the cell culture medium with this viral supernatant for 24 h. The infection procedure was repeated for a second time after a 12 h recovery period in cell culture medium without virus. Quantification of sub-G1 population was performed by FACS analysis of propidium iodide-stained nuclei, as previously described.72
RNA isolation and qRT-PCR
Total mRNA was isolated using the RNeasy mini kit (Qiagen) following manufacturer recommendations. Total RNA was quantified using a NanoDrop Spectophotometer (Thermo Scientific) and used for cDNA synthesis using SuperScript Reverse Transcriptase (Promega), according to the manufacturer’s protocol. cDNA was subsequentially used for real-time PCR analysis (qRT-PCR). Each 25 μl reaction contained 2X SYBR-Green PCR Master Mix (Applied Biosystems), 0.125 μl MultiScribe Reverse Transcriptase (Applied Biosystems), 2 μl cDNA and the appropriate specific primers (0.5 μM, sequences available upon request). Amplification and fluorescence detection according to the manufacturer's instructions was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The expression of each gene was defined from the threshold cycle (Ct), and relative expression levels were calculated using the 2-ΔΔCt method. The following primers were used: for hGLS2 5′-ACACCCTCAGCCTCATGCAT-3′; rev hGLS2 5′-ATGGCTCCTGATACAGCTGACTT-3′; for hGLS1 5′-CACTGCCCTCCCATTACCTAG-3′, rev hGLS1 5′-GAAGCTCAAGCATGGGAACAG-3′; for hActin 5′GTTGCTATCCAGGCTGTGCTA-3′; rev hActin 5′-AATGTCACGCACGATTTCCCG-3′; for hK10 5′- AGGAGGAGTGTCATCCCTAAG-3′, rev hK10 5′-AAGCTGCCTCCATAACTCCC-3′; for hInvolucrin 5′-CAGGTCCAAGACATTCAACC-3′, rev h Involucrin 5′- CAAGTTCACAGATGAGACGG-3′; F TAp63 TCAGAAGATCGTGCGACAAAC, R TAp63 5′-GTTCAGGAGCCCCAGGTTCG-3′; for hΔNp63 5′-GAAGAAAGGACAGCAGCATTG-3′, rev hΔNp63 5′-GGGACTGGTGGACGAGGAG-3′; for h TBP 5′-TCAAACCCAGAATTGTTGTCC-3′, rev hTBP 5′-CCTGAATCCCTTTAGAATAGG-3′. P-value was computed using the Student’s t-test.
Immunoblot analysis and antibodies
Immunoblot analysis was performed using whole-cell extracts obtained by lysing cell pellets with Triton Buffer (50 mM Tris-Hcl pH 7.5, 250 mM NaCl, 50 mM NaF, 1mM EDTA 1 pH 8, 0.1% Triton), supplemented with protease and phosphatase inhibitors. Proteins were separated by SDS-PAGE, transferred onto PVDF membranes and blocked with PBS-T (phosphate-buffered saline and 0,1%Tween-20) containing 5% non-fat dry milk for 1 h at room temperature (RT). The incubation with primary antibodies was performed for 2 h at RT, followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibody. Detection was performed with ECL Western Blot Reagent (Perkin Elmer). Mitochondria were purified using the Qproteome Mitochondria Purification System (Qiagen) following manufacturer recommendations. Mitochondrial proteins were extracted by lysing mitochondria pellets with Triton Buffer. Mouse monoclonal antibodies were from Covance (anti-HA), Sigma (actin, cyclofilin D), Neomarkers (anti-p63). Rabbit polyclonal antibody were from Covance (keratin 10). Rabbit polyclonal against GLS2 was kindly provided by Dr. Zhaohui Feng (Cancer Institute of New Jersey). Anti-rabbit IgG or anti-mouse IgG horseradish peroxidase-conjugated antibodies were purchased by Perkin Elmer.
Chromatin immunoprecipitation (ChIP) analysis.
ChIP experiments were performed in TAp63α Saos-2 inducible cell line after 24 h of doxyciclin treatment using MAGnify™ Chromatin Immunoprecipitation system (Invitrogen). Briefly, cells were crosslinked for 10 min at room temperature with 1% formaldehyde (Merck). The crosslinking reaction was stopped by addition of 125 mM glycine for 5 min followed by a washing step with PBS. The pellet was lysed and sonicated using sonicator (Bioruptor™ UCD-200, diagenode), shearing the chromatin into 500-1,000 bp fragments. The chromatin extract was incubated with Dynabeads® Protein A/G coupled to 10 µg mouse anti-HA antibody or mouse IgG (negative control) at 4°C with rotation for 2 h. The immunocomplexes were washed and treated with proteinase K (20 mg/ml) at 55°C for 15 min to reverse the crosslinking. DNA was purified with the DNA purification magnetic beads, dissolved in elution buffer and used for PCR analysis. The following oligos were used for GLS2 promoter: for 5′-GGCCTCCCAAGTCACCAGTTCA-3′ rev 5′-TGTTTTTGCTTGTTTTCGCCTTCT-3′. The p53 RE located on hMDM2, used as positive control, was amplified with one set of primers: forward 5′-GGTTGACTCAGCTTTTCCTCTTG-3′ and reverse 5′-GGAAAATGCATGGTTTAAATAGCC-3′ (119 bp). PCR products were analysed by electrophoresis on agarose gels.
Luciferase assay
For luciferase assay, pGL3 basic luciferase reporter vector (Promega) containing human GLS2 promoter was co-transfected with the pRL-CMV Renilla luciferase vector (Promega) and HA-tagged TAp63α, TAp63β, TAp63γ and ∆Np63 expression vector into HEK293E cells using Effectene (Qiagen). Twenty-four hours after transfection, luciferase activity was measured with the Dual Luciferase Reporter Assay system (Promega). Luciferase and Renilla luminescence were measured over 10 seconds using OPTOCOMP I luminometer. Relative luciferase activity was determined after normalization with the Renilla activity for each sample.
Measurement of energy metabolism
Glutamate levels were measured by using the Amplex Red Glutamine Acid/Glutamate oxidase assay kit (Invitrogen). α-Ketoglutarate levels were measured by using the α-ketoglutarate assay kit (Biovision). ATP levels were measured by using the ATP Bioluminescence assay kit (Roche). ROS levels were measured by CM-H2DCFDA (Invitrogen) staining in a flow cytometry assay.
Data analysis
Human sample data was downloaded from the GEO database, accession numbers GSE12452,73 GSE19188,74 GSE11969.75 Colorectal cancer data was obtained from the TCGA portal (gene expression, level 3 data, as available on 11/01/12) and additional colorectal cancer data was obtained from a previously described cohort of patients treated at MSKCC between 1992 and 2004. Median of probes per gene was computed. Samples were divided into normal versus tumor in each set and variability of expression was assessed using the two-sample Kolmogorov-Smirnov test (as in some cases the distribution of expression values for tumor samples was not normal). Tests were performed in matlab (kstest2). Pearson correlation was also computed in matlab using the routine corr.
Supplementary Material
Acknowledgments
This work has been supported by the Medical Research Council, UK; grants from “Alleanza contro il Cancro” Grant (ACC12), MIUR/PRIN (20078P7T3K_001)/FIRB (RBIP06LCA9_0023, RBIP06LCA9_0C), AIRC grant (#5471), (2011-IG11955), Telethon Grant GGPO9133 to G.M., MIUR/PRIN 2008MRLSNZ_004, AIRC 5xmille (#9979), RF06 c.73, RF08 c.15, RF07 c.57 awarded to G.M., and the AIRC grant 9202 awarded to F.B.
Glossary
Abbreviations:
- GLS2
glutaminase 2
- GLS1
glutaminase 1
- qRT-PCR
quantitative real-time PCR
- NHEK
normal human epidermal keratinocytes
- ChIP
chromatin immunoprecipitation
- HDAC
histone deacetylase
- TSA
trichostatin
- ROS
reactive oxygen species
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24478
References
- 1.Amelio I, Grespi F, Annicchiarico-Petruzzelli M, Melino G. p63 the guardian of human reproduction. Cell Cycle. 2012;11:4545–51. doi: 10.4161/cc.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18:1815–24. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levrero M, De Laurenzi V, Costanzo A, Gong J, Melino G, Wang JY. Structure, function and regulation of p63 and p73. Cell Death Differ. 1999;6:1146–53. doi: 10.1038/sj.cdd.4400624. [DOI] [PubMed] [Google Scholar]
- 4.Tissir F, Goffinet AM. p73 and p63: Estranged relatives? Cell Cycle. 2011;10:1351. doi: 10.4161/cc.10.9.15383. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16. doi: 10.1016/S1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 6.Petitjean A, Ruptier C, Tribollet V, Hautefeuille A, Chardon F, Cavard C, et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–81. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- 7.Seitz SJ, Schleithoff ES, Koch A, Schuster A, Teufel A, Staib F, et al. Chemotherapy-induced apoptosis in hepatocellular carcinoma involves the p53 family and is mediated via the extrinsic and the intrinsic pathway. Int J Cancer. 2010;126:2049–66. doi: 10.1002/ijc.24861. [DOI] [PubMed] [Google Scholar]
- 8.Vandenabeele P, Melino G. The flick of a switch: which death program to choose? Cell Death Differ. 2012;19:1093–5. doi: 10.1038/cdd.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition mechanism of p63 by the E3 ligase Itch: novel strategy in the study and inhibition of this interaction. Cell Cycle. 2012;11:3638–48. doi: 10.4161/cc.21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghioni P, Bolognese F, Duijf PH, Van Bokhoven H, Mantovani R, Guerrini L. Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol Cell Biol. 2002;22:8659–68. doi: 10.1128/MCB.22.24.8659-8668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helton ES, Zhu J, Chen X. The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem. 2006;281:2533–42. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- 12.Duijf PH, Vanmolkot KR, Propping P, Friedl W, Krieger E, McKeon F, et al. Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Hum Mol Genet. 2002;11:799–804. doi: 10.1093/hmg/11.7.799. [DOI] [PubMed] [Google Scholar]
- 13.Peschiaroli A, Scialpi F, Bernassola F. El Sherbini el S, Melino G. The E3 ubiquitin ligase WWP1 regulates DeltaNp63-dependent transcription through Lys63 linkages. Biochem Biophys Res Commun. 2010;402:425–30. doi: 10.1016/j.bbrc.2010.10.050. [DOI] [PubMed] [Google Scholar]
- 14.Leonard MK, Kommagani R, Payal V, Mayo LD, Shamma HN, Kadakia MP. ΔNp63α regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ. 2011;18:1924–33. doi: 10.1038/cdd.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, et al. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–96. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–36. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 17.Candi E, Rufini A, Terrinoni A, Giamboi-Miraglia A, Lena AM, Mantovani R, et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA. 2007;104:11999–2004. doi: 10.1073/pnas.0703458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci USA. 2012;109:1133–8. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–47. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 20.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 21.Candi E, Terrinoni A, Rufini A, Chikh A, Lena AM, Suzuki Y, et al. p63 is upstream of IKK alpha in epidermal development. J Cell Sci. 2006;119:4617–22. doi: 10.1242/jcs.03265. [DOI] [PubMed] [Google Scholar]
- 22.Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–61. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 23.Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–99. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–41. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 25.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 26.Park BJ, Lee SJ, Kim JI, Lee SJ, Lee CH, Chang SG, et al. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 2000;60:3370–4. [PubMed] [Google Scholar]
- 27.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, et al. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab. 2012;16:511–25. doi: 10.1016/j.cmet.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 30.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peschiaroli A, Giacobbe A, Formosa A, Markert EK, Bongiorno-Borbone L, Levine AJ, et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32:797–802. doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 33.Markert EK, Levine AJ, Vazquez A. Proliferation and tissue remodeling in cancer: the hallmarks revisited. Cell Death Dis. 2012;3:e397. doi: 10.1038/cddis.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyssiotis CA, Vander-Heiden MG, Muñoz-Pinedo C, Emerling BM. Emerging concepts: linking hypoxic signaling and cancer metabolism. Cell Death Dis. 2012;3:e303. doi: 10.1038/cddis.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qie S, Liang D, Yin C, Gu W, Meng M, Wang C, et al. Glutamine depletion and glucose depletion trigger growth inhibition via distinctive gene expression reprogramming. Cell Cycle. 2012;11:3679–90. doi: 10.4161/cc.21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes LC, Di Benedetto G, Scorrano L. Essential amino acids and glutamine regulate induction of mitochondrial elongation during autophagy. Cell Cycle. 2011;10:2635–9. doi: 10.4161/cc.10.16.17002. [DOI] [PubMed] [Google Scholar]
- 37.Martín-Rufián M, Tosina M, Campos-Sandoval JA, Manzanares E, Lobo C, Segura JA, et al. Mammalian glutaminase Gls2 gene encodes two functional alternative transcripts by a surrogate promoter usage mechanism. PLoS One. 2012;7:e38380. doi: 10.1371/journal.pone.0038380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aledo JC, Gómez-Fabre PM, Olalla L, Márquez J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm Genome. 2000;11:1107–10. doi: 10.1007/s003350010190. [DOI] [PubMed] [Google Scholar]
- 39.Campos-Sandoval JA, López de la Oliva AR, Lobo C, Segura JA, Matés JM, Alonso FJ, et al. Expression of functional human glutaminase in baculovirus system: affinity purification, kinetic and molecular characterization. Int J Biochem Cell Biol. 2007;39:765–73. doi: 10.1016/j.biocel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Gómez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sánchez P, et al. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–42. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner A, McGivan JD. Glutaminase isoform expression in cell lines derived from human colorectal adenomas and carcinomas. Biochem J. 2003;370:403–8. doi: 10.1042/BJ20021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 44.Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24:2458–71. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browne G, Cipollone R, Lena AM, Serra V, Zhou H, van Bokhoven H, et al. Differential altered stability and transcriptional activity of ΔNp63 mutants in distinct ectodermal dysplasias. J Cell Sci. 2011;124:2200–7. doi: 10.1242/jcs.079327. [DOI] [PubMed] [Google Scholar]
- 46.Masse I, Barbollat-Boutrand L, Molina M, Berthier-Vergnes O, Joly-Tonetti N, Martin MT, et al. Functional interplay between p63 and p53 controls RUNX1 function in the transition from proliferation to differentiation in human keratinocytes. Cell Death Dis. 2012;3:e318. doi: 10.1038/cddis.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;109:15312–7. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayan BS, Yang AL, Conforti F, Bernardini S, Tucci P, Vasa-Nicotera M, et al. Induction of TAp63 by histone deacetylase inhibitors. Biochem Biophys Res Commun. 2010;391:1748–51. doi: 10.1016/j.bbrc.2009.12.147. [DOI] [PubMed] [Google Scholar]
- 49.Newbold A, Vervoort SJ, Martin BP, Bots M, Johnstone RW. Induction of autophagy does not alter the anti-tumor effects of HDAC inhibitors. Cell Death Dis. 2012;3:e387. doi: 10.1038/cddis.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matés JM, Segura JA, Martín-Rufián M, Campos-Sandoval JA, Alonso FJ, Márquez J. Glutaminase Isoenzymes as Key Regulators in Metabolic and Oxidative Stress against Cancer. Curr Mol Med. 2012 doi: 10.2174/1566524011313040005. In press. [DOI] [PubMed] [Google Scholar]
- 51.Hsiao CH, Li W, Lou TF, Baliga BS, Pace BS. Fetal hemoglobin induction by histone deacetylase inhibitors involves generation of reactive oxygen species. Exp Hematol. 2006;34:264–73. doi: 10.1016/j.exphem.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Ben-Sahra I, Dirat B, Laurent K, Puissant A, Auberger P, Budanov A, et al. Sestrin2 integrates Akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ. 2013;20:611–9. doi: 10.1038/cdd.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nayak G, Cooper GM. p53 is a major component of the transcriptional and apoptotic program regulated by PI 3-kinase/Akt/GSK3 signaling. Cell Death Dis. 2012;3:e400. doi: 10.1038/cddis.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tucci P. Caloric restriction: is mammalian life extension linked to p53? Aging (Albany NY) 2012;4:525–34. doi: 10.18632/aging.100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–33. doi: 10.1158/0008-5472.CAN-03-0846. [DOI] [PubMed] [Google Scholar]
- 56.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–85. [PubMed] [Google Scholar]
- 57.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 58.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–80. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 59.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 60.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–13. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–40. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 62.Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26:2009–14. doi: 10.1101/gad.197640.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/S1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 64.Yan W, Chen X. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J Biol Chem. 2006;281:7856–62. doi: 10.1074/jbc.M512655200. [DOI] [PubMed] [Google Scholar]
- 65.Szeliga M, Obara-Michlewska M, Matyja E, Łazarczyk M, Lobo C, Hilgier W, et al. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57:1014–23. doi: 10.1002/glia.20825. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA. 2010;107:7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hampton RY. ER stress response: getting the UPR hand on misfolded proteins. Curr Biol. 2000;10:R518–21. doi: 10.1016/S0960-9822(00)00583-2. [DOI] [PubMed] [Google Scholar]
- 69.Finkel T. Redox-dependent signal transduction. FEBS Lett. 2000;476:52–4. doi: 10.1016/S0014-5793(00)01669-0. [DOI] [PubMed] [Google Scholar]
- 70.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell. 2002;9:387–99. doi: 10.1016/S1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 71.Peschiaroli A, Skaar JR, Pagano M, Melino G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene. 2010;29:1384–93. doi: 10.1038/onc.2009.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peschiaroli A, Figliola R, Coltella L, Strom A, Valentini A, D’Agnano I, et al. MyoD induces apoptosis in the absence of RB function through a p21(WAF1)-dependent re-localization of cyclin/cdk complexes to the nucleus. Oncogene. 2002;21:8114–27. doi: 10.1038/sj.onc.1206010. [DOI] [PubMed] [Google Scholar]
- 73.Sengupta S, den Boon JA, Chen IH, Newton MA, Dahl DB, Chen M, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 74.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–88. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.