Abstract
Accurate mitotic spindle positioning is essential for the regulation of cell fate choices, cell size and cell position within tissues. The most prominent model of spindle positioning involves a cortical pulling mechanism, where the minus end-directed microtubule motor protein dynein is attached to the cell cortex and exerts pulling forces on the plus ends of astral microtubules that reach the cortex. In nonpolarized cultured cells integrin-dependent, retraction fiber-mediated cell adhesion is involved in spindle orientation. Proteins serving as intermediaries between cortical actin or retraction fibers and astral microtubules remain largely unknown. In a recent genome-wide RNAi screen we identified a previously uncharacterized protein, MISP (C19ORF21) as being involved in centrosome clustering, a process leading to the clustering of supernumerary centrosomes in cancer cells into a bipolar mitotic spindle array by microtubule tension. Here, we show that MISP is associated with the actin cytoskeleton and focal adhesions and is expressed only in adherent cell types. During mitosis MISP is phosphorylated by Cdk1 and localizes to retraction fibers. MISP interacts with the +TIP EB1 and p150glued, a subunit of the dynein/dynactin complex. Depletion of MISP causes mitotic arrest with reduced tension across sister kinetochores, chromosome misalignment and spindle multipolarity in cancer cells with supernumerary centrosomes. Analysis of spindle orientation revealed that MISP depletion causes randomization of mitotic spindle positioning relative to cell axes and cell center. Together, we propose that MISP links microtubules to the actin cytoskeleton and focal adhesions in order to properly position the mitotic spindle.
Keywords: cell adhesion, centrosomal clustering, focal adhesion, mitosis, spindle orientation, centrosome, actin, MISP, spindle positioning
Introduction
Centrosomes act as microtubule-organizing centers and function as mitotic spindle poles during mitosis, directing the formation of bipolar spindles.1,2 Centrosome amplification is frequent in both solid tumors and hematological malignancies and is linked to tumorigenesis and chromosomal instability.3-5 In mitosis, supernumerary centrosomes can lead to the formation of multipolar spindles, which is a hallmark of many tumor types.3,6,7 Multipolar cell division, however, is antagonistic to cell viability.8,9 In order to circumvent lethal multipolar divisions, many cancer cells cluster supernumerary centrosomes into two spindle poles, enabling bipolar division.3,8-12 The mechanisms of centrosomal clustering in tumor cells are incompletely understood. Recent genome-wide RNAi screens in cells with supernumerary centrosomes that have been performed by us and others suggest, among others, the involvement of spindle tension as controlled by the actin cytoskeleton and cell adhesion molecules as well as dynein and NuMA in this process.10,11,13 In our genome-wide RNAi screen we identified a previously uncharacterized protein, MISP (focal adhesion-associated and spindle positioning; C19ORF21) as being involved in centrosome clustering.
Similar to centrosomal clustering, spindle positioning and orientation depend on tension generated by cortically anchored dynein, which exerts forces on astral microtubules by its minus end-directed motor activity, thereby pulling mitotic spindles into their correct position within the cell.14-17 It has been shown that the extracellular matrix, which is connected to the intracellular actin cytoskeleton via focal adhesions, impacts on the orientation of mitotic spindles.18-20 Correspondingly, integrins, which are key receptors involved in the assembly of focal adhesions, have also been demonstrated to play a role in orienting the mitotic spindle parallel to the substrate in tissue culture.21 While cells round up in mitosis, they remain connected to the adhesive substrate through actin-rich retraction fibers. Laser ablation experiments of cells on ECM micropatterns revealed that retraction fibers provide external cues necessary for the proper orientation of mitotic spindles.20 Interaction of astral microtubules with cortical structures is mediated by microtubule plus end-binding proteins (+TIPs), which include EB1, adenomatous polyposis coli (APC) and dynein, with dynein being recruited by a complex containing NuMA.14,22-25
With regard to centrosomes, it has been shown that deletion of focal adhesion kinase (FAK), a tyrosine kinase that is recruited to focal adhesions and activated as an early consequence of integrin clustering upon ligand binding, results in multipolar mitotic spindles in endothelial cells.26,27 Also, depletion or inhibition of integrin-linked kinase (ILK), a serine-threonine kinase and scaffold protein at focal adhesions, leads to mitotic spindle defects and inhibition of centrosomal clustering in cancer cells with supernumerary centrosomes.28,29
In this study, we show that the previously uncharacterized protein MISP is predominantly expressed in adherent cell lines and colocalizes with the actin cytoskeleton and focal adhesions in interphase cells as well as with retraction fibers during mitosis. Furthermore, MISP interacts with FAK, the dynactin subunit p150glued and the +TIP protein EB1 and is phosphorylated during mitosis, most probably by Cdk1. Depletion of MISP caused mitotic arrest and impaired mitotic spindle positioning and orientation. Also MISP knockdown reduced tension across sister kinetochores and led to chromosome misalignment and spindle multipolarity in cancer cells with supernumerary centrosomes. In summary, we propose that MISP links microtubules to the actin cytoskeleton and focal adhesions in order to properly position the mitotic spindle.
Results
MISP is involved in centrosomal clustering
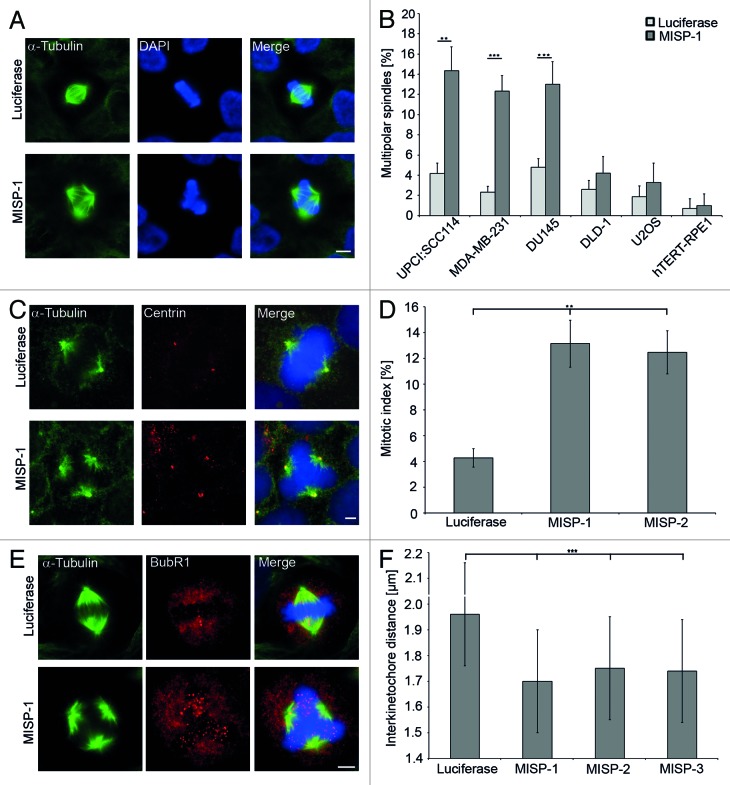
In a genome-wide siRNA screen in human cancer cells containing supernumerary centrosomes, we identified MISP as a protein required for centrosomal clustering.11 In UPCI:SCC114 cells, knockdown of MISP by MISP-1-siRNA resulted in 14.3 ± 3.3% multipolar spindles compared with 4.2 ± 1.0% multipolar spindles in cells treated with luciferase-siRNA (n = 600 mitoses per siRNA, p = 0.002; Fig. 1A and B). To investigate whether multipolar spindles in UPCI:SCC114 cells induced by depletion of MISP are a consequence of centrosomal declustering UPCI:SCC114 cells stably expressing GFP-α-tubulin were immunostained with an antibody to the centriolar protein centrin (Fig. 1C). After MISP-1-siRNA transfection, in 77.2% of cells with multipolar spindles (n = 92), each spindle pole contained two centrin signals. Similarly, 79.5% of multipolar spindles (n = 44) contained two centrin signals at each pole after luciferase-siRNA treatment, arguing for the presence of complete centrosomes at the extra spindle poles. In MDA-MB-231 as well as DU145 cells, both of which also harbor supernumerary centrosomes, knockdown of MISP by MISP-1-siRNA leads to the formation of multipolar mitotic spindles as well (Fig. 1B). In contrast, siRNA-mediated depletion of MISP by MISP-1-siRNA in cells with a regular centrosome content (U2OS, DLD-1, hTERT-RPE-1) does not result in multipolar spindle induction (Fig. 1B). Depletion of MISP by a second siRNA (MISP-2) induced the formation of multipolar spindles in UPCI:SCC114 cells as well (12.7 ± 0.5% vs. 4.2 ± 1.0% multipolar spindles after transfection with luciferase-siRNA, n = 600 mitoses per siRNA, p = 0.0003; Fig. S1). In addition, transfection of a siRNA to the 3′UTR of MISP (MISP-3) also led to the induction of multipolar spindles in UPCI:SCC114 cells (16.3 ± 1.5% vs. 4.2 ± 1% multipolar spindles after transfection with luciferase-siRNA, n = 600 mitoses per siRNA, p = 0.006; Fig. S1) Together, these results demonstrate that MISP is involved in centrosomal clustering.
Figure 1. MISP is required for centrosomal clustering. (A) Depletion of MISP leads to the formation of multipolar mitotic spindles in UPCI:SCC114 cells. Cells were treated with luciferase- or MISP-1-siRNA for 48 h and immunostained with an antibody to α-tubulin. Scale bar, 10 µm. (B) siRNA-mediated depletion of MISP results in multipolar mitotic spindles only in cells harboring supernumerary centrosomes (UPCI:SCC114, MDA-MB-231, DU145) but not in cells with a regular centrosome content (DLD-1, U2OS or hTERT-RPE1). Cells were treated with luciferase- or MISP-1-siRNA for 48 h and immunostained with an antibody to α-tubulin. The graph shows the average of three independent experiments; mean ± SD (C) Depletion of MISP in UPCI:SCC114 cells constitutively expressing GFP-α-tubulin leads to spindle multipolarity with two centrioles at each spindle pole. Cells were transfected with luciferase- or MISP-1-siRNA for 48 h and subsequently immunostained with an antibody to centrin. Scale bar, 5 µm. (D) Depletion of MISP by two different siRNAs leads to mitotic arrest in UPCI:SCC114 cells. Mitotic indices were scored after immunostaining with an antibody to α-tubulin 48 h after siRNA transfection. The graph shows the average of three independent experiments; mean ± SD (E) Depletion of MISP leads to loss of spindle tension in UPCI:SCC114 cells. Cells were immunostained with antibodies to BubR1 and α-tubulin 48 h after transfection with luciferase- or MISP-1-siRNA. Scale bar, 10 µm. (F) Depletion of MISP leads to significantly reduced interkinetochore distances in bipolar mitotic spindles of UPCI:SCC114 cells. Interkinetochore distances were measured 48 h after transfection of indicated siRNAs. Cells were co-immunostained with Hec-1 and Crest antibodies. Luciferase-specific siRNA served as a control. The graph shows the average of three independent experiments; mean ± SD.
Mitotic arrest of MISP-depleted cells is due to loss of spindle tension
In addition to the formation of multipolar mitotic spindles siRNA-mediated depletion of MISP led to an enrichment of UPCI:SCC114 cells in G2/M phase of the cell cycle compared with cells transfected with luciferase-siRNA 72 h after transfection (luciferase-siRNA 16.6 ± 3.4, MISP-1-siRNA 34.5 ± 4.1; p = 0.0006). Accordingly, the mitotic index of UPCI:SCC114 cells was increased to 13.1 ± 1.8% and 12.5 ± 1.7%, 48 h after depletion of MISP using two different siRNAs (n = 1,500 cells per siRNA; Fig. 1D), indicating that the spindle assembly checkpoint (SAC) is activated upon knockdown of MISP. To analyze for activation of the SAC in more detail, we immunostained UPCI:SCC114 cells with an antibody against BubR1 and observed an increase in BubR1-positive kinetochores in MISP- compared with luciferase-depleted cells 48 h after MISP-1-siRNA transfection (Fig. 1E). To confirm loss of tension between corresponding sister kinetochores as a cause for SAC activation, at least 200 interkinetochore distances per siRNA were measured in bipolar mitotic spindles of UPCI:SCC114 cells co-immunostained with Hec1- and Crest-antibodies 48 h after transfection with siRNAs specific for MISP or luciferase, respectively. Interkinetochore distances were significantly decreased after MISP depletion using three different siRNAs (luciferase-siRNA 1.97 ± 0.19 µm, MISP-1-siRNA 1.70 ± 0.22 µm, p = 1.2 × 10−36; MISP-2-siRNA 1.75 ± 0.22 µm, p = 4.7 × 10−28; MISP-3-siRNA 1.74 ± 0.23 µm, p = 1.9 × 10−27; Fig. 1F). We conclude that depletion of MISP leads to mitotic arrest due to loss of tension between sister kinetochores.
Generation and characterization of antibodies against MISP
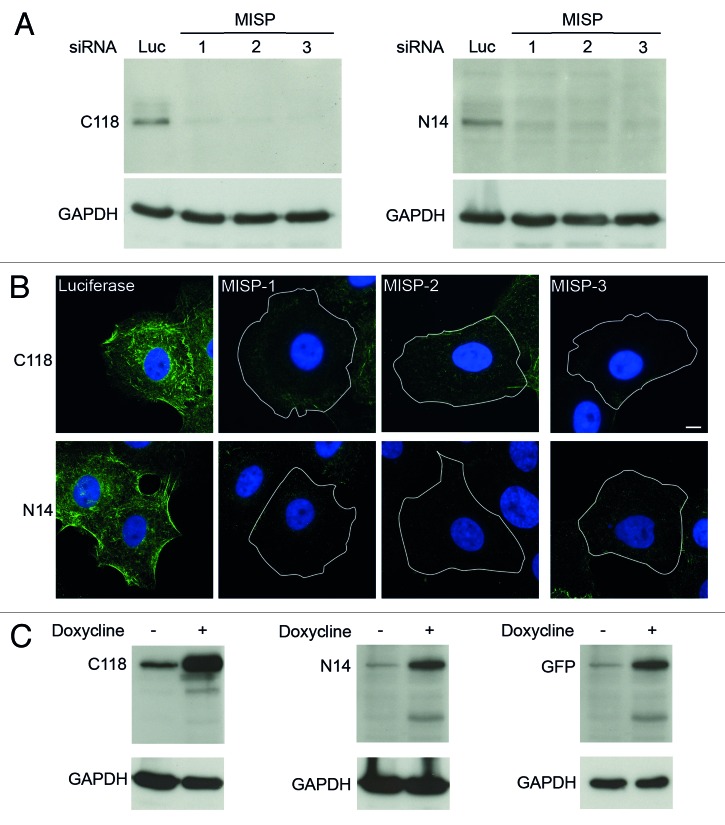
To further elucidate the function of MISP, we generated two mouse monoclonal antibodies against the N- and C-terminus of the protein (N14 and C118; see “Material and Methods”). Both antibodies detect a band with a molecular weight of 75 kDa corresponding to the calculated size of MISP, which is clearly reduced after treatment of UPCI:SCC114 cells with three different MISP-specific siRNAs (Fig. 2A). Also, immunostaining of UPCI:SCC114 cells after treatment with luciferase- and MISP-specific siRNAs with these antibodies led to a clear reduction in MISP signal intensity with both antibodies (Fig. 2B). Furthermore, we generated an U2OS cell line conditionally expressing GFP-MISP (GFP-MISP-U2OS) and detected GFP-MISP by immunoblotting with both MISP antibodies as well as with an antibody against GFP (Fig. 2C). As endogenous MISP levels are very low in U2OS cells (see results below), the endogenous protein is not reliably detectable in this cell line. Therefore, most of the experiments were performed in UPCI:SCC114 cells, which express larger amounts of MISP (see results below). To further determine the specificity of the MISP-siRNAs used, we analyzed for depletion of GFP-MISP by western blotting as well as immunofluorescence microscopy in GFP-MISP-U2OS cells. RNAi-mediated knockdown of MISP in this cell line led to efficient depletion of GFP-MISP with siRNAs MISP-1 and MISP-2 but not with MISP-3-siRNA, which is directed against the 3′UTR of endogenous MISP (Fig. S2A and B).
Figure 2. MISP antibodies detect both endogenous and overexpressed GFP-MISP. (A) A mouse monoclonal MISP antibody directed against the C-terminus of MISP (Clone C118, left panel) detects a band of 75 kDa in UPCI:SCC114 cells transfected with a siRNA against luciferase. Transfection of MISP-specific siRNAs (1–3) for 48 h reduces the MISP signal. GAPDH served as loading control. A monoclonal MISP antibody directed against the N-terminus of the protein (Clone N14, right panel) detects a 75 kDa band in UPCI:SCC114 cells transfected with a siRNA against luciferase as well. Transfection of MISP-specific siRNAs (1–3) for 48 h again reduces the MISP signal using this antibody. GAPDH served as a loading control. (B) Immunostaining with both MISP antibodies (C118 and N14) detects fiber-like structures which are clearly diminished in UPCI:SCC114 cells transfected with MISP-specific siRNAs (MISP-1, MISP-2, MISP-3) for 48 h as compared with cells transfected with luciferase-siRNA. Cell boundaries are depicted as white lines. Scale bar, 10 µm. (C) GFP-MISP can be detected by MISP antibodies C118 (left panel) and N14 (middle panel) as well as by an antibody to GFP (right panel) in GFP-MISP-U2OS cells 24 h after the addition of doxycycline. GAPDH served as loading control.
MISP colocalizes with the actin cytoskeleton and focal adhesions
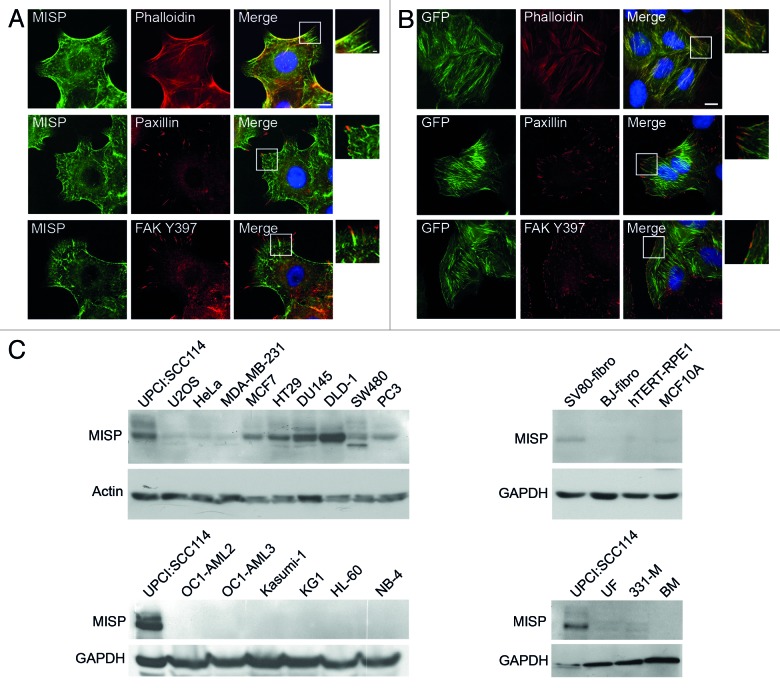
To determine the subcellular localization of MISP, UPCI:SCC114 cells were co-immunostained with an antibody to MISP (C118) and Phalloidin-TRITC to visualize the actin cytoskeleton, demonstrating a partial colocalization of MISP with actin structures, predominantly with the tips of actin filaments (Fig. 3A, upper panel). Co-immunostaining of UPCI:SCC114 cells with antibodies to MISP (C118) and paxillin (Fig. 3A, middle panel) or autophosphorylated focal adhesion kinase (FAKY397; Fig. 3A, lower panel), both of which mark focal adhesion sites, revealed that the filamentous structures of MISP direct toward focal adhesions. Similar results were obtained with GFP-MISP-U2OS cells, in which GFP-MISP demonstrated a more pronounced colocalization with the actin cytoskeleton, with less restriction to the tips of actin filaments (Fig. 3B, upper panel). The fiber-like structures of GFP-MISP, however, were also directed toward focal adhesions immunostained by antibodies to paxillin (Fig. 3B, middle panel) or FAKY397 (Fig. 3B, lower panel).
Figure 3. MISP localizes to actin and focal adhesions and is only expressed in adherent cell types. (A) Co-immunostaining of exponentially growing UPCI:SCC114 cells with an antbody to MISP (C118) and Phalloidin-TRITC (upper panel) shows partial colocalization of MISP with the actin cytoskeleton. Co-immunostaining of MISP with the focal adhesion markers paxillin (middle panel) and autophosphorylated focal adhesion kinase (FAK Y397, lower panel) reveals that fiber-like MISP structures often end in focal adhesions. White rectangles mark the position of the magnified inset. Scale bar, 10 µm; inset 2 µm. (B) GFP-MISP shows prominent colocalization with Phalloidin-TRITC in GFP-MISP-U2OS cells (upper panel). Immunostaining with focal adhesion markers paxillin (middle panel) and FAK Y397 (lower panel) reveals that GFP-MISP fibers also end in focal adhesions. Expression of GFP-MISP was induced by addition of doxycycline for 48 h. White rectangles mark the position of the magnified insets. Scale bar, 10 µm; inset 2 µm. (C) Immunoblots showing expression levels of MISP in various adherent cancer (upper left panels) and non-transformed (upper right panels) as well as non-adherent cancer (lower left panels) and non-transformed (lower right panels) cell lines using an antibody to MISP (C118). Lysates of UPCI:SCC114 cells are loaded as positive controls. Actin and GAPDH served as loading controls.
MISP is specifically expressed in adherent cancer cell lines
Next, we investigated MISP expression levels in various cell lines. Since MISP partially colocalizes with focal adhesions, we first asked whether MISP is differentially expressed in adherent (Fig. 3C, upper panel) vs. non-adherent cell lines (Fig. 3C, lower panel). Whereas MISP expression levels significantly varied between adherent cancer cell types (Fig. 3C, upper left panel), none of the non-adherent cancer cell lines tested contained detectable levels of MISP (Fig. 3C, lower left panel), in line with its localization to focal adhesions. In addition, we found that MISP is predominantly expressed in cancer cell lines but, except for SV80-transformed fibroblasts, not in the other non-transformed cell types tested (Fig. 3C, right panel).
MISP is phosphorylated during mitosis
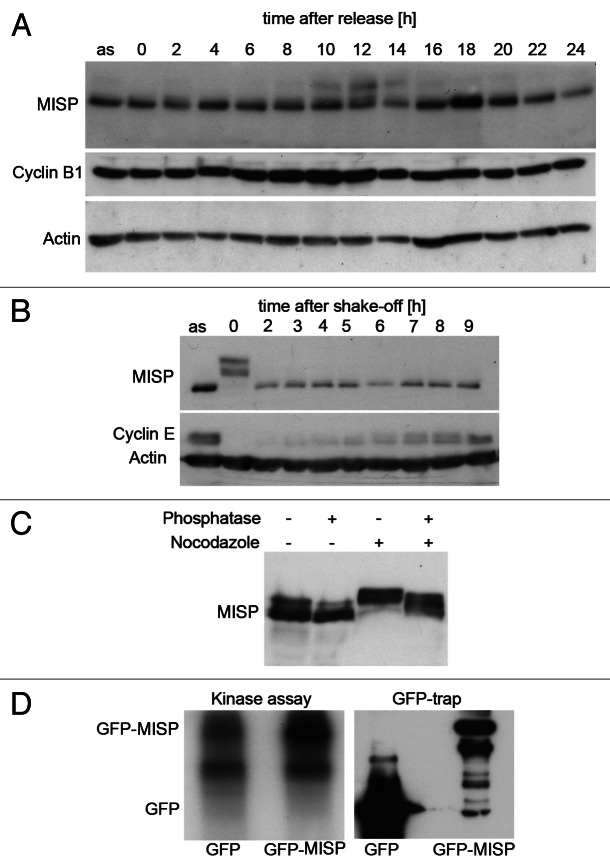
To investigate whether MISP expression levels are regulated during the cell cycle, UPCI:SCC114 cells were synchronized by a double-thymidine block at the G1/S boundary and released for different time points. Cell cycle progression was monitored by cyclin B1 immunoblotting and FACS analysis (data not shown). As shown in Figure 4A, a slower migrating band appears stepwise in G2/M phases (8–12 h after release) and persists until the end of mitosis. To confirm a mitotic modification of MISP, UPCI:SCC114 cells were synchronized in mitosis by nocodazole and collected by mitotic shake-off, thereby confirming the appearance of slower migrating bands in mitotic cells (Fig. 4B). MISP has been found in phosphoproteome analyzes of the mitotic spindle.30,31 Accordingly, incubation of lysates from mitotic UPCI:SCC114 cells with λ-phosphatase led to the disappearance of slower migrating bands, indicating that MISP is phosphorylated during mitosis (Fig. 4C). As Cdk1 is one of the major mitotic kinases, a Cdk1 kinase assay was performed using GFP-MISP from exponentially growing GFP-MISP-U2OS cells as a substrate, thereby demonstrating that GFP-MISP is phosphorylated by Cdk1 in vitro (Fig. 4D). In addition, employing immunofluorescence microscopy, we analyzed whether MISP is present at mitotic spindles. However, by co-immunostaining of both untreated and paclitaxel-treated UPCI:SCC114 cells in order to stabilize microtubules, we were unable to detect MISP at microtubules of the mitotic spindle using both antibodies to C- and N-terminus of the protein (Fig. S3A and B, left panels). Also, GFP-MISP does not localize to mitotic spindles (Fig. S3C, left panel).
Figure 4. MISP is phosphorylated during mitosis. (A) Immunoblotting of UPCI:SCC114 cells synchronized by a double-thymidine block and released for up to 24 h reveals a slower migrating MISP band in G2/M phase of the cell cycle (8–12 h after release). Cyclin B1 expression was used to monitor cell cycle progression. Actin served as loading control. A lysate of UPCI:SCC114 asynchronously growing cells (as) was loaded for comparison. (B) Mitotic modification of MISP (0 h) is more obvious in UPCI:SCC114 cells that have been arrested in mitosis by nocodazole. Cyclin E was used to monitor cell cycle progression (0–9 h) after shake-off of mitotic cells and replating in medium without nocodazole. Actin served as loading control. (C) Immunoblotting of asynchronous (− nocodazole) and mitotic (+ nocodazole) UPCI:SCC114 cell lysates in the absence (− phosphatase) or presence (+ phosphatase) of λ-phosphatase reveals that MISP is phosphorylated during mitosis. (D) GFP-MISP or GFP were isolated from GFP-MISP-U2OS cells or U2OS cells constitutively expressing GFP (right panel) by GFP-trap, incubated with recombinant Cdk1/cyclin B and [γ-32P]-ATP and analyzed by autoradiography (left panel).
In parallel, we also analyzed whether MISP localizes to microtubules during interphase. Untreated or paclitaxel-treated UPCI:SCC114 cells were co-immunostained with an antibody to α-tubulin and either the N14 or C118 antibody against MISP. As in mitotic cells ,we were unable to detect a colocalization of MISP with interphase microtubules (Fig. S3A and B, right panels). Also, no colocalization of GFP-MISP with microtubules was found in GFP-MISP-U2OS cells (Fig. S3C, right panel).
MISP interacts with focal adhesion kinase
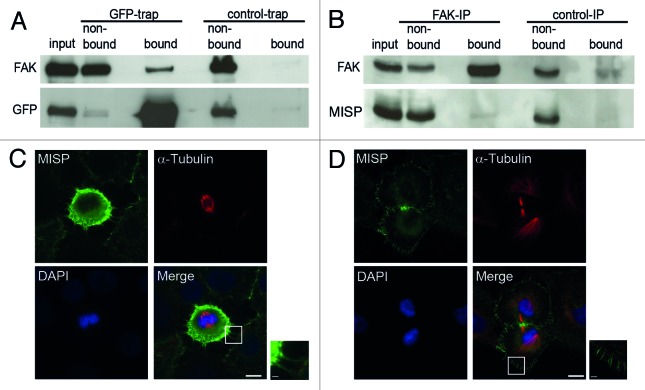
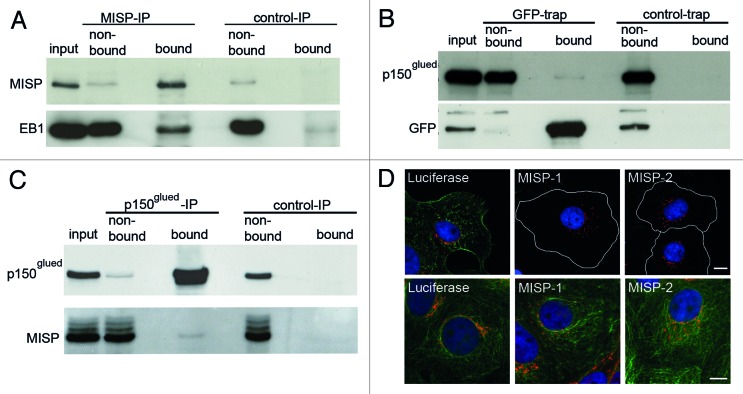
Since we found that the filamentous MISP structures are directed toward focal adhesions by immunofluorescence microscopy, we next analyzed whether MISP interacts with FAK or paxillin. In GFP-trap experiments using GFP-MISP-U2OS cells, FAK clearly co-precipitated with GFP-MISP, whereas no interaction with paxillin was found (Fig. 5A and data not shown). To confirm the interaction of MISP with FAK, we additionally performed co-immunoprecipitation experiments using UPCI:SCC114 cells, demonstrating that endogenous MISP also co-immunoprecipitates with FAK (Fig. 5B).
Figure 5. MISP interacts with FAK and localizes to mitotic retraction fibers. (A) Immunoblot of a GFP-trap showing that FAK co-precipitates with GFP-MISP (bound, GFP-trap) but not with control beads (bound, control-trap). (B) Co-immunoprecipitation (IP) demonstrating that endogenous MISP co-precipitates with FAK (bound), whereas no MISP is detectable in the bound fraction of the control IP. (C) Co-immunostaining with antibodies to MISP (C118) and α-tubulin demonstrates that MISP is present at retraction fibers in mitotic UPCI:SCC114 cells. White rectangles mark the position of the magnified inset. Scale bar, 10 µm; inset 2 µm. (D) In UPCI:SCC114 cells that re-attach during cytokinesis, MISP localizes to membrane protrusions at the cell cortex. White rectangles mark the position of the magnified inset. Scale bar, 10 µm; inset 2 µm.
Further supporting the association of MISP with focal adhesions, immunofluorescence microscopy revealed that MISP is present in retraction fibers, which are formed at former adhesion sites during mitosis18,32 (Fig. 5C). In addition, we observed that MISP is present at spicular membrane protrusions, most pronounced in re-attaching cytokinetic cells (Fig. 5D).
MISP is necessary for directed migration and centrosome orientation
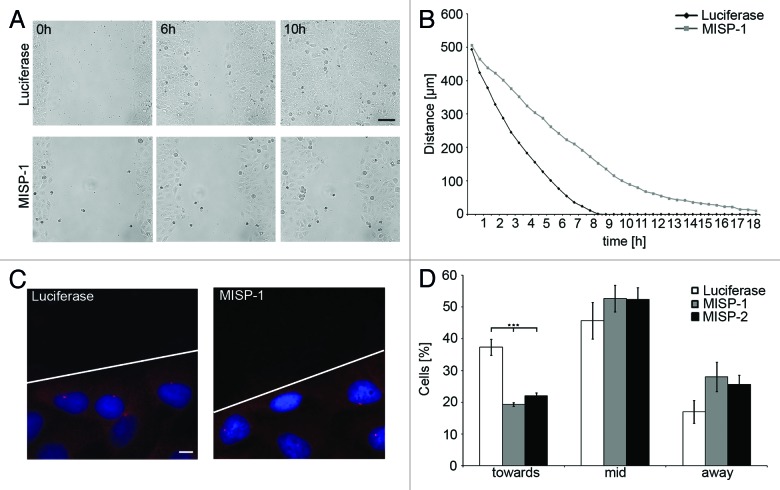
Since focal adhesions are involved in cell migration, and focal adhesion turnover requires dynamic microtubules,33,34 we analyzed whether MISP plays a role in directed migration of cells employing a wound-healing assay. UPCI:SCC114 cells were reverse transfected with MISP-1- or luciferase-siRNA for 24 h and subsequently imaged by live cell video microscopy. A difference between cells transfected with luciferase-siRNA and MISP-depleted cells was already obvious after 6 h (Fig. 6A). The gap between the opposing cell borders was clearly wider in cells after MISP depletion. At 10 h, luciferase-siRNA-transfected cells had completely closed the wound gap, whereas a clear gap between the cell borders was still evident in MISP-depleted cells. Luciferase-siRNA-transfected control cells closed the 500 µm gap within approximately 8 h, whereas cells transfected with MISP-1-siRNA need more than 18 h to close the gap, as revealed by measurement of the gap width at single time points (Fig. 6B).
Figure 6. MISP is involved in directed cell migration and centrosome re-orientation. (A) Live cell video microscopy of UPCI:SCC114 cells in a wound-healing assay shows that MISP-depleted cells close the wound gap more slowly than cells transfected with a siRNA against luciferase. Cells were transfected with luciferase- or MISP-1-siRNA for 24 h and imaged subsequently. Images were taken every 30 min. Scale bar, 100 µm. (B) Determination of the distances between opposing cell borders in UPCI:SCC114 cells transfected with siRNAs against luciferase or MISP from 24 to 42 h after transfection. The graph shows the average of two independent experiments. (C) Immunostaining of centrosomes by an antibody to pericentrin in UPCI:SCC114 cells forced to migrate in a defined direction reveals a defect in centrosome positioning to the side of the nucleus facing the migration direction upon MISP depletion. Cells were transfected with indicated siRNAs for 48 h. The scratch was set 2 h prior fixation. White line depicts scratch. Scale bar, 10 µm. (D) Graph showing centrosome positions relative to the wound edge in UPCI:SCC114 cells forced to migrate in a defined direction demonstrates a reduced frequency of cells in which the centrosome is positioned at the side of the nucleus facing the wound edge (toward) and an increased percentage of cells with centrosomes positioned at (mid) or behind the nucleus (away) after MISP depletion for 48 h. The graph shows the average of three independent experiments; mean ± SD.
Next, we investigated, whether MISP-depleted interphase UPCI:SCC114 cells show defects in centrosome repositioning as a possible reason for impaired directed cell migration. If cells are forced to migrate in a defined direction—as is the case when using the wound-healing assay—centrosomes are reoriented toward the direction of migration in many cell types in a microtubule- and dynein-dependent manner.35,36 To analyze for centrosome repositioning, coverslips with wound-edged UPCI:SCC114 cells forced to migrate in a defined direction were fixed and stained with an antibody against pericentrin to mark centrosomes 2 h after wounding by manual scratching (Fig. 6C). In roughly one-third of the cells (37.3 ± 2.5%) transfected with luciferase-siRNA, the centrosome was repositioned in front of the nucleus relative to the direction of migration. Using two different siRNAs, in MISP-depleted cells, the centrosome was repositioned toward the migration direction in only about 20% of the cells (MISP-1-siRNA 19.3 ± 0.6%, p = 0.0003; MISP-2-siRNA 22.0 ± 1.0%, p = 0.0006). Correspondingly, the number of cells in which the centrosome was located at the level of the nucleus (luciferase-siRNA 45.7 ± 5.8%, MISP-1-siRNA 52.7 ± 4.2%, MISP-2-siRNA 52.3 ± 2.8%) or even behind the nucleus (luciferase-siRNA 17.0 ± 3.6%, MISP-1-siRNA 28 ± 4.6%, MISP-2-siRNA 25.7 ± 2.9%) was increased in MISP-depleted cells compared with cells transfected with a siRNA to luciferase (n = 300 cells per siRNA, Fig. 6D).
MISP is involved in spindle orientation and positioning
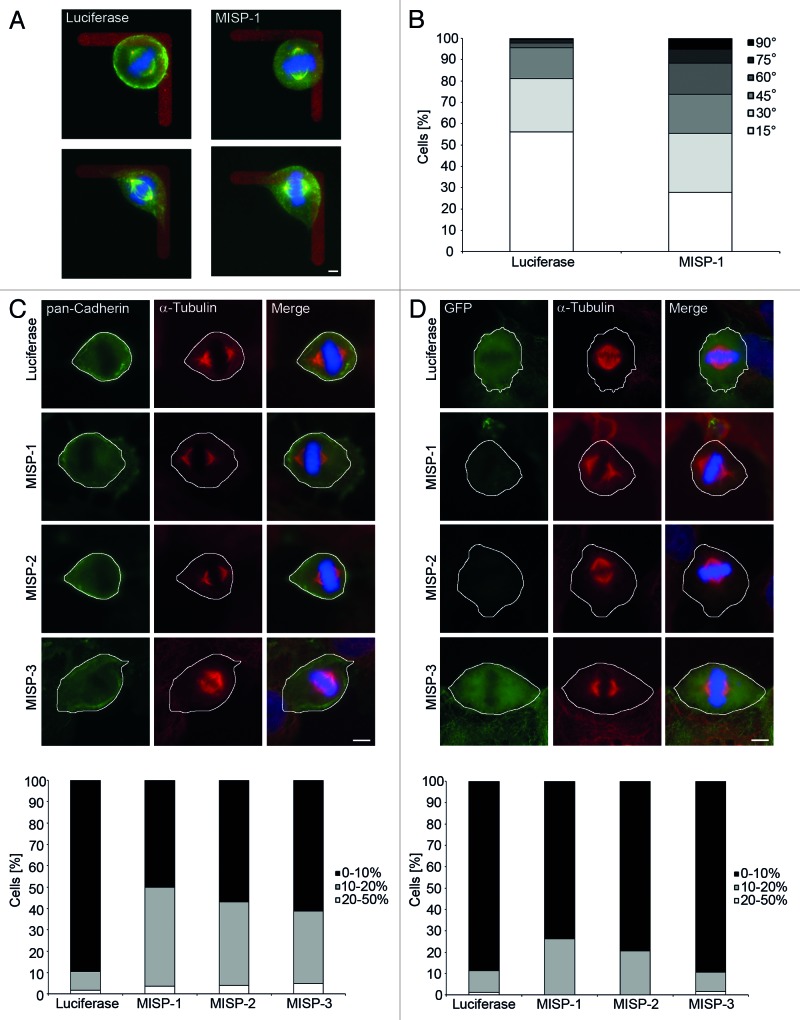
Based on its localization and role in the generation of spindle tension, we next asked whether MISP might also play a role in spindle orientation. To test for this assumption, we made use of fibronectin-coated L-shaped micropatterns on which cells orient their mitotic spindles along the hypotenuse, corresponding to longest cell axis, of the L-shaped pattern.18 UPCI:SCC114 cells were transfected with MISP-1- or luciferase-siRNA, plated onto micropatterns and evaluated for spindle orientation 48 h after plating. At least 130 bipolar spindles were evaluated per siRNA. Cells were grouped according to the angular deviation of their spindle axis away from the hypotenuse. More than half of the cells transfected with luciferase-siRNA (56%; n = 132 cells) oriented their mitotic spindle along the hypotenuse with a deviation of less than 15° (Fig. 7A and B). Orientation of mitotic spindles was more randomized in cells transfected with MISP-1-siRNA, with only 28% of cells (n = 137) having their mitotic spindle oriented along the hypotenuse with a deviation of less than 15°. Accordingly, significantly more mitotic spindles in MISP-depleted cells deviate for more than 15° from the hypotenuse of the L-pattern compared with cells transfected with luciferase-siRNA (p = 9 × 10−12).
Figure 7. MISP is involved in spindle orientation and positioning. (A) Representative examples of mitotic UPCI:SCC114 cells transfected for 48 h with a siRNA against luciferase (left) or MISP (right) on an L-shaped fibronectin micropattern, stained with antibodies against pericentrin (red) and α-tubulin (green); DNA (Hoechst 33342) is shown in blue. Luciferase-siRNA-transfected cells orient their mitotic spindles along the hypotenuse of the L-shaped micropattern. In contrast, in MISP-1-siRNA-transfected cells the spindle orientation deviates from this orientation. Scale bar, 5 µm. (B) Frequency of angular deviations of spindle orientation away from the hypotenuse of L-shaped micropatterns for cells treated with siRNAs to luciferase or MISP. The graph represents the sum of all mitotic cells evaluated in three independent experiments. (C) Representative images (upper panel) of UPCI:SCC114 cells transfected with indicated siRNAs for 48 h and stained with antibodies against pan-cadherin to mark cell boundaries and α-tubulin reveal that mitotic spindles in MISP-depleted cells are located outside the cell center. White lines depict cell boundaries. Scale bar, 10 µm. A graph depicting the distribution of mitotic spindle positions relative to the cell center after transfection of the indicated siRNAs is shown in the lower panel. The graph represents the sum of mitotic cells evaluated in three independent experiments. (D) Representative images (upper panel) of GFP-MISP-U2OS cells transfected with indicated siRNAs stained with an antibody to α-tubulin and Hoechst 33342. The mitotic spindle is displaced from the cell center upon depletion of MISP by siRNAs MISP-1 and MISP-2, both of which deplete endogenous as well as overexpressed GFP-MISP. In cells transfected with MISP-3-siRNA targeting only endogenous MISP mitotic spindles are positioned at the cell center. Expression of GFP-MISP was induced 24 h after siRNA transfection for further 24 h. White lines depict cell boundaries. Scale bar, 10 µm. Determination of spindle positions (lower panel) shows that mitotic spindles in cells transfected with siRNAs MISP-1 and MISP-2 are displaced from the cell center, whereas overexpression of GFP-MISP rescues spindle positioning to the cell center. The graph shows the sum of mitotic cells evaluated in three independent experiments.
In line with these defects in spindle orientation on micropatterns, we found that mitotic spindles in MISP-depleted UPCI:SCC114 cells are not located at the geometric center of the cells. In 90% of luciferase-siRNA-treated cells the mitotic spindle was located at the geometric cell center, whereas depletion of MISP resulted in a significantly increased percentage of cells with mitotic spindles whose localization deviated > 10% from the cell center with all three siRNAs used (MISP-1-siRNA 50%, p = 3 × 10−20; MISP-2-siRNA 43%, p = 5 × 10−13; MISP-3-siRNA 39%, p = 2 × 10−11; n ≥ 50 cells per siRNA, Fig. 7C). Similar results were obtained when GFP-MISP-U2OS cells were used. Whereas in 91% of luciferase-siRNA-treated GFP-MISP-U2OS cells, the mitotic spindle was located at the geometric cell center, after depletion of MISP by MISP-1- and MISP-2-siRNAs, targeting the coding region, localization of the mitotic spindle with less than 10% deviation from the geometric cell center was found in only 74% (p = 1 × 10−7) and 71% (p = 1 × 10−7) of cells, respectively (n ≥ 60 cells per siRNA, Fig. 7D). In contrast, in GFP-MISP-U2OS cells transfected with MISP-3-siRNA, which targets the 3′UTR of endogenous MISP, spindle positioning was rescued with 91% (p = 0.1) of the mitotic spindles being localized at the geometric cell center (Fig. 7D).
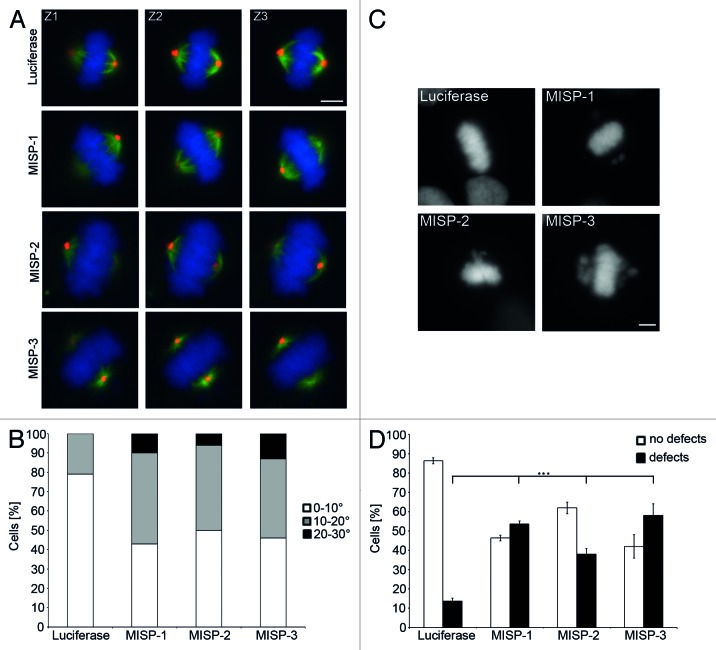
As in nonpolarized cells in tissue culture, integrin-dependent, retraction fiber-mediated cell adhesion orients the spindle parallel to the substrate;21 we next determined whether MISP-depleted cells are also defective in orienting their spindle axis parallel to the tissue culture dish. Almost 80% of luciferase-siRNA-transfected GFP-α-tubulin-expressing UPCI:SCC114 cells harbored properly oriented mitotic spindles with less than 10° deviation relative to the substrate plane (79%, Fig. 8A and B). In contrast, depletion of MISP caused misorientation of spindles. In about half of MISP-depleted cells the angle of mitotic spindles deviated more than 10° relative to the substrate plane (MISP-1-siRNA 57%, p = 1 × 10−20; MISP-2-siRNA 50%, p = 1 × 10−12, MISP-3-siRNA 54%, p = 5 × 10−16; n = 100 cells per siRNA, Fig. 8A and B). Together, these experiments demonstrate that MISP is important for correct spindle positioning and orientation.
Figure 8. MISP impacts on spindle positioning and chromosome alignment. (A) Representative z-stack images (Z1, Z2, Z3 are 1 µm apart from each other) of GFP-α-tubulin-expressing UPCI:SCC114 cells immunostained with an antibody to pericentrin (red) and Hoechst 33342 (blue). Whereas centrosomes of luciferase-siRNA-transfected cells localize to the same focal plane, centrosomes in MISP-depleted cells are found in different focal planes. Cells were transfected with indicated siRNAs for 48 h. Scale bar, 10 µm. (B) The frequency distribution of the angles between substrate plane and the spindle axis is shown. The graph depicts the sum of mitotic cells evaluated in three independent experiments. Angles were determined using inverse trigonometric functions. (C) DNA staining by Hoechst 33342 of UPCI:SCC114 cells transfected with luciferase-siRNA or MISP-siRNAs for 48 h exhibits chromosomes that are not aligned at the metaphase plate in MISP-depleted cells. Scale bar, 10 µm. (D) Quantification of defects in chromosome alignment in UPCI:SCC114 cells 48 h after transfection with indicated siRNAs. The graph shows the average of three independent experiments; mean ± SD.
As MISP knockdown delays mitotic progression and causes defects in spindle tension, orientation and positioning, we analyzed MISP-depleted cells for further mitotic defects. Whereas 48 h after luciferase-siRNA transfection in 86.3 ± 1.5% of UPCI:SCC114 cells in metaphase all chromosomes were correctly aligned, MISP depletion led to significantly increased chromosome misalignment rates with all three MISP-specific siRNAs used. Accordingly, mitoses with unperturbed metaphase chromosome alignment were reduced to 46.3 ± 1.5% (MISP-1-siRNA, p = 5.6 × 10−6), 62.0 ± 3.0% (MISP-2-siRNA, p = 0.0002) and 42.0 ± 6.0% (MISP-3-siRNA, p = 0.0003), respectively (n = 300 mitoses per siRNA, Fig. 8C and D).
MISP interacts with p150glued
As the dynein/dynactin complex and EB1 are known to be critically important for spindle positioning and orientation as well as for directed migration and centrosome reorientation,14,21,22,24,35,37-39 we next analyzed whether MISP interacts with components of the dynein/dynactin complex or EB1. An interaction between endogenous MISP with EB1 could be clearly demonstrated by co-immunoprecipitation experiments in lysates from UPCI:SCC114 cells (Fig. 9A). Also, a weak interaction between GFP-MISP and p150glued, a component of the dynein/dynactin complex, was detectable by GFP-trap technology in GFP-MISP-U2OS cells (Fig. 9B). In co-immunoprecipitation experiments using an antibody against p150glued for precipitation, this weak interaction could be confirmed (Fig. 9C). In line with these results, we also detected an impact of MISP on the intracellular distribution of the Golgi complex stained by the Golgi marker GM130. Compared with luciferase-siRNA-transfected UPCI:SCC114 cells, cytoplasmic dispersal of the Golgi complex with Golgi stacks scattered within the cytoplasm was significantly more frequent after MISP depletion using two different MISP-siRNAs (luciferase-siRNA 24 ± 0.5%, MISP-1-siRNA 43.3 ± 2.5%, p = 0.0002; MISP-2-siRNA 41.8 ± 2.4%, p = 0.0002; n = 600 cells per siRNA; Fig. 9D). As it has been described that an impaired dynein/dynactin function results in Golgi dispersal,40 these findings point to a role of MISP on dynein/dynactin function as well.
Figure 9. MISP interacts with p150glued and EB1. (A) Immunoblot of a co-immunoprecipitation showing that EB1 co-precipitates with MISP (MISP-IP, bound). (B) Immunoblot of a GFP-trap from GFP-MISP-U2OS cells showing a weak interaction of GFP-MISP with p150glued (GFP-trap, bound). (C) Co-immunoprecipitation demonstrating that endogenous MISP co-precipitates with p150glued (p150glued-IP, bound). (D) Co-immunostaining of UPCI:SCC114 cells with antibodies to either MISP (C118, green) and the Golgi component GM130 (red, upper panel) or α-tubulin (green) and GM130 (red, lower panel) 48 h after transfection with indicated siRNAs demonstrating that the Golgi complex is scattered in MISP-depleted cells. Cell boundaries are depicted as white lines. Scale bar, 10 µm.
Discussion
In this study, we show that the previously uncharacterized protein MISP (C19ORF21) plays an important role in spindle positioning and orientation by its association with the actin cytoskeleton and focal adhesions on the one hand and EB1 and the dynein/dynactin complex on the other hand, thereby allowing for the generation of spindle tension. In addition, MISP is required for clustering of supernumerary centrosomes into a bipolar spindle array in cancer cells via the same mechanism.
We identified MISP in a genome-wide RNAi screen as a protein involved in centrosomal clustering in cancer cells with extra centrosomes.11 Centrosomal clustering allows cancer cells harboring supernumerary centrosomes to successfully divide in a bipolar manner and to avoid lethal multipolar divisions.8,10,12 Bipolar spindle formation via centrosomal clustering, however, is associated with an increased frequency of lagging chromosomes during anaphase, thereby explaining the link between extra centrosomes and chromosomal instability.8,41 Mechanistically, centrosomal clustering depends on the SAC, which provides the additional time required for the clustering process and spindle tension as controlled by the cortical actin cytoskeleton, cell adhesion molecules and kinetochore components.11,13,42 Accordingly, dynein, NuMA and various other proteins involved in cell adhesion or associated with the actin or microtubule cytoskeleton have been identified to be required for centrosomal clustering in Drosophila S2 and human cancer cells.10,11,13
Here, we show that MISP is predominantly expressed in adherently growing cancer cell lines, and that depletion of MISP leads to spindle multipolarity only in cells with extra centrosomes, with complete centrosomes being present at each spindle pole. In addition, we found that MISP depletion causes a mitotic arrest by activation of the SAC. Importantly, this arrest was not restricted to cells harboring supernumerary centrosomes, pointing to a function of MISP in mitotic progression independent of the centrosome content of cells. Accordingly, interference with MISP caused SAC activation as a consequence of reduced spindle tension, as evidenced by persistent kinetochore BubR1 staining and reduced interkinetochore distances, irrespective of spindle polarity in both multipolar spindles after inhibition of centrosomal clustering as well as in bipolar spindle arrays in cells with a regular centrosome content. Together, these data demonstrate that MISP is required for spindle tension in general and centrosomal clustering into a bipolar spindle array in cancer cells with supernumerary centrosomes.
Despite that MISP has been found in phosphoproteome screens of the mitotic spindle,30,31 we were unable to detect the protein—albeit heavily phosphorylated during mitosis—at either interphase or mitotic microtubules. Instead, MISP colocalizes with both the actin cytoskeleton and focal adhesions during interphase and retraction fibers during mitosis and interacts with FAK.
In addition, we have found interactions between MISP and EB1 as well as the dynactin subunit p150glued. Astral microtubules are tethered to the cell cortex via microtubule +TIP proteins including CLASPs, EB1 and the dynein/dynactin complex.22,25,43,44 In yeast, disruption of astral microtubule attachment to the cell cortex leads to activation of the SAC and a delay in anaphase onset as well as defects in chromosome congression.45 Insufficient attachment of astral microtubule plus ends to the cell cortex leads to spindle misorientation.17,18,21 In line with these findings, in addition to induction of spindle multipolarity in cells with extra centrosomes, ablation of MISP also causes defects in spindle orientation and positioning, processes known to be controlled by integrin-mediated cell adhesion to the extracellular matrix via actin-rich mitotic retraction fibers.18,20,21 Attachment of astral microtubules to the cell cortex as well as EB1 and the dynein/dynactin complex have also been shown to be important for spindle positioning and orientation in mammalian cells.14,15,18,21,24 Thus, our findings suggest that spindle positioning and orientation defects after MISP depletion are a consequence of deficient attachment of astral microtubules to the cell cortex, although by immunofluorescence microscopy we did not observe obvious abnormalities of astral microtubules (data not shown).
Directed cell migration and centrosome repositioning to the leading edge of migrating cells, processes that are focal adhesion- and microtubule- as well as dynein-dependent, respectively,33-36,38 were defective after interference with MISP function as well. These findings therefore corroborate the proposed linker function of MISP between cell cortex and microtubule cytoskeleton, not only in mitosis but also during interphase.
In conclusion, we provide evidence that MISP localizes to cortical structures of adherent cells to bridge actin filaments and focal adhesions with astral spindle microtubules, thereby explaining how the interphase adhesion pattern of cells might impact on spindle positioning and orientation in rounded cells during mitosis. As, via the same mechanism, MISP, which is predominantly expressed in transformed adherent cell types, seems to be involved in centrosome clustering in cancer cells, targeting MISP might be a novel strategy to selectively eradicate malignancies with supernumerary centrosomes.
Materials and Methods
Plasmid generation
The cDNA of MISP was purchased from Imagenes (Clone ID: IRATp970D1068D). The cDNA was cloned into pEGFP-C1 (Clontech, 6084-1; for-primer: ctcgagtggaccgcgtgacc, rev-primer: ggatcctcagtcatcctcctcactg). To generate an inducible cell line these fusion construct was cloned into pTRE2hyg (Clontech, 631014; for-primer: ggatccatggtgagcaagggcgaggag, rev-primer: gcggccgctcagtcatcctcctcactg).
Cell culture
UPCI:SCC114 cells (S.M. Gollin; oral squamous cell carcinoma) and U2OS cells (ATCC HTB-96, osteosarcoma) were cultured in DMEM (Invitrogen, 31966-021) supplemented with 10% FCS (Biochrom AG, S0415). An inducible GFP-MISP cell line was generated using the U2OS-Tet-On-System (GFP-MISP-U2OS) from Clontech (U2-OS Tet-On, 631143). The cells were cultured in DMEM (Invitrogen) supplemented with 10% Tetracylin-free FCS (Clontech, 631106) in the presence of 200 µg/ml G418 (PAA Laboratories, P11-012) and 100 µg/ml Hygromycin B (Invitrogen, 10687010). UPCI:SCC114 cells stably expressing GFP-α-tubulin were cultured in DMEM supplemented with 10% FCS and 500 µg/ml G418.9 MDA-MB-231 (ATCC HTB-26; breast adenocarcinoma), DLD-1 (ATCC, CCl-221; colorectal adenocarcinoma) and DU145 cells (ATCC, HTB-81; prostate carcinoma) were cultured in RPMI 1640 (Invitrogen, 61870-010) supplemented with 10% FCS (Biochrom AG), and hTERT-RPE-1 cells (ATCC, CRL-4000; retinal pigmented epithelium) were cultured in DMEM-F12 (Invitrogen) supplemented with 10% FCS (Biochrom AG) and 7.5% sodium bicarbonate (Pan Biotech, P04-441000). When indicated, cells were treated with paclitaxel (Calbiochem, 580555) or nocodazole (Calbiochem, 487928). Cell lines or lysates only used for immunoblotting were kindly provided by M. Matuszewska, M. Schmidt-Zachmann and F. Liu.
Generation of mouse monoclonal antibodies against MISP
Antibodies were raised against N-terminal (aa 1–250) and C-terminal (aa 430–679) sequences of MISP. The corresponding sequences (N-for-primer: tatataggatccatggaccgcgtgacc; N-rev-primer: tatatactcgagctggggcttgatggg; C-for-primer: tatataggatccatgagccccgggac; C-rev-primer: tatatactcgaggtcatcctcctcactg) were cloned into the pET21a-plasmid (Novagen, 69740-3) containing a 6×Hisidin-tag. Bacterially expressed and affinity-purified His-tagged proteins were used to immunize mice according to a standard immunization protocol.46 Fusions were screened by immunoblotting as well as immunofluorescence microscopy. Positive clones were subcloned and the subclones used (N14/1 and C118/6) were typed as mouse IgG1.
Plasmid and siRNA transfection
siRNAs were purchased from either Eurofins MWG Operon or Dharmacon (Thermo Scientific) and reverse transfected into the indicated cell lines using Dharmafect-1 (Thermo Scientific, T-2001) according to manufacturer’s instructions in a final concentration of 50 nM. Luciferase-siRNA: cuuacgcugaguacuucga; MISP-1-siRNA: cgacccagcuccagaagug; MISP-2-siRNA: gcacaaagcaagaggcauc; MISP-3-siRNA: ggucaaagaggguggcaca. Plasmids were transfected using Fugene 6 (Promega, E2691) according to manufacturer's instructions.
Antibodies
The following antibodies were used: mouse monoclonal antibodies to α-tubulin (Sigma-Aldrich, T6199), centrin-1 (J.L. Salisbury), cyclin B1 (Santa Cruz, sc-245), cyclin E (Santa Cruz, sc-198), GAPDH (Santa Cruz, sc-47724), GFP (Santa Cruz, sc-9996) and Hec-1 (Novus Biologicals, NB100-338) as well as rabbit polyclonal antibodies to actin (Santa Cruz, sc-1616-R), α-tubulin (Abcam, ab52866), FAK (Cell Signaling, 3285), FAKY397 (Abcam, ab4803), GM130 (Abcam, ab52649), paxillin (Sigma-Aldrich, SAB4300592), pan-cadherin (Abcam, ab16505), pericentrin (Abcam, ab4448) and p150glued (Bethyl Laboratories, A303-072A-1). The actin cytoskeleton was visualized by Phalloidin-TRITC (Sigma-Aldrich, P1951).47 In addition, we used a human antiserum against Crest (ANA, Euroimmun, CA1611-0101) and a rat monoclonal antibody against EB1 (Abcam, ab53358). A sheep polyclonal antibody against BubR1 was kindly provided by S.S. Taylor.
Immunofluorescence microscopy
Immunofluorescence staining was performed as described before.48 The following fluorochrome-conjugated secondary antibodies were used: anti-mouse Alexa-488 (Invitrogen, A11029), anti-rabbit Alexa-488 (Invitrogen, A11034), anti-human Alexa-488 (Invitrogen, A11013), anti-mouse Cy3 (Dianova, 115-165-166), anti-rabbit Cy3 (Dianova, 111-165-144) and anti-sheep Cy3 (Dianova, 313-165-045). For DNA counterstaining, Hoechst 33342 (Invitrogen, H1399) was used. Immunostained cells were examined using an Axiovert 200 M microscope (Zeiss) or a Cell Observer (Zeiss) equipped with LD-Plan neofluar 40x/0.6 NA, EC-Plan Neofluar 40x/1.30 NA, EC-Plan Neofluar 100x/1.30 NA and Plan-Apochromat 63x/1.4 NA objectives (Zeiss). Images were processed with AxioVision (Zeiss), Zen 2011 (Zeiss), ImageJ (NIH) and Photoshop software (Adobe).
Spindle tension measurement
Spindle tension measurement was performed as described before.11 Briefly, cells were reverse transfected on coverslips and fixed after 48 h. Cells were stained with Hec-1, Crest and Hoechst, and the distance between Hec-1 signals connected by Crest was measured.
Centrosome positioning
Centrosome repositioning was analyzed as described.49 Cells were reverse transfected on coverslips for 48 h. Two hours before fixation, a scratch using a sterile 10 µl pipette tip was set in the middle of the coverslip. Cells were stained with pericentrin and Hoechst to determine centrosome position in relation to the nucleus. Centrosome positions were grouped into toward (centrosome located at the side of the nucleus facing the scratch), away (centrosome located away from scratch relative to the nucleus) or mid (centrosome located at the height of the nucleus).
Spindle orientation assay using CYTOOchips
Spindle orientation was determined as described by Théry and coworkers.18 UPCI:SCC114 cells were reverse transfected with indicated siRNAs. Forty-five hours post transfection cells were trypsinized, and 60,000 cells were plated onto L-shaped, fibronectin-coated CYTOOchips in a 35 mm culture dish. To allow initial attachment of the cells to the micropattern, culture dishes were incubated under the hood for 15 min and then moved to the incubator for 1 h. Floating cells that had not attached to micropattern within this hour were removed by gently washing the micropattern with medium. After an additional hour cells were fixed using 4% PFA.
Spindle positioning
For determination of the spindle position relative to the cell center, UPCI:SCC114 cells were reverse transfected with indicated siRNAs. After 48 h, cells were fixed and stained for α-tubulin and pan-cadherin to mark spindles and cell boundaries. GFP-MISP-U2OS cells were also transfected with indicated siRNAs for 48 h, and the expression of GFP-MISP was induced 24 h after siRNA transfection. After fixation, GFP-MISP-U2OS cells were stained with α-tubulin. To determine the spindle position, cell diameters were measured along the spindle pole axes. Along the same axis, the distance from the cell cortex to the mid of the metaphase plate was measured. From these values, the location of mitotic spindles was calculated, and a deviation of less than 10% was counted as centered position.
For determination of the spindle position relative to the substrate plane, GFP-α-tubulin-expressing UPCI:SCC114 cells were reverse transfected with indicated siRNAs. After 48 h, cells were fixed and immunostained for pericentrin to mark centrosomes. To determine the spindle position, z-stack (1 µm apart) images were taken and the angle of the mitotic spindles relative to the substrate plane was determined using inverse trigonometric functions. A deviation of less than 10° was counted as parallel to the substrate.
Flow cytometry
Cell cycle analysis was performed by propidium iodide (BD biosciences, 556463) staining according to manufacturer’s instructions. Measurements and analyzes were performed using a FACScan (BD Biosciences) and Cell Quest software (BD Biosciences). At least 25,000 cells per experiment were measured.
Wound-healing assay
Healing assays were performed using culture inserts (Ibidi, μ-dishes, 80206). Cells were reverse transfected and plated at a density of 5 × 105 cells/ml. Inserts were removed 24 h after transfection and cells were imaged subsequently at 37°C in a humidified atmosphere containing 5% CO2 using a Cell Observer (Zeiss). Pictures were taken every 30 min at eight positions per experiment for up to 20 h. For analysis, the distance between the opposing cell fronts at each position was measured at every time point using AxioVision (Zeiss).
GFP-trap
For GFP-trap (chromotek, gta-100) assays,50 GFP-MISP-U2OS cells were induced by 0.5 µg/ml doxycycline (Sigma-Aldrich, 44577) for 48 h. GFP-trap was performed according to the manufacturer’s instructions (Chromotek). Negative controls were generated by trap with blocked agarose beads (Chromotek, bab-20). All washing steps were performed using dilution buffer (10mM TRIS-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA) supplemented with Complete Protease Inhibitor Cocktail Ultra (Roche, 04693116001) and Phos-Stop (Roche, 04906837001).
Immunoprecipitation
For immunoprecipitation, precipitating antibodies were preincubated with cell lysates for 1 h at room temperature. Afterwards the antibody/lysate-mixture was added to agarose beads coupled with protein G or protein A (50% slurry; Roche, protein A: 11719408001; protein G: 11719416001) and incubated overnight at 4°C on an end-over-end rotator. Negative controls were generated by immunoprecipitation with either mouse or rabbit IgG (Santa Cruz, mouse: sc-2025; rabbit: sc-2027). After three washing steps for 5 min each, proteins were analyzed by immunoblotting. All washing steps were performed using dilution buffer (10mM TRIS-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA) containing Complete Protease Inhibitor Cocktail Ultra (Roche) and Phos-Stop (Roche).
Immunoblotting
Cell protein extracts were prepared by lysis of cells in an appropriate volume of RIPA buffer (50 mM TRIS-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with Complete Protease Inhibitor Cocktail (Roche) and Phos-Stop (Roche), followed by mechanical homogenization and collection of the supernatant after 15 min centrifugation at 20,000 × g. Immunoblotting was performed according to standard protocols. All used horseradish-peroxidase coupled antibodies were obtained from Santa Cruz.
Kinase assay
Following GFP-trap using GFP-MISP-U2OS cells or U2OS cells stably expressing GFP as a control, the beads were incubated with 20 U Cdk1/cyclin B (NEB, P6020S) as well as 10 µCurie [γ-32P]-ATP for 30 min at 30°C in protein kinase buffer (NEB, B6022). After incubation, the sample was loaded on a SDS gel. Then, the gel was dried and examined by autoradiography.
Statistical analysis
If not indicated otherwise, results are given as mean ± standard deviation (SD) of at least three independent experiments. Significances were determined using the two-tailed Student’s t-test or the ×2-test.
While this paper was in the review process, another paper appeared51 that also characterized C19ORF21 and designated it MISP (mitotic interactor and substrate of Plk1).
Supplementary Material
Acknowledgments
Support of the DKFZ Light Microscopy Facility and the Carl Zeiss Application Center Heidelberg is gratefully acknowledged. We thank S. Gollin for providing UPCI:SCC114 cells. We are grateful to S.S. Taylor and J.L. Salisbury for providing antibodies against BubR1 and Centrin-1, respectively. We are indebted to M. Schmidt-Zachmann, M. Matuszewska and F. Liu for providing cell lysates. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, KR-1981/3-1) granted to A.K. and financial support by the University of Heidelberg.
Glossary
Abbreviations:
- Cdk
cyclin-dependent kinase
- ECM
extracellular matrix
- GFP
green fluorescent protein
- FAK
focal adhesion kinase
- MISP
mitotic interactor and substrate of Plk1
- RNAi
RNA interference
- SAC
spindle assembly checkpoint
- SD
standard deviation
- siRNA
small interfering RNA
- +TIP
microtubule plus end-binding proteins
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24602
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24602
References
- 1.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–63. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 2.Lüders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–7. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 3.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–25. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 4.Krämer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia. 2002;16:767–75. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- 5.Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, et al. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–85. [PubMed] [Google Scholar]
- 6.Krämer A, Neben K, Ho AD. Centrosome aberrations in hematological malignancies. Cell Biol Int. 2005;29:375–83. doi: 10.1016/j.cellbi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci. 2011;7:1122–44. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebacz B, Larsen TO, Clausen MH, Rønnest MH, Löffler H, Ho AD, et al. Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res. 2007;67:6342–50. doi: 10.1158/0008-5472.CAN-07-0663. [DOI] [PubMed] [Google Scholar]
- 10.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 11.Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, et al. Proteins required for centrosome clustering in cancer cells. Sci Transl Med. 2010;2:33ra38. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- 12.Krämer A, Maier B, Bartek J. Centrosome clustering and chromosomal (in)stability: a matter of life and death. Mol Oncol. 2011;5:324–35. doi: 10.1016/j.molonc.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotak S, Busso C, Gönczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–74. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–5. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Gönczy P. Mechanisms of spindle positioning: focus on flies and worms. Trends Cell Biol. 2002;12:332–9. doi: 10.1016/S0962-8924(02)02306-1. [DOI] [PubMed] [Google Scholar]
- 18.Théry M, Racine V, Pépin A, Piel M, Chen Y, Sibarita JB, et al. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–53. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 19.Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–9. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Fink J, Carpi N, Betz T, Bétard A, Chebah M, Azioune A, et al. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771–8. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- 21.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–98. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuyler SC, Pellman D. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 2001;105:421–4. doi: 10.1016/S0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 23.Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol. 2010;30:3519–30. doi: 10.1128/MCB.00394-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–7. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–84. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park AY, Shen TL, Chien S, Guan JL. Role of focal adhesion kinase Ser-732 phosphorylation in centrosome function during mitosis. J Biol Chem. 2009;284:9418–25. doi: 10.1074/jbc.M809040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto S, Teramoto H, Coso OA, Gutkind JS, Burbelo PD, Akiyama SK, et al. Integrin function: molecular hierarchies of cytoskeletal and signaling molecules. J Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fielding AB, Dobreva I, McDonald PC, Foster LJ, Dedhar S. Integrin-linked kinase localizes to the centrosome and regulates mitotic spindle organization. J Cell Biol. 2008;180:681–9. doi: 10.1083/jcb.200710074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fielding AB, Lim S, Montgomery K, Dobreva I, Dedhar S. A critical role of integrin-linked kinase, ch-TOG and TACC3 in centrosome clustering in cancer cells. Oncogene. 2011;30:521–34. doi: 10.1038/onc.2010.431. [DOI] [PubMed] [Google Scholar]
- 30.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011;10:004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nousiainen M, Silljé HH, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Théry M, Bornens M. Cell shape and cell division. Curr Opin Cell Biol. 2006;18:648–57. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Stehbens S, Wittmann T. Targeting and transport: how microtubules control focal adhesion dynamics. J Cell Biol. 2012;198:481–9. doi: 10.1083/jcb.201206050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–90. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 35.Yvon AM, Walker JW, Danowski B, Fagerstrom C, Khodjakov A, Wadsworth P. Centrosome reorientation in wound-edge cells is cell type specific. Mol Biol Cell. 2002;13:1871–80. doi: 10.1091/mbc.01-11-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23:579–88. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–40. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–11. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gierke S, Wittmann T. EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr Biol. 2012;22:753–62. doi: 10.1016/j.cub.2012.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–84. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–42. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–50. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- 44.Laan L, Roth S, Dogterom M. End-on microtubule-dynein interactions and pulling-based positioning of microtubule organizing centers. Cell Cycle. 2012;11:3750–7. doi: 10.4161/cc.21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tournier S, Gachet Y, Buck V, Hyams JS, Millar JB. Disruption of astral microtubule contact with the cell cortex activates a Bub1, Bub3, and Mad3-dependent checkpoint in fission yeast. Mol Biol Cell. 2004;15:3345–56. doi: 10.1091/mbc.E04-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 47.Mattison CP, Stumpff J, Wordeman L, Winey M. Mip1 associates with both the Mps1 kinase and actin, and is required for cell cortex stability and anaphase spindle positioning. Cell Cycle. 2011;10:783–93. doi: 10.4161/cc.10.5.14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krämer A, Mailand N, Lukas C, Syljuåsen RG, Wilkinson CJ, Nigg EA, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–91. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 49.Euteneuer U, Schliwa M. Mechanism of centrosome positioning during the wound response in BSC-1 cells. J Cell Biol. 1992;116:1157–66. doi: 10.1083/jcb.116.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–9. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Zhu M, Settele F, Kotak S, Sanchez-Pulido L, Ehret L, Pointing CP, et al. MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. J Cell Biol. 2013;200:773–87. doi: 10.1083/jcb.201207050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.