Increasing evidence for the role of histone modifying enzymes on non-histone substrates, including those in the cytoplasm, has been emerging in recent years. Some chromatin modifying proteins, such as the histone methyl transferase EZH2, compartmentalize to both the nucleus and cytoplasm; whereby EZH2 can methylate histones and actin, respectively.1 Alternatively, the mammalian nuclear membrane, during mitosis, disintegrates and the structural boundary separating nuclear and cytoplasmic proteins breaks down. This allows nuclear proteins to diffuse into the cytoplasm and potentially catalyze non-nuclear substrates. Evidence suggests that the Gcn5 containing histone acetyl transferase complex ATAC controls mitotic progression through the modification on non-histone targets during mitosis.2 And more recently, studies have shown that the HIPK2 kinase controls cytokinesis through the phosphorylation of histone H2B localized at the midbody, the site of cell abscission at the end of mitosis.3 What these and other studies suggest is that coordination and crosstalk between the chromatin and cytoskeletal structures is much more intertwined than previously appreciated.
In a recent issue of Cell Cycle, we show that the histone ubiquitin ligase RNF8, which orchestrates the mammalian DNA damage response (DDR) following DNA double strand break (DSB) formation, also controls mitotic progression through the ubiquitylation of septins in cells4 (Fig. 1). Furthermore, we provide evidence that this novel role of RNF8 in mitosis is conserved from budding yeast to humans. On the contrary, support for a direct role of the yeast RNF8 orthologs Dma1 and Dma2 in the DDR or in histone ubiquitylation remains lacking. This is partly due to the compartmentalization of yeast Dma proteins to the cytoplasm,5,6 and partly because Dma null yeast strains lack documented hypersensitivity to DNA damaging agents.5 Our preliminary studies suggest that Dma1 and Dma2 double-null yeast strains are hypersensitive to the DNA damaging agents hydroxyurea and bleomycin but these have been attributed to a defect in Swe1 protein levels or efficiency of drug uptake (cells might be more permeable) and not necessarily to a defect in the DDR per se (Gravel S, unpublished). Since the phosphorylation of the histone variant H2AX and the subsequent recruitment of the MDC1 adaptor protein is required for mammalian RNF8 localization to DSB sites—and since these proteins have conserved homologs in yeast—it is possible that the lack of localization of yeast Dma proteins to the nucleus might be a critical hindrance from them acting on histones. This is especially pertinent because budding yeast cells, unlike higher eukaryotic cells, do not disintegrate their nuclear membrane during mitosis. In light of this, it would be interesting to see whether adding a nuclear localization signal to yeast Dma proteins could promote their activity on nucleosomes following DNA damage.
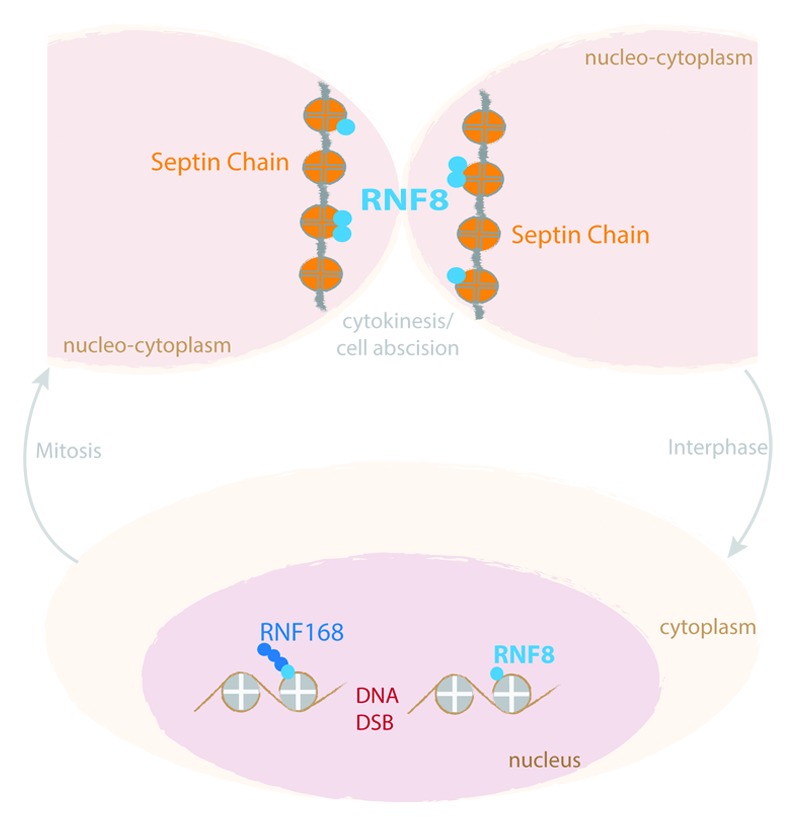
Figure 1. Cartoon depicting the differential localization and function of RNF8 at nucleosomal and cytoskeletal higher order structures during interphase and mitosis, respectively.
Taken together, our study suggests that RNF8 may have originally arisen to mediate functions in mitosis and was only later co-opted to act on histones. Remarkably, in both the nucleus and cytoplasm, RNF8 seems to target proteins that form poly-octameric higher order structures (Fig. 1). These are the histones, which constitute nucleosomes, and septins, which form cytoskeletal filaments. With their ability to form higher order chains, septins have been described as the “fourth component of the cytoskeleton.”7 Septin filaments have also been compared with nucleosomes not only because of their structural similarity but also due to their long half-life, heavy and reversible post-translational modifications, and their mode of inheritance during cell division.8 It is worth noting that our work suggests that, in some particular cases, the “histone code” might have evolved to mimic post-translational modifications occurring in cytoskeletal structures rather than the other way around. It would be intriguing, in the future, to address such a possibility and ascertain whether RNF168, a ubiquitin E3 ligase that works in synergy with RNF8 during the DDR, can recognize and mediate subsequent septin ubiquitylation events during mitosis. More broadly, it will be interesting to determine whether and how nucleosomal and cytoskeletal post-translational modifications are coordinated, and what the functions for such coordination might be.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24402
References
- 1.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–36. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Orpinell M, Fournier M, Riss A, Nagy Z, Krebs AR, Frontini M, et al. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010;29:2381–94. doi: 10.1038/emboj.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinaldo C, Moncada A, Gradi A, Ciuffini L, D’Eliseo D, Siepi F, et al. HIPK2 controls cytokinesis and prevents tetraploidization by phosphorylating histone H2B at the midbody. Mol Cell. 2012;47:87–98. doi: 10.1016/j.molcel.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 4.Chahwan R, Gravel S, Matsusaka T, Jackson SP. Dma/RNF8 proteins are evolutionarily conserved E3 ubiquitin ligases that target septins. Cell Cycle. 2013;12:1000–8. doi: 10.4161/cc.23947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. http://www.yeastgenome.org/
- 6.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–7. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 7.Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–94. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 8.McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]


