To deal with DNA lesions, cells activate the DNA damage response (DDR) network.1 DDR activation results in cell-cycle delay to allow time for DNA repair. Upon excessive damage, the cell-cycle checkpoint arrest will become permanent (a state known as senescence) or cells will undergo cell death (apoptosis). A key player controlling these cell fate decisions is the transcription factor p53. In response to genotoxic stress, p53 becomes modified and stabilized by different pathways and initially induces the expression of genes involved in DNA repair and cell-cycle arrest. Prolonged p53 stabilization, however, also induces the expression of pro-apoptotic genes. This, together with the prominent role of p53 in tumor suppression2 highlights the importance of a tight control of p53 activity. An exciting new aspect of p53 regulation in response to DNA double-strand breaks (DSBs), the most toxic DNA lesions,1 has now been reported by Moumen et al.3
DSBs activate three of the phosphatidylinositol 3-kinase-related kinases (PIKKs), namely ataxia telangiectasia mutated (ATM), ataxia and rad3-related (ATR) as well as the DNA-dependent protein kinase (DNA-PK). These kinases phosphorylate a large number of proteins involved in DNA repair, cell-cycle checkpoints and transcription.1 ATM phosphorylates p53 on serine 15 upon DNA damage, thereby contributing to p53 activation. ATM also phosphorylates HDM2, an ubiquitin E3-ligase that regulates p53 protein levels by proteasomal degradation. Upon DNA damage HDM2 ubiquitylation of p53 is greatly reduced, leading to p53 accumulation and p53-dependent transcription.2
Heterogeneous nuclear ribonucleoprotein K (hnRNPK) was originally characterized as a protein of the ribonucleoprotein complex with a strong binding preference for poly(C)-sequences in RNA.4 hnRNPK has diverse functions, including a role as a transcriptional co-activator of p53.5 Previous work by Moumen et al. showed that hnRNPK is stabilized upon ionizing radiation (IR) due to a reduction in its HDM2-dependent ubiquitylation, thus enhancing p53-dependent transcription after DNA damage.5 To gain deeper mechanistic insight into hnRNPK stabilization, Moumen et al. have now shown that upon IR treatment ATM also phosphorylates hnRNPK, on 4 serine/threonine residues (Fig. 1).3 Phosphorylation by ATM reduced hnRNPK ubiquitylation, suggesting that such phosphorylation prevents HDM2-dependent ubiquitylation and subsequent degradation of hnRNPK. The importance of ATM-dependent phosphorylation of hnRNPK is illustrated by the DNA-damage induced p53-dependent transcription of the cell-cycle inhibitor p21 (Fig. 1). Downregulation of hnRNPK with RNAi and complementation of its deficiency revealed that unlike wild-type hnRNPK, the mutant hnRNPK protein that cannot be phosphorylated by ATM does not facilitate p21 expression upon DNA damage.3
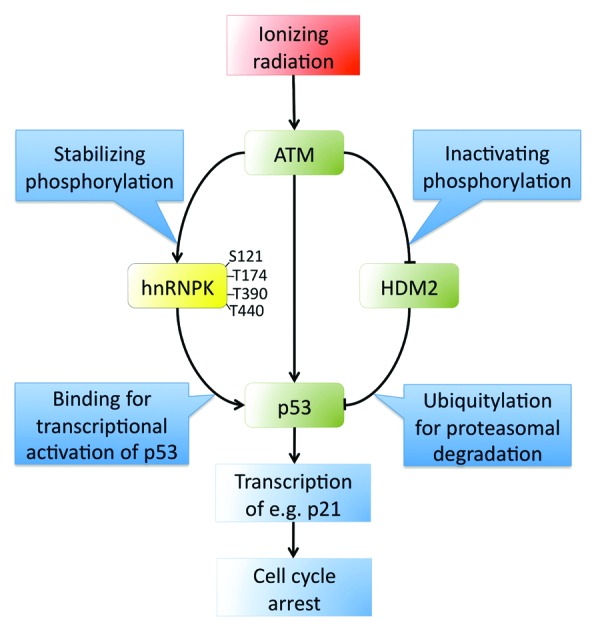
Figure 1. Pathways through which ATM activates p53-dependent transcription in response to ionizing radiation. ATM-targeted phosphorylation sites of hnRNPK identified by Moumen et al.3 are indicated.
Despite the long-term focus on p53 biology, many aspects of p53 regulation remain unexplained at the molecular level. It remains unclear whether ATM-dependent phosphorylation of p53 directly disrupts the p53-HDM2 interaction. Also, while p53 is modified by several post-translational modifications upon DNA damage, the significance of a potential crosstalk between these modifications in regulation of the abundance and activity of p53 remains largely unexplored. For example, Aurora-A, a mitotic protein kinase, phosphorylates both p53 and hnRNPK in the absence of DNA damage, leading to a destabilization of their interaction. Reduced Aurora-A activity upon DNA damage therefore stabilizes the hnRNPK-p53 interaction, thus increasing p53 transcriptional activity.6 hnRNPK is also SUMOylated upon DNA damage, a modification that is required for p53 transcriptional activation,7 and it is methylated on several arginine residues, which also seems crucial for p53 activity.8 Do all of these modifications work in an additive manner or is each of them sufficient for a full p53-dependent cell-cycle arrest?
To further complicate things, there are different cellular pools and isoforms of hnRNPK performing various functions9 and it is unclear whether ATM phosphorylates all hnRNPK molecules and which of hnRNPK’s functions are affected by this modification. We also do not know whether the hnRNPK that is phosphorylated by ATM is bound to RNA, or whether such phosphorylation affects hnRNPK binding to RNA and vice versa. It also remains to be tested if hnRNPK becomes phosphorylated on the same residues in response to other types of genotoxic stress and whether this is then mediated by different protein kinases such as ATR.
The new study by Moumen et al.3 opens the way to answer these questions and help better understand the protein network surrounding p53, its physiological roles and malfunction in cancer cells.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24485
References
- 1.Jackson SP, Bartek J. Nat. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Moumen A, Magill C, Dry KL, Jackson SP. ATM-dependent phosphorylation of heterogeneous nuclear ribonucleoprotein K promotes p53 transcriptional activation in response to DNA damage. Cell Cycle. 2013;12:698–704. doi: 10.4161/cc.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matunis MJ, Michael WM, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992;12:164–71. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–78. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 6.Hsueh KW, Fu SL, Huang CY, Lin CH. Aurora-A phosphorylates hnRNPK and disrupts its interaction with p53. FEBS Lett. 2011;585:2671–5. doi: 10.1016/j.febslet.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Pelisch F, et al. JBC. 2012;287:30789–99. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W, et al. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett. 2008;582:1761–5. doi: 10.1016/j.febslet.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 9.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–38. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]


