Abstract
Hexameric complexes of the six related Mcm2–7 proteins form the core of the replicative helicase. Two other proteins, Mcm8 and Mcm9, with significant homology to Mcm2–7 were first shown to play distinct roles during DNA replication in Xenopus laevis egg extract. Recent work has revealed that Mcm8 and 9 form a complex that plays a role during homologous recombination in human, chicken and mouse cells. We have therefore re-examined the behavior of the Xenopus homologs of these proteins. We show that Mcm8 and Mcm9 form a dimeric complex in Xenopus egg extract. They both associate with chromatin at later stages of DNA replication, and this association is stimulated by DNA damage, suggesting that their function is analogous to the one described in higher eukaryotes. In contrast to previous reports, we do not find Mcm9 essential for loading of Mcm2–7 complex onto chromatin during origin licensing nor detect its interaction with Cdt1 origin licensing factor. Altogether, we conclude that the role Mcm8 and Mcm9 play in Xenopus egg extract is not different from recent findings in higher eukaryotes, consistent with an evolutionary conservation of their function.
Keywords: DNA replication, Xenopus, DNA damage, MCM, helicase, Mcm8, Mcm9
Introduction
Every dividing cell must duplicate its genome before cell division. Failure to replicate the whole genome perfectly can lead to cell death or genomic instabilities. The MCM (minichromosome maintenance) proteins play a central role in DNA replication. Mcm2–7 hexamers form the core of the replication machinery (the replisome) during S-phase, while the regulated loading of Mcm2–7 onto DNA during G1 phase of the cell cycle defines the location of replication origins.1-7 Two additional members of this family have been identified: Mcm8 and Mcm9, which, unlike the other MCM proteins, are present only in higher eukaryotes due to their evolutionary loss in some fungi and some animals.8-12 Both proteins were reported to play a role during DNA replication, but there is controversy as to their function in different organisms.
Human Mcm8 was suggested to be involved in the assembly of pre-replicative complexes (pre-RC; origin licensing),13,14 while Xenopus laevis Mcm8 was shown to play a role during the elongation stage of DNA replication.15 There are also contradicting reports about the interaction of Mcm8 with Mcm2–7 complexes and Cdc6.8,9,14,15 In Drosophila Mcm8 appears to act during meiotic recombination,16,17 although its depletion was also shown to reduce the number of replication forks during DNA replication.18
Xenopus laevis Mcm9 has also been proposed to play an essential function in loading Mcm2–7 onto origins of replication, through its interaction with the licensing factor Cdt1.19 However, in mice, Mcm9 was shown to be dispensable for origin licensing but important for germ-line stem-cell maintenance, proliferation and genome stability.20 Moreover, the process of origin licensing has been previously reconstituted with purified proteins without the addition of Mcm9.4
Interestingly, a comparative genomics analysis showed that Mcm8 and Mcm9 paralogs are typically either both present or both absent in genomes, with the exception of Drosophila, which contains only Mcm8.12 This suggested that the two proteins may have associated functions, and indeed, two recent reports provided elegant evidence for existence of Mcm8/Mcm9 complexes in human, chicken and mouse cells.21,22 The reported complex was shown to function during homologous recombination and to play an essential role in generation of germ cells in mice21 and resistance to DNA damaging agents in chicken cell lines.22
As these recent findings are in conflict with original investigations of Mcm8 and Mcm9 functions in Xenopus laevis egg extract system, we decided to re-examine the role of Mcm9 and Mcm8 during DNA replication in this system. We aimed to establish whether they evolved a unique function in this early embryonic system. We generated a new set of antibodies against XlMcm8 and XlMcm9, and here we show that Mcm8 and Mcm9 form a stable dimeric complex in Xenopus egg extract. They both associate with chromatin at later stages of S-phase and depletion of Mcm9, which co-depletes Mcm8, does not significantly affect the loading of Mcm2–7 complexes and does not block DNA replication but slows down its progression. Finally, both proteins bind chromatin at higher levels in response to DNA damage, consistent with a conserved function in higher eukaryotes associated with recombination.
Results
Xenopus Mcm8 and Mcm9 interact with each other
We have raised an antibody against XlMcm8 and two antibodies against XlMcm9 (one in rabbit and one in sheep) to confirm our findings with two independent antibodies. All antibodies specifically recognized recombinant proteins purified from E. coli (not shown) and proteins of the expected size in Xenopus egg extract (Fig. 1A). Using these antibodies we investigated if Mcm8 and Mcm9 interact with each other. Both Mcm9 antibodies efficiently immunoprecipitated Mcm9 and co-immunoprecipitated Mcm8 (Fig. 1B, lanes 10 and 12). Similarly, the Mcm8 antibody efficiently immunoprecipitated Mcm8 and co-immunoprecipitated Mcm9 (Fig. 1B, lane 9).

Figure 1. Mcm8 and Mcm9 form a complex in egg extract. (A) Western blot of 0.5 µl of Xenopus interphase and metaphase arrested extracts with preimmune and Mcm9-specific rabbit serum or sheep affinity purified antibodies anainst Mcm8 and Mcm9. (B) Mcm8 and Mcm9 were immunoprecipitated from egg extract using rabbit (rb) and sheep (sh) antibodies. Commercial sheep IgG and rabbit pre-immune serum (PI) were used as control. Input (extract), depleted input (flow through) and immunoprecipitation samples were analyzed by western blotting with indicated antibodies. (C) Cdt1 was immunoprecipitated from interphase egg extract. Immunoprecipitated samples were analyzed as in (B).
The sheep Mcm9 antibody was able to deplete all Mcm9 and Mcm8 from the extract (Fig. 1B, lane 4), while extracts in which Mcm8 was entirely depleted retained a fraction of Mcm9 (~30–50%). This suggests that in the Xenopus egg extract all of Mcm8 forms a complex with Mcm9, while a proportion of Mcm9 is either monomeric or present in another complex. Alternatively, all Mcm8 and Mcm9 interact with each other and the Mcm8 antibody used for immunoprecipitation disrupts the complex leaving a proportion of Mcm9 behind.
We have also re-examined the previously reported interaction of Mcm9 with Cdt1.19 We were unable to substantially co-immunoprecipitate Cdt1 with either Mcm9 or Mcm8 (Fig. 1B) or Mcm8 and Mcm9 with Cdt1 (Fig. 1C). As a positive control we showed that geminin, a known interacting partner of Cdt1,23 could be immunoprecipitated with Cdt1. We therefore conclude that while Mcm8 and 9 interact with one another, they do not strongly interact with Cdt1. Neither of the proteins interacted with Mcm2–7 subunits in our experiments (Fig. 1B and C).
Mcm8 and Mcm9 form a dimer
To establish the stoichiometry of the Mcm8/Mcm9 complex, interphase egg extract was separated by gel filtration (Fig. 2A) or glycerol gradient sedimentation (Fig. 2B), and fractions were examined by western blotting. Mcm8 co-fractionated with Mcm9 with both separation methods, while Cdt1 was clearly separated from Mcm8 and 9 by glycerol gradient sedimentation. Similarly, Mcm2–7 subunits formed distinct complex.
Figure 2. Mcm8 and Mcm9 form a dimer. (A) Interphase extract was separated through Superose 6 gel filtration column and fractions analyzed by western blotting with the indicated antibodies. Molecular mass and Stokes radius of marker proteins used for column calibration are indicated above the blots. (B) Interphase extract was separated by 20–40% glycerol gradient centrifugation and fractions analyzed by western blotting with the indicated antibodies. Molecular mass and sedimentation coefficients of marker proteins used to calibrate the gradient are indicated above the blots. (C) Stokes radiuses and sedimentation coefficients of indicated proteins were used to calculate the molecular sizes of native complexes.
The native molecular weight of proteins and protein complexes can be calculated using their Stokes radius (derived from gel filtration) and sedimentation coefficient (derived from glycerol gradients) using the equation of Siegel and Monty24 (Fig. 2C). We calculated the native molecular weight of Mcm8 in Xenopus egg extract as 186 kD. As all of the Xenopus Mcm8 present in the extract interacts with Mcm9, this is likely to represent the mass of the Mcm8/Mcm9 complex. Since the molecular weight of monomeric Mcm8 is 92.5 kD and of monomeric Mcm9 is 126.6 kD, they have a combined mass of 219.1 kD, suggesting that the Mcm8/9 complex exists as a heterodimer in egg extract. In the same experiments, Mcm2–7 subunits formed a hexamer as expected, while the pre-RC proteins Cdt1 and Cdc6 behaved approximately as monomers. We cannot, however, exclude the possibility that a larger Mcm8/9 complex exists and is not stable when fractionated using the methods above.
Mcm8 and Mcm9 bind to chromatin at later stages of DNA replication
Mcm9 has been shown previously to bind to chromatin during origin licensing in parallel to Cdt1 and interact with chromatin throughout S-phase.19 Mcm8, on the other hand, was shown to interact with chromatin after initiation of DNA synthesis and accumulate on chromatin during the course of the S-phase and G2.15 These results are not consistent with the data presented here that Mcm8/Mcm9 form a dimeric complex. We therefore re-examined the timing of chromatin binding of Mcm8 and 9 with respect to other replication factors. Figure 3A shows the typical profile of DNA synthesis in egg extract, with an initial lag period of ~20 min, during which time origins are licensed, and template DNA is assembled into an interphase nucleus. S-phase lasts for ~30 min and then extracts enter G2. As shown in Figure 3B, Mcm8 and Mcm9 both associate with chromatin in late S and G2, binding most strongly after the peak of replication fork proteins on chromatin and after most of the DNA synthesis has been completed. The observed chromatin binding is in agreement with the data shown previously for Mcm8, but not Mcm9, and is consistent with them being constitutively present as a heterodimer.
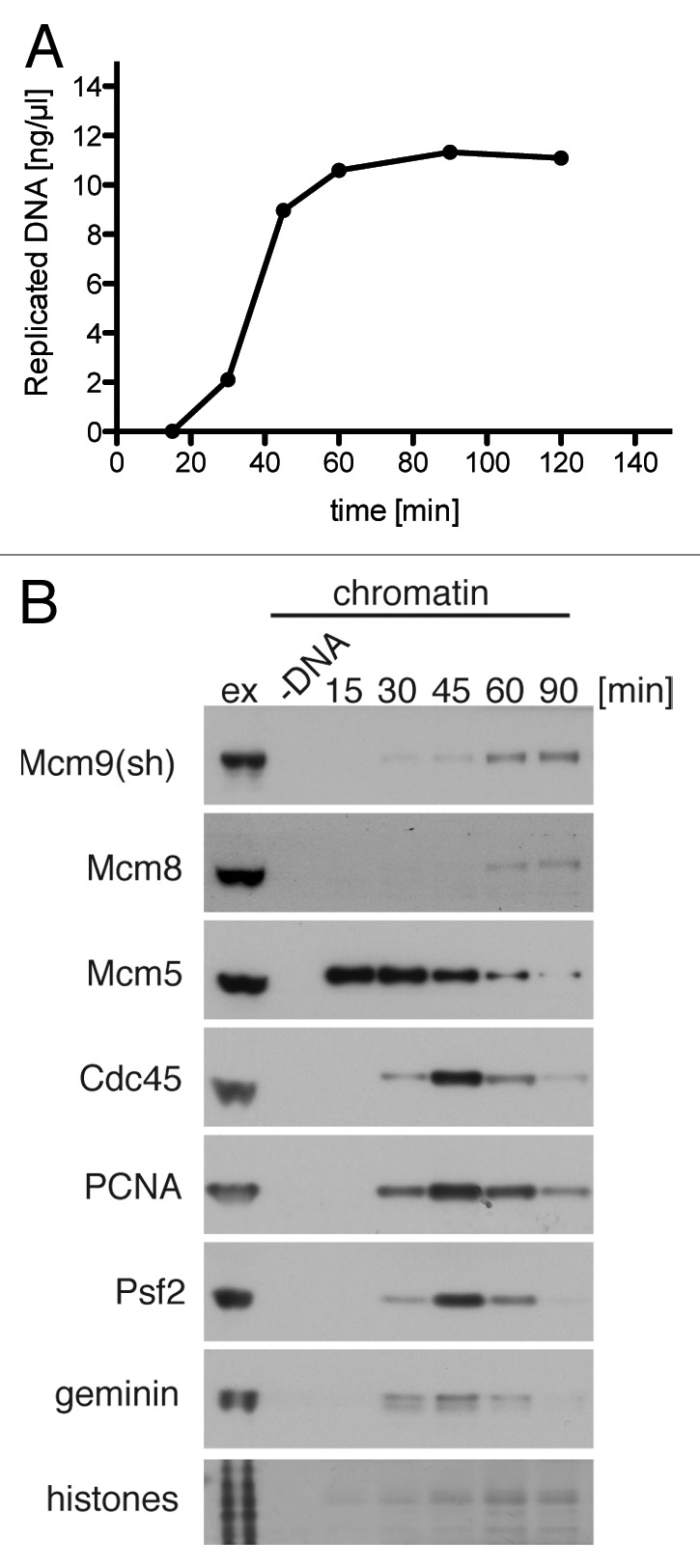
Figure 3. Mcm8 and Mcm9 bind chromatin at the late stages of DNA replication. Sperm nuclei were incubated at 10 ng DNA/µl in interphase egg extract. (A) Extract was supplemented with α-[32P]dATP. At the indicated times DNA synthesis was assessed by TCA precipitation and scintillation counting. (B) Chromatin was isolated from 10 µl aliquots at the indicted times and immunoblotted alongside 0.5 µl of egg extract with the indicated antibodies. Chromatin isolation of a sample without addition of sperm DNA was performed as a control for chromatin specificity of the assay.
Mcm9 is dispensable for origin licensing
As a partner of Cdt1, Mcm9 was proposed to act as a loading factor for the core of the replicative helicase (Mcm2–7 complex) before the onset of S-phase.19 As we failed to detect a robust Mcm9/Cdt1 complex formation and no significant association of Mcm9 with chromatin during the licensing period, we examined whether we can reproduce this essential replication phenotype. We immunodepleted Mcm9 from Xenopus egg extract using the antibodies raised in rabbit. Two rounds of immunodepletion were sufficient to remove > 99% Mcm9, as estimated using both Mcm9 antibodies (Fig. 4A). This efficient removal of Mcm9 also co-depleted Mcm8 down to < 2% but failed to significantly co-deplete Cdt1 or geminin (Fig. 4A).

Figure 4. Mcm9 is required for efficient DNA replication but not Mcm2–7 loading. Egg extract prior to depletion was diluted and the indicated percentage of original extract volume analyzed by western blotting with the indicated antibodies alongside samples of control and Mcm9-depleted extracts. (B) Control-depleted and Mcm9-depleted extract were supplemented with sperm nuclei at 7.5 ng DNA/µl and α-[32P]dATP. At the indicated times, aliquots were taken, and DNA synthesis assessed by TCA precipitation and scintillation counting. The mean value with the standard error of the mean (SEM) are plotted in three sets of independently depleted mock and Mcm9-depleted extracts normalized to the maximum DNA synthesis observed in each control-depleted extract. (C) Control-depleted and Mcm9-depleted extracts were supplemented with sperm nuclei at 7.5 ng DNA/µl. Chromatin was isolated at the indicated times and immunoblotted with the indicated antibodies. A representative of three performed experiment is presented.
We next assessed the replication potential of extract depleted of the Mcm8/Mcm9 complex in comparison to extract depleted with non-immune antibodies. Figure 4B shows mean efficiency of DNA replication in three independently depleted sets of extracts. All three extracts were depleted to less than 1% of Mcm9 remaining, and DNA synthesis was judged by the level of incorporation of radioactive dATP into newly synthetized DNA. Mcm9-depleted extracts progressed slightly slower through S-phase; on average, the replication rate (between 30 and 120 min) of Mcm9-depleted extract was 38% ± 10.8 (SEM) lower than control extract depleted with non-immune antibodies. Mcm9-depleted extracts were, however, able to replicate considerable amounts of DNA; the average percentage of replicated DNA at 180 min in Mcm9-depleted extract equals 78% ± 8.3 (SEM) of DNA replicated in control-depleted extract. We were unable to restore the replication rate defect of Mcm9-depleted extract by addition of recombinant Mcm9 or Mcm8/9, due to difficulty with expression of recombinant Mcm8/9.
Five to ten times more Mcm2–7 complexes are loaded onto chromatin than are required for efficient DNA replication under normal circumstances. The excess Mcm2–7 complexes provide additional dormant origins that can be activated if neighboring replication forks irreversibly stall.25-27 This means that the ability of Mcm9-depleted extract to replicate DNA efficiently does not prove that they licensed a similar number of origins. We therefore examined the level of chromatin loaded Mcm2–7 complexes in Mcm9-depleted extract. The process of immunodepletion causes nuclear assembly to occur more slowly, and because nuclear assembly turns off the licensing period by reactivating geminin,28-30 this extends the period of time when Mcm2–7 are loaded onto DNA (Fig. 4C, control extract). However, the rate and extent of Mcm2–7 loading onto DNA (as revealed by Mcm7 and Mcm5 subunits, Fig. 4C) was essentially the same in control-depleted and Mcm9-depleted extracts. At the same time, we observed reduced levels of replisome components Cdc45 and Psf2 bound to chromatin in Mcm9-depleted extract. This suggests fewer assembled active forks or reduced stability of replicative helicase, in agreement with a slower replication phenotype. We confirmed these data using the second (sheep) Mcm9 antibody (not shown). We conclude, therefore, that the defect in DNA replication observed is not due to reduced Mcm2–7 loading.
More Mcm9 and Mcm8 interact with chromatin after DNA damage
We next considered the possibility that the reduced rate of DNA replication seen in Mcm9-depleted extract might be due to the Mcm8/9 dimer being involved in resolving DNA damage or fork stalling that occurs during S-phase. If this is the case, it might be anticipated that Mcm8 and 9 recruitment to chromatin may increase when DNA damage or fork stalling is induced during S-phase. We therefore challenged extracts at the start of S-phase (at 30 min after sperm addition) with a spectrum of inhibitors: aphidicolin, an inhibitor of family B polymerases; camptothecin, a topoisomerase I inhibitor; mitomycin C, an inter-strand crosslinker; etoposide, a topoisomerase II inhibitor; and EcoRI, which causes double-strand DNA breaks. As seen in Figure 5A all these inhibitors considerably reduced the total amount of DNA that had been replicated 60 min later. In parallel, we isolated chromatin and analyzed levels of chromatin-bound proteins by western blotting. By the time of chromatin isolation (90 min from sperm addition) most sperm DNA is replicated in untreated control sample (Fig. 3A), and so replication fork proteins are no longer detectable on chromatin (Fig. 5B). Treatment with DNA damaging agents, especially aphidicolin, led to accumulation of Cdc45, PCNA and Psf2 on chromatin, while induction of DNA double-strand breaks by EcoRI led to accumulation of the phosphorylated form of histone H2AX. Interestingly, Mcm9 and Mcm8 bound chromatin at a higher level after treatment with all of the inhibitors used (Fig. 5C). These data are in agreement with the reported sensitivity of cells lacking Mcm9 and/or Mcm8 to a variety of DNA damaging agents and replication inhibitors and their involvement in homologous recombination21,22 and suggest that the function of the Mcm8/Mcm9 complex is also conserved through the evolution.
Figure 5. Mcm8 and Mcm9 bind chromatin at higher level after DNA damage. Sperm nuclei were incubated at 10 ng DNA/µl in interphase egg extract, and after 30 min, aliquots were supplemented with the indicated DNA damaging agents. (A) Extract was also supplemented with α-[32P]dATP at 30 min, and the total DNA synthesized at 90 min was determined. (B) Chromatin was isolated at 90 min after sperm DNA addition and immunoblotted with the indicated antibodies. (C) The intensity of the Mcm8, Mcm9 and ATR bands were quantified in immunoblots from three independent experiments, normalized to the quantity of histone loaded and the mean fold of increased chromatin binding of indicated proteins was determined for all tested DNA replication inhibitors. Standard error of the mean is also presented.
Discussion
Recent genomic analysis suggested that Mcm8 and Mcm9 have associated functions due to their joint presence or absence in eukaryotic genomes.12 However, the reported functions of Mcm8 and Mcm9 in various model systems were divergent, including the functions of Mcm8 and Mcm9 in the Xenopus egg extract system. Finally, recent work in mouse, chicken and human cell lines elegantly showed the existence of an Mcm8/Mcm9 complex and its essential role in homologous recombination.21,22 These reports, however, did not address the emerging discrepancy with Xenopus laevis system.15,19 We have therefore set out to ask if Xenopus system evolved divergent and separate functions for these two proteins, or whether Mcm8/Mcm9 indeed functions as a complex in the extract.
We have shown that all of Mcm8 interacts in a stable complex with Mcm9 in interphase extract, suggesting their common function as predicted.12 Using a combination of gel filtration and glycerol gradient sedimentation, we obtained an estimated molecular weight of 186 kD for this complex, close to the combined molecular weights of both proteins (219 kD). We could not detect the previously reported interaction between Mcm9 and Cdt1,19 though a proportion of Xenopus Mcm9 may not be engaged in the Mcm8/Mcm9 complex and thus has a potential to interact with other factors. Alternatively, it is possible that the Mcm9/Cdt1 interaction is stabilized under specific conditions not reproduced in our experiments.
Interestingly, it was reported recently that Trypanosoma brucei Mcm8 interacts with TbMcm-BP protein. TbMcm-BP also interacts with Mcm4–7 and is required for repression of gene expression of subtelomeric variant surface glycoprotein (VSG) and genes transcribed by RNA polymerase I.31 In our experiments, however, Mcm8 and Mcm9 do not co-immunoprecipitate Mcm-BP (data not shown), suggesting that unlike Trypanosoma they do not form a stable complex with Mcm-BP in Xenopus egg extract.
Consistent with our data that Mcm9 does not interact with Cdt1, we could not detect any significant defect in loading of Mcm2–7 complexes onto chromatin in Mcm9 depleted extract despite depleting Mcm9 to undetectable level of less than 1% remaining protein. The discrepancy with previously published data may arise from difference in the antibodies used, although both studies used antibodies raised against the C-terminal end of Mcm9.19 Instead, the data presented here more closely matches the previously published phenotype of depletion of Mcm8,15 which results in less efficient DNA replication. The most likely reason for the observed defect in DNA replication efficiency is the accumulation of replication stress due to lack of Mcm8/Mcm9.
The previously reported replication defect in Mcm8-depleted extract could be rescued with recombinant Mcm8.15 Taking into account our data showing potential excess of Mcm9 which is not co-depleted with Mcm8, it seems likely that Mcm9 remaining in the Mcm8-depleted is sufficient to restore complete replication after addition of recombinant Mcm8.
Our data also agrees with the non-essential roles of Mcm8 and Mcm9 in other model systems: Mcm8−/− and Mcm9−/− mice are viable, but somatic cells obtained from these mice and mcm8KO and mcm9KO DT40 chicken cells exhibit growth defects, genetic instability and higher levels of DNA damage.20-22
Cells lacking Mcm8 or Mcm9 were shown to be defective in responding to DNA damage and sensitive to DNA damaging agents.21,22 We also detect higher level of Mcm8 and Mcm9 bound to chromatin after exposure to DNA damage, suggesting an analogous function of these proteins in Xenopus extract. While this manuscript was under revision, another study was published reporting the sensitivity of Mcm8 and/or Mcm9 knockdown cells to cisplatin.32 The same study also showed accumulation of Mcm8 and Mcm9 in the vicinity of inter-strand crosslinks in Xenopus egg extract, supporting a role for these proteins in DNA damage repair.32
Nishimura et al.22 suggested that the Mcm8/Mcm9 complex acts a hexameric helicase based on its gel filtration profile in DT40 whole-cell extract. The calculated molecular size of Mcm8/Mcm9 complex in Xenopus egg extract suggests the existence of a dimeric complex rather than hexamer. It is possible, however, that once chromatin bound, the dimers combine to form hexamers. Additionally, we cannot exclude the possibility that a hexameric Xenopus Mcm8/Mcm9 complex is less stable during gel filtration and glycerol gradient sedimentation than its chicken homolog, even in conditions gentle enough to preserve the Mcm2–7 hexamer. Mcm8 was previously shown to display helicase activity in vitro,15 but it remains to be tested whether Mcm8/Mcm9 complexes also possess such an activity, or if Mcm9 functions as a loader for an Mcm8 helicase.
Altogether, our data show that in contrast to previous evidence from the Xenopus egg extract system, Mcm8/Mcm9 forms a heterodimeric complex that is recruited to chromatin during DNA replication, possibly to deal with various types of DNA damage. This is consistent with recent data in mice, chicken and human cells and suggests that Mcm8 and 9 have a conserved function throughout evolution.
Materials and Methods
Xenopus egg extract
Metaphase-arrested Xenopus laevis egg extracts were prepared and replication reactions assembled with demembranated Xenopus sperm at 10–15 ng DNA/µl as previously described.33 DNA synthesis was assessed in extract supplemented with α-[32P]dATP (PerkinElmer) by TCA precipitation as described.33 Immunodepletion of interphase extract with Mcm9 rabbit antibody was performed in analogous way to Mcm3 immunoprecipitation described in Chong et al.,34 performing two rounds of depletion using 60% v:v beads. Because of the extract dilution this causes, sperm nuclei were incubated in immunodepleted extract at 7.5 ng DNA/µl. Chromatin isolation was performed in ANIB/100 buffer as described.33
Antibodies and reagents
Antibodies against XlMcm9 were raised in rabbit and in sheep by immunization with a HIS6-tagged recombinant peptide of 843–1143 aa of Xenopus laevis Mcm9 expressed and purified from Rosetta 2(DE3)pLysS E.coli strain (Novagen) under native conditions according to the standard Ni2+-NTA purification protocol (Qiagen). Antibodies against Mcm8 were raised in sheep against 1–400 aa of XlMcm8 expressed in Rosetta 2(DE3)pLysS E.coli strain (Novagen) and purified under denaturing conditions (Qiagen). Both sheep antibodies were further affinity purified and used at 1:1,000 concentration for western blotting. Cdc45 and Psf2 antibodies were previously described.3 PCNA PC10 antibody was from Sigma, and Cdt1 and geminin antibodies were previously described.23 Aphidicolin (Sigma) was dissolved in DMSO at 10 mM and used at 40 µM. Mitomycin C (Calbiochem) was dissolved in water at 5 mM in water and used at 500 µM. Etoposide (Calbiochem) was dissolved in DMSO at 30 mM and used at 400 µM. Camptothecin was dissolved in DMSO at 50 mM and used at 500 µM. EcoRI (Roche) was used at 0.05 U/µl of egg extract.
Immunoprecipitation
Interphase egg extract was diluted five times with IP buffer (40 mM Hepes-KOH pH 7.5, 50 mM KCl, 10% sucrose, 25 mM sodium β-glycerophosphate pH 7.5, 0.5 mM EGTA, 0.1 mM sodium vanadate, 1 µg/ml leupeptin, 1 µg/ml pepstatin, 1 µg/ml aprotinin, 0.1 mM PMSF) and spun 15 min at 18,500 g at 4°C. One hundred µl aliquots were supplemented with 1 µg affinity purified sheep antibodies or IgG from sheep serum (I5131 Sigma) as a control, or 5 µl of pre-immune or third bleed rabbit serum. After an hour-long incubation on ice immunoprecipitation samples were mixed with 30 µl washed Protein A or Protein G Dynabeads (Invitrogen) and incubated with mixing at 4°C for 1 h. Beads were washed twice with buffer as above, once with the buffer supplemented with 0.1% triton X-100 and again twice with the buffer alone. Immunoprecipitated proteins were eluted off beads by boiling in 1× NuPAGE LDS loading buffer (Invitrogen).
Gel filtration and glycerol gradient
Gel filtration, glycerol gradient and calculaction of native molecular sizes of complex were performed as previously described.3
Acknowledgments
This work was funded by CRUK grants C303/A8102 and C303/A7399. A.G. was supported by a Sir Henry Wellcome Fellowship 082716/B/07/Z and Birmingham Cancer Research Centre Award DF/270312.
Glossary
Abbreviations:
- MCM
minichromosome maintenance
- pre-RC
pre-replicative complex
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24310
References
- 1.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 2.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–30. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem. 2011;286:11855–64. doi: 10.1074/jbc.M110.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–5. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–58. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson EM, Kinoshita Y, Daniel DC. A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13. Nucleic Acids Res. 2003;31:2915–25. doi: 10.1093/nar/gkg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gozuacik D, Chami M, Lagorce D, Faivre J, Murakami Y, Poch O, et al. Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res. 2003;31:570–9. doi: 10.1093/nar/gkg136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutzmann M, Maiorano D, Méchali M. Identification of full genes and proteins of MCM9, a novel, vertebrate-specific member of the MCM2-8 protein family. Gene. 2005;362:51–6. doi: 10.1016/j.gene.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida K. Identification of a novel cell-cycle-induced MCM family protein MCM9. Biochem Biophys Res Commun. 2005;331:669–74. doi: 10.1016/j.bbrc.2005.03.222. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Richards TA, Aves SJ. Ancient diversification of eukaryotic MCM DNA replication proteins. BMC Evol Biol. 2009;9:60. doi: 10.1186/1471-2148-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkening M, Hoffmann I. Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin. Mol Cell Biol. 2005;25:1560–8. doi: 10.1128/MCB.25.4.1560-1568.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinoshita Y, Johnson EM, Gordon RE, Negri-Bell H, Evans MT, Coolbaugh J, et al. Colocalization of MCM8 and MCM7 with proteins involved in distinct aspects of DNA replication. Microsc Res Tech. 2008;71:288–97. doi: 10.1002/jemt.20553. [DOI] [PubMed] [Google Scholar]
- 15.Maiorano D, Cuvier O, Danis E, Méchali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–28. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Matsubayashi H, Yamamoto MT. REC, a new member of the MCM-related protein family, is required for meiotic recombination in Drosophila. Genes Genet Syst. 2003;78:363–71. doi: 10.1266/ggs.78.363. [DOI] [PubMed] [Google Scholar]
- 17.Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 2005;1:e40. doi: 10.1371/journal.pgen.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crevel G, Hashimoto R, Vass S, Sherkow J, Yamaguchi M, Heck MM, et al. Differential requirements for MCM proteins in DNA replication in Drosophila S2 cells. PLoS ONE. 2007;2:e833. doi: 10.1371/journal.pone.0000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutzmann M, Méchali M. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol Cell. 2008;31:190–200. doi: 10.1016/j.molcel.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Hartford SA, Luo Y, Southard TL, Min IM, Lis JT, Schimenti JC. Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression. Proc Natl Acad Sci USA. 2011;108:17702–7. doi: 10.1073/pnas.1113524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, et al. MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination. Mol Cell. 2012;47:523–34. doi: 10.1016/j.molcel.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 22.Nishimura K, Ishiai M, Horikawa K, Fukagawa T, Takata M, Takisawa H, et al. Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks. Mol Cell. 2012;47:511–22. doi: 10.1016/j.molcel.2012.05.047. [DOI] [PubMed] [Google Scholar]
- 23.Tada S, Li A, Maiorano D, Méchali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–13. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–62. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 25.Mahbubani HM, Chong JP, Chevalier S, Thömmes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–35. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S-phase checkpoint activation. J Cell Biol. 2004;165:181–90. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodward AM, Göhler T, Luciani MG, Oehlmann M, Ge X, Gartner A, et al. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–83. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgson B, Li A, Tada S, Blow JJ. Geminin becomes activated as an inhibitor of Cdt1/RLF-B following nuclear import. Curr Biol. 2002;12:678–83. doi: 10.1016/S0960-9822(02)00778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li A, Blow JJ. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 2005;24:395–404. doi: 10.1038/sj.emboj.7600520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisielewska J, Blow JJ. Dynamic interactions of high Cdt1 and geminin levels regulate S-phase in early Xenopus embryos. Development. 2012;139:63–74. doi: 10.1242/dev.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HS, Park SH, Günzl A, Cross GA. MCM-BP Is Required for Repression of Life-Cycle Specific Genes Transcribed by RNA Polymerase I in the Mammalian Infectious Form of Trypanosoma brucei. PLoS ONE. 2013;8:e57001. doi: 10.1371/journal.pone.0057001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, et al. MCM8-9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol. 2013 doi: 10.1128/MCB.01503-12. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie PJ, Gambus A, Blow JJ. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods. 2012;57:203–13. doi: 10.1016/j.ymeth.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong JP, Thömmes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Methods Enzymol. 1997;283:549–64. doi: 10.1016/S0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]




