Abstract
The transcription factor E2F1 belongs to the E2F family and plays a crucial role during cell cycle progression and apoptosis. Ser/Arg-Rich (SR) proteins are a family of RNA-binding phosphoproteins that control both constitutive and alternative pre-mRNA splicing events. We previously identified the SR protein SRSF2 as a new transcriptional target of E2F1 and demonstrated that both proteins cooperate to induce apoptosis in non-small cell lung carcinoma. In this study, we postulated that SRSF2 is also involved in the proliferative functions of E2F1. Using IHC, we first demonstrate that SRSF2 and its phosphorylated form (P-SRSF2) are overexpressed in neuroendocrine lung tumors that are highly proliferative tumors expressing high levels of E2F1. Importantly, we show a direct correlation between cyclin E, an E2F1-target gene controlling S phase, and P-SRSF2 proteins levels (p = 0.0083), suggesting a role of SRSF2 in E2F1-mediated cellular proliferation. Accordingly, using neuroendocrine lung carcinoma cell lines, we demonstrate that SRSF2 is a cell cycle-regulated protein involved in entry and progression into S phase. We also provide evidence that SRSF2 interacts with E2F1 and stimulates its transcriptional control of cell cycle target genes such as cyclin E. Finally, we show that inhibition of AKT signaling pathway prevents SRSF2 phosphorylation and activity toward E2F1 transcriptional function. Taken together, these results identify a new role of SRSF2 in the control of cell cycle progression and reinforce the functional link between SRSF2 and E2F1 proteins.
Keywords: AKT, cellular proliferation, E2F1, neuroendocrine lung tumors, p45SKP2, SRSF2
Introduction
A central aspect of development and disease such as cancer is the control of cellular proliferation through regulation of the cell cycle. A key step in this regulation is the transition from the G1 to S phase of the cell cycle. This critical passage is tightly coupled to the transcriptional control of genes involved in growth and DNA replication. In mammalian cells, this temporal control is achieved mostly by the E2F family of transcription factors.1,2 E2F transcription factors were originally identified as activators of adenovirus transcription. They belong to the E2F protein family, encompassing eight members called E2F1–E2F8. E2Fs are classified into different groups based on domain conservation and transcriptional activity.3,4 Among the E2F1–E2F5 proteins, E2F1, E2F2 and E2F3a primarily activate transcription, whereas E2F3b, E2F4 and E2F5 primarily repress transcription.5 It is now well-known that interaction of E2F1–3 proteins with members of the retinoblastoma (Rb) ”pocket” protein family pRb, p107 and p130, inhibits E2Fs transcriptional activity in G0 or in early G1 phase of the cell cycle. As cells progress through the G1 phase, Rb is sequentially phosphorylated by Cdks, causing the release of the activator E2Fs and the transcriptional activation of genes important for passage into the S phase.5 As E2Fs deregulation frequently occurs in human cancer, the characterization of additional partners of E2Fs involved in the control of cellular proliferation is critical to improve our knowledge regarding the contribution of each of these E2Fs to the tumorigenic process.
The SR protein family comprises a number of phylogenetically conserved and structurally related proteins that play a crucial role in the control of constitutive and alternative pre-mRNA splicing. Members of the SR family have a modular structure containing one or two copies of a RNA recognition motif (RRM) at the N-terminus that provides RNA-binding specificity and a C-terminal RS domain enriched in alternating serine and arginine residues that promotes protein-protein interactions and facilitates the recruitment of the spliceosome. Owing to these serine residues, the RS domain is highly regulated by phosphorylation that controls SR proteins interaction. Studies in mice depleted for individual SR proteins have provided evidence that these proteins are not redundant and are absolutely required for cell viability and/or animal development.6-8 To date, a few evidences exist regarding a role of the SR proteins in the regulation of components of the cell-division machinery in distinct organisms.9-11 However, the molecular mechanisms underlying these effects remain largely unknown.
Large cell neuroendocrine lung carcinoma (LCNEC) and small cell lung carcinoma (SCLC) are considered as the most malignant lung tumors. We previously demonstrated that the transcription factor E2F1 is upregulated in these tumors while being expressed at a very low level in low grade neuroendocrine tumors (typical and atypical carcinoids) as well as in lung adenocarcinoma and squamous lung carcinoma.12 Furthermore, and consistent with a proliferative role of E2F1 in LCNEC and SCLC, we also showed a direct correlation between E2F1 protein status and the expression of some of its transcriptional targets involved in S phase progression, namely the cyclin E and p45SKP2 genes.13 More recently, we identified SRSF2 as a new target of E2F1 in various human lung carcinoma cell lines, including neuroendocrine lung carcinoma, and demonstrated that both proteins cooperate to induce apoptosis in lung adenocarcinoma cells.14 In this study, we postulated that SRSF2 contributes to the proliferative function of E2F1 in neuroendocrine lung tumors.
Results
SRSF2 and P-SRSF2 proteins are overexpressed in neuroendocrine lung tumors
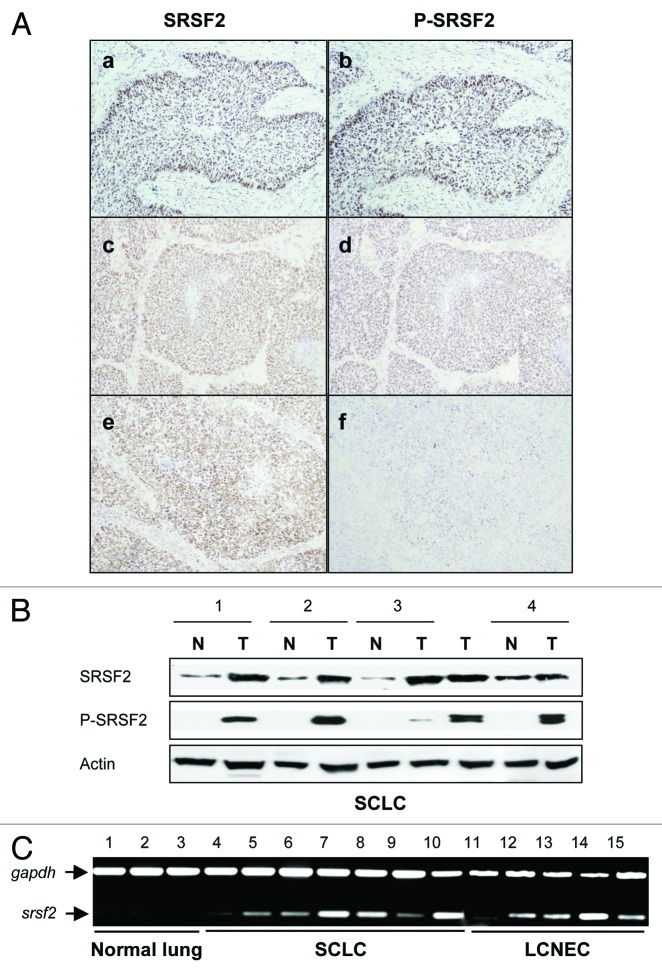
We first analyzed the status of SRSF2 and its phosphorylated form P-SRSF2 in a series of 27 neuroendocrine (NE) lung tumors and their associated normal lung tissues by immunohistochemistry as previously described.15 Compared with normal lung tissues, SRSF2 and P-SRSF2 proteins were overexpressed and accumulated in the nucleus in 89% (24/27) and 78% (21/27) of NE lung tumors, respectively (Fig. 1A). By using western blotting (Fig. 1B) and RT-PCR (Fig. 1C), we confirmed the increase of SRSF2 expression in human tumors. We previously observed a direct correlation between E2F1 and cyclin E status in NE lung tumors.13 Interestingly, we also found here a direct relationship between P-SRSF2 and cyclin E status (p = 0.0083; Table S1). By contrast, we did not find a significant correlation between E2F1 and P-SRSF2 immunostaining. Altogether, these results provide the first evidence that SRSF2 and its phosphorylated form are overexpressed in NE lung tumors and closely connected with proliferative E2F1-target genes.
Figure 1. SRSF2 and P-SRSF2 proteins are overexpressed in human neuroendocrine lung tumors. (A) Representative immunostainings of SRSF2 and P-SRSF2 proteins in NE lung tumors. (a and b) A small cell lung carcinoma displaying a strong staining of both SRSF2 (score 300) and P-SRSF2 (score 300); (c and d) A large cell neuroendocrine carcinoma displaying a strong overexpression of both SRSF2 (score 300) and P-SRSF2 (score 300); (e and f) A small cell lung carcinoma exhibiting a strong SRSF2 staining (score 300) and a faint P-SRSF2 staining (score 40). (B) SRSF2 and P-SRSF2 protein levels were analyzed by western blotting in NE lung tumors (T) and their normal counterparts (N). Actin was used as a loading control. (C) RT-PCR analysis of SRSF2 mRNA level in three normal lung tissues, seven small cell lung carcinoma (SCLC) and five large cell neuroendocrine carcinoma (LCNEC). Gapdh was used as an internal control.
SRSF2 is a cell cycle-regulated protein involved in entry and progression into S phase
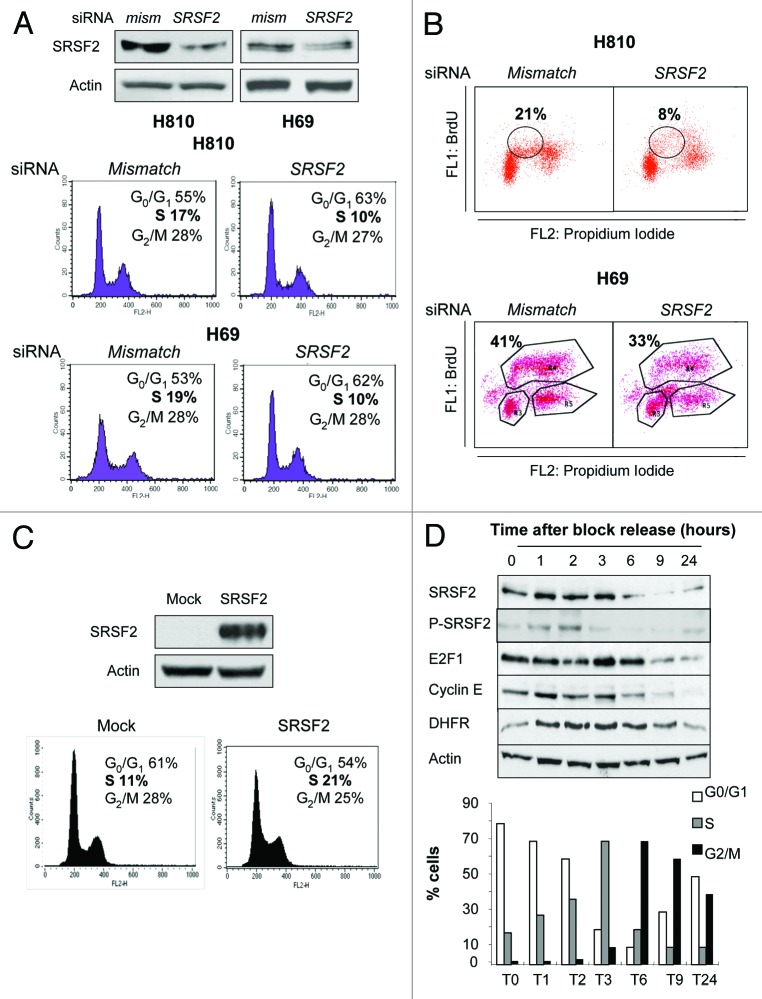
To analyze whether SRSF2 could play a role during cell cycle progression of NE lung tumors, we took advantage of two NE lung carcinoma cell lines, namely the H69 and H810 cells, that are highly proliferative and express high level of both SRSF2 and E2F1 proteins.14 First, we asked whether SRSF2 knockdown affects the cell cycle distribution of these cells. Upon co-transfection with a combination of two distinct siRNAs specifically targeting SRSF2 mRNA, the SRSF2 protein level was efficiently downregulated (Fig. 2A, upper panel). Compared with control cells transfected with mismatch siRNA, the neutralization of SRSF2 significantly decreased the proportion of cells in S phase (Fig. 2A, lower panel). In addition, in both cell lines, the number of cells incorporating bromodeoxyuridine (BrdU) significantly decreased upon transfection with SRSF2 siRNA compared with mismatch siRNA (Fig. 2B), indicating that neutralization of SRSF2 decreases S phase entry. Conversely, the transient overexpression of SRSF2 in H1299 cells that express NE features (neuromedin B) but a low level of SRSF2 protein promoted the accumulation of cells in S phase (Fig. 2C). As numerous proteins that control the cell cycle, including E2F1, are cell cycle-regulated, we next studied whether SRSF2 expression fluctuates during cell cycle progression. H69 and H810 cells cannot be easily synchronized. Thus, we used the H1299 model to synchronize cells in late G1 using hydroxyurea. At time 0, the block was released, and the cell cycle distribution was analyzed by fluorescence-activated cell sorting (FACS) after DNA staining using propidium iodide. Cells synchronized in G1 began to enter in S phase 1 h after the block release, progressed into the G2/M phases between 6–9 h and then returned in G1 following 24 h (Fig. 2D, left panel). We observed that the SRSF2 protein level transiently peaks between 1 and 3 h after the block release (Fig. 2D, right panel). Interestingly, transient accumulation of phosphorylated SRSF2 (P-SRSF2) was also detected 1 and 2 h after the block release. SRSF2 accumulation was concomittant with the upregulation of both E2F1 and cyclin E proteins (Fig. 2D). Similar results were obtained in U2OS cells that were synchronized in G1 by the use of a double thymidine block (Fig. S1). Taken together, these results demonstrate that expression and phosphorylation of the SRSF2 protein are regulated during cell cycle progression, and show that accumulation of SRSF2 correlates with S phase entry and progression.
Figure 2. SRSF2 is required for efficient S phase entry and progression. (A and B) H810 and H69 cells were transfected for 72 h with either mismatch or a combination of two distinct SRSF2 siRNAs. (A) Upper panel: Efficiency of SRSF2 neutralization was assessed by western blotting. Lower panel: Cell cycle distribution was analyzed by flow cytometry after DNA labeling with propidium iodide. Percentages of cells in the different phases are indicated. (B) BrdU incorporation was studied by FACS. The percentage of cells having incorporated BrdU is indicated in each condition. (C) H1299 cells were transfected for 48 h with an expression vector encoding the SRSF2 protein (SRSF2) or with a control plasmid (Mock). Upper panel: SRSF2 overexpression was assessed by western blotting. Lower panel: cell cycle distribution was studied by flow cytometry following propidium iodide staining. The percentages of cells in the different phases are indicated. (D) H1299 human lung adenocarcinoma cells were treated for 18h with 1 mM hydroxyurea, then washed and released in hydroxyurea-free complete medium. Cells were harvested at the indicated time points after block release. Left panel: western blot analysis of the indicated proteins. Actin was used as a loading control. Right panel: Percentages of cells in the different phases of cell cycle are indicated for each time.
SRSF2 controls the expression of E2F1-target genes involved in S phase
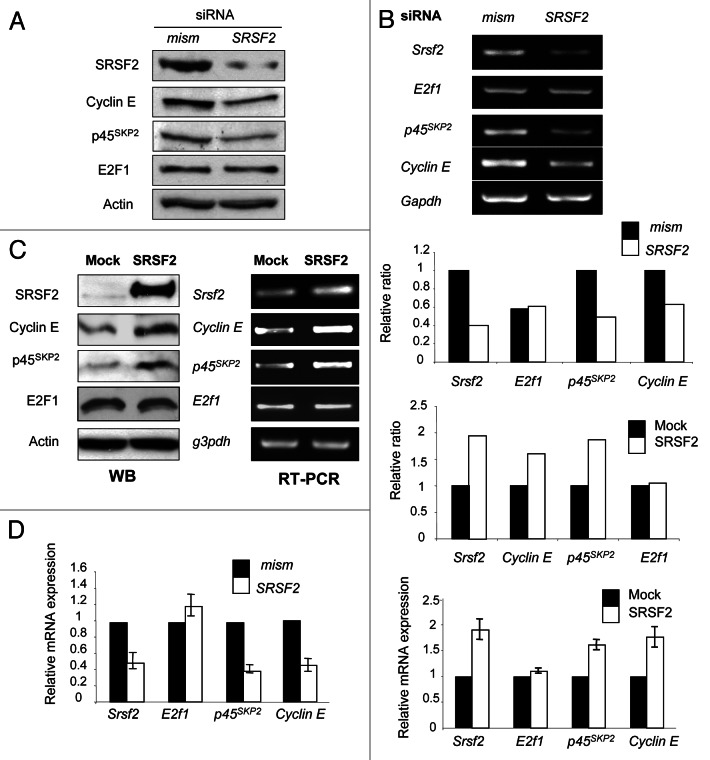
The above observations demonstrating that SRSF2 is involved in S phase entry (Fig. 2A&B) and that SRSF2 and E2F1 proteins accumulate at the same time during cell cycle (Fig. 2D) were consistent with the idea that SRSF2 contributes to the proliferative functions of E2F1. We previously reported that the cyclin E and p45SKP2 E2F1-target genes controlled S phase entry in NE lung tumor cells.13 These data prompted us to investigate whether SRSF2 regulates expression of these genes. Compared with mismatch H69 transfected cells, the knockdown of SRSF2 significantly decreased the expression level of both cyclin E and p45SKP2 proteins and transcripts, whereas E2F1 protein and mRNA levels did not vary (Fig. 3A and B). Conversely, overexpressing SRSF2 protein in H1299 cells induced the accumulation of both cyclin E and p45SKP2 proteins (Fig. 3C, left panel) and mRNAs (Fig. 3C, right panel). Again, E2F1 expression did not vary. Results of RT-PCR were confirmed by RT-qPCR analyses (Fig. 3D). Of note, modulation of SRSF2 protein amount was also associated with variations of other E2F1-target genes involved in S phase, such as dhfr, thymidilate synthase or DNA polymerase α (data not shown). Overall, these results demonstrate that SRSF2 regulates the expression of S phase-controlling E2F1-target genes.
Figure 3. SRSF2 controls the expression of E2F1-target genes required for S phase. (A and B) H69 human lung carcinoma cells were transfected for 72 h with either mismatch or a combination of two distinct SRSF2 siRNAs. (A) Western blot analysis of the indicated proteins. Actin was used as a loading control. (B) Upper panel: RT-PCR analysis of srsf2, e2f1, p45SKP2 and cyclin E mRNAs level. Gapdh was used as an internal control. Middle panel: a densitometric analysis of the specific signals was performed using ImageJ software. The value for each specific signal was normalized according to gapdh signal. A value of 1 was arbitrarily assigned to the ratio obtained in cells transfected with mismatch siRNA. (C) H1299 cells were transfected for 48 h with an expression vector encoding the SRSF2 protein (SRSF2) or with a control plasmid (Mock). Left panel: total protein extracts were subjected to western blotting using the indicated antibodies. Actin was used as a loading control. Middle panel: total RNAs were extracted and subjected to RT-PCR analysis using the indicated primers. Gapdh was used as an internal control. Right panel: a densitometric analysis of the specific signals was performed using ImageJ software. The value for each specific signal was normalized according to gapdh signal. A value of 1 was arbitrarily assigned to the ratio obtained in mock tranfected cells. (D) RT-qPCR analysis of srsf2, e2f1, p45SKP2 and cyclin E mRNAs level in H69 (lef panel) or H1299 (right panel) cells transfected either with mismatch/SRSF2 siRNA or SRSF2-encoding vector, respectively. Gapdh was used as an internal control.
SRSF2 and E2F1 interact
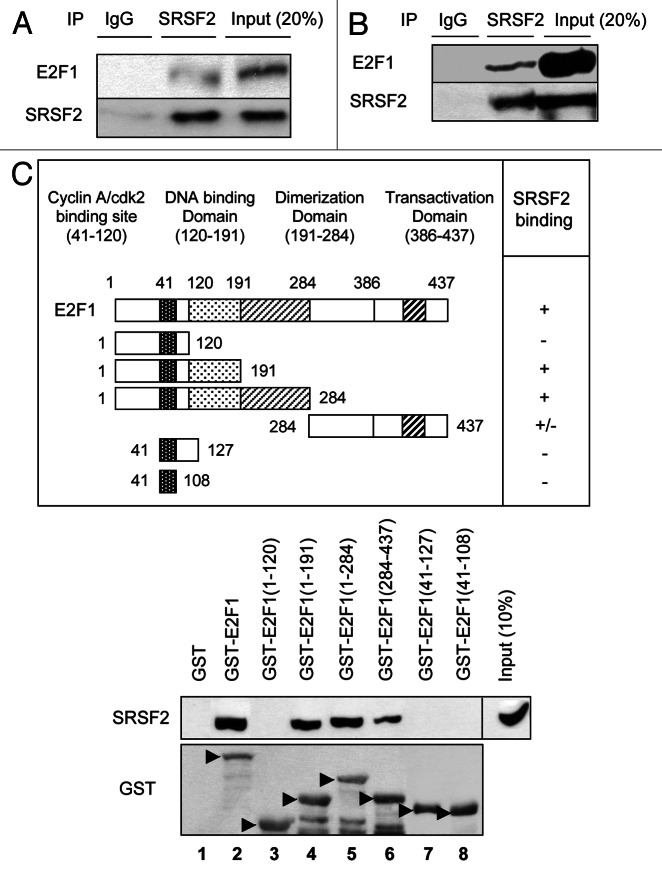
As our results suggested a functional link between SRSF2 and E2F1, we investigated whether both proteins interact. Because H69 and H810 cells are hard to transfect, we used H1299 cells to transiently overexpress E2F1 and SRSF2 using expression vectors. Immunoprecipitation experiments using a specific anti-SRSF2 antibody followed by immunodetection of E2F1 were carried out in whole-cell extracts. As shown in Figure 4A, the E2F1 protein was detected in SRSF2 immunoprecipitates. To demonstrate that these E2F1/SRSF2 complexes are not artifacts generated by the overexpression of both proteins, similar immunoprecipitation experiments were repeated in H69 cells that physiologically express high levels of both E2F1 and SRSF2. Again, co-precipitation of both proteins was detected indicating that the interaction also occurred between the two endogenous proteins (Fig. 4B). To go further, SRSF2-E2F1 binding was investigated in a cell-free system by incubating an in vitro-translated SRSF2 protein with a GST-E2F1 fusion protein. Binding between both proteins was analyzed after recovery of GST-E2F1 complexes and immunoblotting using an anti-SRSF2 antibody. The results indicated that the proteins have the ability to physically interact (Fig. 4C, lane 2). In order to map the binding sites on E2F1, several GST-E2F1 truncated mutants were generated, and the same experiments were repeated. The data demonstrated that SRSF2 interacts within the E2F1 DNA binding domain, between amino acids 127–191 (Fig. 4C, lanes 3 and 4). Of note, SRSF2 was still able, albeit less efficiently, to bind to an E2F1-truncated mutant exhibiting only the C-terminal domain (amino acids 284–437; Fig. 4C, lane 6), suggesting the existence of an additional SRSF2 binding site at the C-terminus of E2F1. Taken together, these results provide the first evidence of a direct interaction between E2F1 and SRSF2 proteins.
Figure 4. SRSF2 and E2F1 proteins interact. (A) H1299 cells were co-transfected for 48 h with SRSF2 and E2F1 expression vectors. Whole cellular extracts were immunoprecipitated with anti-SRSF2 antibody or with an irrelevant IgG as a negative control, and immunoblotting was performed with anti-E2F1 or anti-SRSF2 antibody. (B) Endogenous SRSF2 protein was immunoprecipitated from H69 nuclear extract with an anti-SRSF2 antibody. Western blotting was then performed using either anti-E2F1 or anti-SRSF2 antibody. Immunoprecipitation with irrelevant IgG was used as a negative control. (C) Upper panel: A schematic representation of E2F1 truncated proteins used in this study. The ability of each fragment to interact with an in vitro-translated SRSF2 protein is reported. Lower panel: In vitro-translated recombinant SRSF2 protein was subjected to a GST pull-down assay using GST-E2F1 wild-type or various truncated fragments of E2F1 fused to GST as a bait. The interaction between recombinant GST-E2F1 proteins and SRSF2 proteins was detected by western blotting using an anti-SRSF2 antibody. Immunoblotting with anti-GST antibody was performed to verify the expression level of GST-E2F1 fusion proteins.
SRSF2 stimulates E2F1 transcriptional activity
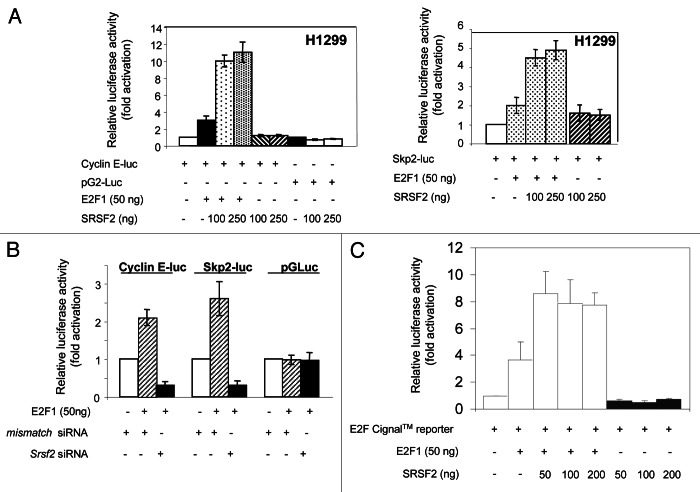
Having demonstrated that E2F1 and SRSF2 proteins physically interact, and that SRSF2 modifies the expression of E2F1-target genes, we next postulated that SRSF2 controls the transcriptional activity of E2F1. To answer, reporter vectors in which the expression of the luciferase is under the control of either the cyclin E (cyclin E-luc) or the p45SKP2 (Skp2-luc) promoter13 were transiently transfected in H1299 cells in the presence or absence of increasing amounts of E2F1 and/or SRSF2 encoding vectors. The luciferase activity was measured 48 h post-transfection. As shown in Figure 5A, SRSF2 strongly stimulated the transcriptional activity of E2F1 toward both cyclin E (left panel) and p45SKP2 (right panel) promoters. In contrast, SRSF2 did not affect the activity of a control pGL2-Luc vector. To confirm these results, the reverse experiments were performed. H1299 cells were co-transfected with either cyclin E-luc, Skp2-luc or control pGL2-Luc plasmid together with mismatch or SRSF2 siRNAs, in the presence of a plasmid encoding E2F1. We observed that the knockdown of SRSF2 significantly decreased the ability of E2F1 to transactivate the cyclin E and p45SKP2 promoters, while it did not affect the activity of the control pGL2-Luc vector (Fig. 5B). To test whether the effect of SRSF2 could take place in a more general context, we used the Cignal™ E2F reporter assay that monitors the transcriptional activity of E2F by using an E2F-responsive luciferase construct encoding the luciferase under the control of tandem repeats of the E2F transcriptional response element (TRE). As shown in Figure 5C, SRSF2 also stimulated the transcriptional activity of E2F1 in that case, thereby confirming, using another E2F reporter assay, that SRSF2 is a positive regulator of E2F1 transcriptional activity. Of note, SRSF2 did not enhance the binding of E2F1 to its consensus binding site, whereas in the same conditions the co-factor DP1 strongly stimulated E2F1 binding (Fig. S2).
Figure 5. SRSF2 stimulates the transcriptional activity of E2F1. (A) Luciferase experiments were conducted in H1299 cells co-transfected for 48 h with 1 µg pGL2-cyclin E-luc (left panel) or pGL2-p45SKP2-luc (right panel) encoding the luciferase under the control of the cyclin E or p45SKP2 promoter respectively, or with 1 µg pGL2-Luc as a control, in the presence or absence of 50 ng pCMV-E2F1 and increasing amounts of pcDNA3-SRSF2 (100 and 250 ng). The luciferase activity obtained in cells transfected with pGL2-cyclin E-luc or pGL2-p45SKP2-luc or pGL2-Luc was normalized to 1, and a relative luciferase activity was then calculated for each condition. (B) H1299 cells were transfected with either mismatch or SRSF2 siRNAs. Twenty-four hours later, cells were co-transfected with either pGL2-cyclin E-luc, Skp2-luc or pGL2-luc plasmid, in the presence or absence of 50 ng pCMV-E2F1 as indicated. The luciferase activity was measured 48 h later. The luciferase activity obtained in cells transfected with either pGL2-cyclin E-luc, Skp2-luc or pGL2-Luc alone was normalized to 1, and a relative luciferase activity was then calculated for each condition. (C) Luciferase assays were performed in H1299 cells co-transfected for 48 h with E2F Cignal™ Reporter (SuperArray) in the presence or absence of 50 ng pCMV-E2F1 and increasing amounts of pcDNA3-SRSF2 (50, 100 and 200 ng). The luciferase activity obtained in cells transfected with E2F Cignal™ Reporter alone was normalized to 1, and a relative luciferase activity was then calculated for each condition. Of note, SRSF2 does not enhance the activity of E2F Cignal™ reporter in the absence of E2F1.
SRSF2 is required for efficient E2F1 recruitment to the Skp2 or cyclin E promoter
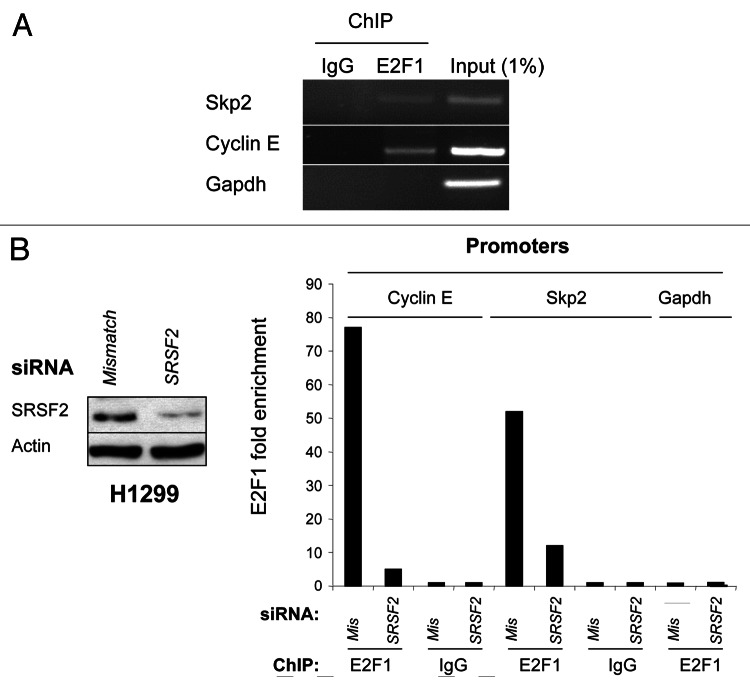
Having provided evidence that SRSF2 is a co-activator of E2F1 transcriptional function, we next wanted to demonstrate the binding of both E2F1 and SRSF2 proteins to cyclin E or Skp2 promoter by performing chromatin immunoprecipitation assays. In H1299 cells, binding of E2F1 to the proximal Skp2 or cyclin E promoter was clearly detected, whereas no binding was observed within the Gapdh-negative control promoter (Fig. 6A). Similar results were obtained in H810 cells (data not shown). Importantly, silencing of SRSF2 in H1299 cells using siRNA (Fig. 6B, left panel) strongly reduced the recruitment of E2F1 on the cyclin E and Skp2 promoters as demonstrated using Q-ChIP experiments (Fig. 6B, right panel). Overall, these results indicated that SRSF2 is required for the binding of E2F1 to the cyclin E and Skp2 promoters. Unfortunately, in similar conditions, we were not able to immunoprecipitate SRSF2 protein from cyclin E or Skp2 promoters, whatever the anti-SRSF2 (SRSF2–4F11; SRSF2-H55) or P-SRSF2 antibody used.
Figure 6. SRSF2 is required for E2F1 recruitment to cyclin E or Skp2 promoter. (A) H1299 cells were processed for ChIP analysis using C-20 antibody for E2F1. The co-precipitated chromatin DNA was analyzed by semi-quantitative PCR using pair of primers that amplify the Skp2, cyclin E or Gapdh promoter, respectively. IgG was used as an irrelevant antibody. Input lane corresponds to PCRs containing 1% of total amount of chromatin used in immunoprecipitation reactions. (B) H1299 cells were transfected for 72 h with either mismatch or SRSF2 siRNA. Left panel: immunoblot analysis of SRSF2 protein level. Actin was used as a loading control. Right panel: chromatin immunoprecipitation experiments were performed using an anti-E2F1 (E2F1) or an irrelevant IgG (IgG) antibody. The genomic DNA regions encompassing E2F1 binding site of the cyclin E or Skp2 promoter were amplified by qPCR. The gapdh promoter was used as a negative control. Results were normalized to input and expressed as fold enrichment compared with irrelevant antibody.
The PI3K/AKT signaling pathway controls SRSF2 phosphorylation and activity toward E2F1
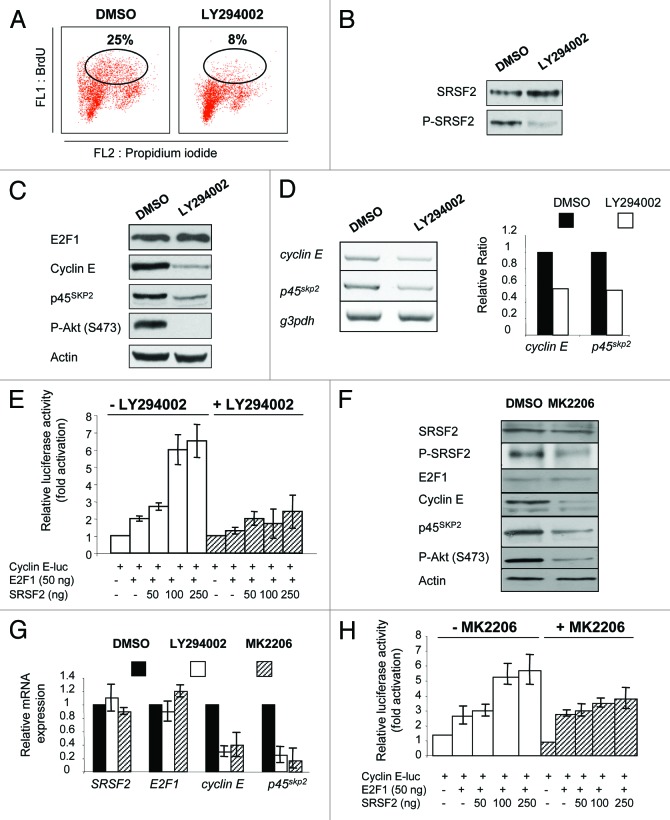
It is well-known that phosphorylation of SR proteins controls their function.16 Here we showed that P-SRSF2 accumulates in synchronized cells entering and progressing in S phase (Fig. 2D; Fig. S1). We also found that P-SRSF2 and cyclin E status are directly correlated in high-grade NE lung tumors (Table S1). Together, these data strongly suggested that phosphorylation of SRSF2 controls E2F1 activity. Several kinases have been reported to phosphorylate SR proteins, including AKT.16 In addition, it has been shown that the PI3K/AKT signaling pathway regulates E2F1-mediated cellular proliferation.17 Therefore, we postulated that the PI3K/AKT pathway controls SRSF2 phosphorylation and function toward E2F1. We first noticed that phosphorylated AKT concomittantly accumulated with P-SRSF2 during cell cycle progression of synchronized cells (Fig. S1). In these conditions, the level of total AKT protein did not vary. In addition, treating H69 cells with LY294002, a well-characterized PI3K inhibitor, significantly inhibited BrdU incorporation (Fig. 7A) and strongly decreased SRSF2 phosphorylation (Fig. 7B) as well as cyclin E and p45SKP2 proteins (Fig. 7C) and mRNAs (Fig. 7D) levels compared with DMSO-treated cells. Moreover, LY294002 treatment significantly prevented SRSF2-dependent activation of E2F1 transcriptional function toward cyclin E (Fig. 7E) or p45SKP2 (data not shown) promoter. In order to confirm these results, we used MK-2206, a more selective inhibitor of AKT1, AKT2 and AKT3. As LY294002, MK-2206 decreased P-SRSF2, cyclin E and p45SKP2 protein (Fig. 7F) and mRNA (Fig. 7G) levels compared with DMSO-treated H69 cells. In addition, MK-2206 prevented the ability of SRSF2 to stimulate E2F1 transcriptional function toward cyclin E (Fig. 7H) or p45SKP2 (data not shown) promoter. In these conditions, MK-2206 did not significantly affect the luciferase activity of a control pGL2-Luc plasmid, indicating that it did not impair translation of the luciferase (data not shown). Overall, these results highly suggest that AKT signaling pathway plays a role in SRSF2 phosphorylation and effect toward E2F1.
Figure 7. The PI3K/AKT signaling pathway phosphorylates SRSF2 and controls its function toward E2F1 transcriptional activity. H69 (A–D, F and G) or H1299 (E and H) cells were incubated for 24 h with DMSO as a control or 50 µM LY294002 or 3 µM MK-2206 as indicated. (A) BrdU incorporation was studied by FACS. The percentage of cells having incorporated BrdU is indicated in each condition. (B) Western blotting was performed for the detection of SRSF2 and its phosphorylated form (P-SRSF2). (C) Western blotting was performed for the detection of the indicated proteins. Anti-P-AKT (S473) antibody was used to control AKT inactivation. Actin was used as a loading control. (D) Upper panel: RT-PCR analysis of cyclin E and p45SKP2 mRNA levels. Lower panel: Each specific signal was quantified after densitometric analysis using ImageJ software and normalized according to the signal obtained for gapdh. A value of 1 was arbitrarily assigned to the ratio obtained in DMSO-treated cells and a relative ratio was then calculated for LY294002 treated cells. (E) Cells were co-transfected for 48 h with 1 µg pGL2-cyclin E-luc in the presence or absence of 50 ng pCMV-E2F1 and increasing amounts of pcDNA3-SRSF2 (50, 100 and 250 ng), and treated or not with 50 µM LY294002. The luciferase activity obtained in cells transfected with pGL2-cyclin E-luc alone was normalized to 1, and a relative luciferase activity was then calculated for each condition. (F) Western blotting was performed for the detection of the indicated proteins. (G) RT-qPCR analysis of the indicated transcripts was performed in H69 cells treated with LY294002 or MK-2206 as mentioned. For each transcript, the mRNA level was normalized according to gapdh. A value of 1 was arbitrarily assigned to the relative gene expression obtained in DMSO-treated cells. (H) Cells were co-transfected for 48 h with 1 µg pGL2-cyclin E-luc in the presence or absence of 50 ng pCMV-E2F1 and increasing amounts of pcDNA3-SRSF2 (50, 100 and 250 ng), and treated or not with 3 µM MK-2206. The luciferase activity obtained in cells transfected with pGL2-cyclin E-luc alone was normalized to 1, and a relative luciferase activity was then calculated for each condition.
Discussion
To date, the role of individual SR proteins in the control of specific cellular processes or diseases remains poorly understood. In this study, we demonstrate that SRSF2 is a cell cycle-regulated protein involved in S phase transition, and show that SRSF2 stimulates the transcriptional activity of E2F1 toward S phase target genes, such as cyclin E and p45SKP2. Therefore, besides its ability to control E2F1-induced apoptosis,14 these results demonstrate that SRSF2 is also able to regulate E2F1 proliferative functions.
We previously demonstrated that SRSF2 is a direct transcriptional target of E2F1 in various cell lines treated with genotoxic agents.14 Here, we show that SRSF2 protein and mRNA amounts fluctuate in the same way that E2F1 does during the cell cycle progression, rising sharply at the G1 to S phase transition and remaining elevated as cells progress into S phase. These data suggest that E2F1 could also control SRSF2 expression during the cell cycle. Importantly, we provide evidence that the SRSF2 protein is required for a proper S phase entry and progression in lung carcinoma cell lines. It has been shown that loss of SRSF2 in mouse embryonic fibroblasts (MEFs) induces a G2/M cell cycle arrest that results, at least in part, from p53 activation and further p21WAF1 accumulation.11 In contrast, downregulation of SR protein expression decreases S phase entry in SV40-immortalized MEF.9 In Drosophila, the SR protein B52 is a positive regulator of the repressive dE2F2 protein that prevents the G1 to S phase transition.10 Moreover, a role of SRSF2 in the control of pre-mRNA splicing during the mitosis has recently been reported in human epithelial tumor cells.18 Altogether, these data demonstrate that various SR proteins control cell cycle progression, and suggest that their distinct ability to affect either positively or negatively different phases of the cell cycle could depend on the cell type (primary vs. immortalized) or on the upstream stimuli.
Functions of SR proteins that are not related to the control of pre-mRNA splicing are emerging. As an example, in vivo depletion of SRSF1 and SRSF2 proteins dramatically attenuates the production of nascent RNA, and nuclear run-on assays provide evidence for an active role of SRSF2 in transcriptional elongation.19 It is known that the promoter choice exerts a strong influence on alternative splicing.20-22 Conversely, mRNA splicing exerts a stimulatory effect on transcription initiation. Indeed, it has been shown that basal transcription initiation factors are recruited to promoters that are closed to 5′ splice site via their docking with constitutive splicing factors such as U1 small nuclear RNA.23-25 In this study, we provide evidence that SRSF2 directly interacts with the DNA binding domain of E2F1 and stimulates E2F1 transcriptional activity toward cell cycle-regulating genes. Therefore, besides controlling elongation, SRSF2 could also play a role during transcriptional initiation through its binding to transcription factors. Of note, SRSF2 was able to stimulate E2F1 transcriptional function toward a promoter containing E2F1-consensus binding sites, suggesting that SRSF2 regulates the expression of numerous E2F1-target genes besides cyclin E or p45SKP2. How does SRSF2 act to stimulate E2F1 function? The transcriptional activity of E2F1 is tightly regulated by its stability, DNA binding capacity or interaction with pocket proteins of the retinoblastoma protein family that sequester E2F1 inside inactive transcriptional complexes.4 In our cellular models, SRSF2 does not control E2F1 protein amount. In contrast, qChIP experiments demonstrate that SRSF2 is required for E2F1 recruitment to cyclin E or p45SKP2 promoter. Unfortunately, in similar conditions, we were not able to immunoprecipitate SRSF2 from both promoters. Although we cannot exclude the possibility that SRSF2 is not recruited together with E2F1 on these promoters, these negative results could reflect technical problems. Indeed, SRSF2 is not known to directly bind DNA, and the commercially available anti-SRSF2 antibodies are not very efficient in immunoprecipitation experiments. Therefore, the amount of SRSF2 cross-linked to DNA through its interaction with E2F1 or immunoprecipitated could be too low to allow further detection. Interestingly, we observed that SRSF2 is able to interact, albeit less efficiently, with the C-terminal domain of E2F1 (amino acids 284–437) that contains the pocket protein-binding domain. Another possibility is that SRSF2 competes with pocket proteins for E2F1 binding, thereby releasing E2F1 from sequestration and allowing transcriptional activation. However, we also obtained the same positive effect of SRSF2 on E2F1 transcriptional activity toward the cyclin E and p45SKP2 promoters in Saos-2 cells that are pRB-null (data not shown), thereby indicating that SRSF2 can regulate E2F1 transcriptional function in a pRB-independent manner. Finally, another possibility relies on our recent publication unraveling the interaction between SRSF2 and the histone acetyl transferase TIP60.26 Whether SRSF2 tethers chromatin remodeling enzymes to E2F1-target genes promoters to facilitate E2F1 transcriptional function remains to be determined.
Phosphorylation of SR proteins is known to control their activity as well as their subcellular localization. Various kinases, such as AKT, CLK, SRPK1, SRPK2 or topoisomerase 1 have been shown to phosphorylate SR proteins. Recently, we demonstrated that SRPK2-dependent phosphorylation of SRSF2 contributes to cisplatin-induced apoptosis in human lung adenocarcinoma cell lines.26 In neuroendocrine lung carcinoma cell lines, SRPK1 or SRPK2 do not stimulate SRSF2 phosphorylation (Edmond, unpublished results). In contrast, we provide evidence that inhibition of the PI3K/AKT signaling pathway decreases P-SRSF2 expression. In addition, we show that AKT signaling pathway regulates SRSF2 activity toward E2F1 transcriptional function on cyclin E and p45SKP2 promoters. It has been shown that the PI3K/AKT pathway is activated in NE lung tumors,27 and we previously demonstrated that cyclin E and E2F1 proteins are aberrantly expressed and directly correlated in these tumors.13 Therefore, although we do not demonstrate a direct control of P-SRSF2 by AKT, the observation that P-SRSF2 status directly correlates with cyclin E protein level in NE lung tumors fits well with a model in which phosphorylation of SRSF2 by the PI3K/AKT signaling pathway could control E2F1 transcriptional activity toward cyclin E, likely contributing to the high proliferative index of NE lung tumors.
Materials and Methods
Cell lines, cell treatments, plasmids and transfection
H1299 human large cell NSCLC with neuroendocrine features, H810 and H69 human lung neuroendocrine carcinoma cell lines were cultured as previously described.13,28 U-2OS osteosarcoma cells were cultured in McCoy medium supplemented with 10% FCS. Transient transfections were performed using Fugene 6 (Roche Diagnostic) according to the manufacturer’s instructions. Plasmids used in transient transfections were pcDNA3.1, pcDNA3.1-SRSF2, pCMV-E2F1, pGL2-Luc, pGL2-cyclin E encoding the luciferase protein under the control of the cyclin E promoter, pGL2-Skp2 encoding the luciferase under the control of the human Skp2 promoter region spanning from –272 to +244 residues and pCMV-DP1. An inducible E2F-responsive construct encoding the firefly luciferase reporter gene under the control of a basal promoter element (TATA box) joined to tandem repeats of a specific E2F transcription response element (TRE) was purchased from SuperArray (Tebu-bio). Hydroxyurea was purchased from Sigma and LY294002 from Ozyme. MK-2206 was purchased from Selleckchem and used at a final concentration of 3 µM. BrdU solution was purchased from Roche. For synchronization of cells, 1 mM hydroxyurea was added to 30% confluent H1299 cells for 18 h. Cells were then washed 3× with PBS, released in hydroxyurea-free complete medium and collected at the indicated time points after block release. For the double block of thymidine, 30% confluent U-2 OS cells were treated with 2.5 mM thymidine for 17 h, then washed 3× in PBS and released in thymidine free complete medium for 8 h. Thymidine was added again at 2.5 mM for 17 h. Cells were released and collected at the indicated time points.
Antibodies
The antibodies anti-E2F1 (KH95), anti-E2F1 (C-20) and anti-SRSF2 (H55) were purchased from Santa Cruz, the anti-cyclin E (13A3) from Novocastra, the anti-DHFR from BD Biosciences, the anti-actin (A2066) from Sigma, the anti-SRSF2 (4F11) from Euromedex, the anti-p45SKP2 from Zymed (Invitrogen) and the anti-phosphorylated SRSF2 from Abcam. The anti-pan-AKT and anti-phosphorylated AKT (S473) were purchased from Cell Signaling (Ozyme). The FITC-conjugated BrdU antibody was purchased from Roche. Immunoprecipitation and western blotting experiments were performed as previously described.28
Transfection of siRNA oligonucleotides
The two sequences designed to specifically target human SRSF2 RNAs were purchased from Eurogentec and were as follows: 5′-UCGAAGUCUCGGUCCCGCACUCG-3′ and 5′-GAGGACGCUAUGGAUGCCAUGGACG-3′. In all experiments, a 50:50 mixture of both siRNAs was used, leading to a final concentration of 100nM siRNA/Petri dish. The mismatch siRNA oligonucleotide used as a control was 5′-UCGGCUCUUACGCAUUCAA-3′. Cells were transfected with siRNA oligonucleotides duplex using Oligofectamine reagent according to the manufacturer’s instructions (Invitrogen). The cells were analyzed 72 h post-transfection.
FACS analyses
For DNA content analysis, cells were fixed with 70% cold ethanol for 30 min on ice, treated with RNase A (20 µg/ml) for 20 min and stained with propidium iodide (10 µg/ml). Flow cytometric analysis of 10,000 cells was performed on a FACScan flow cytometer (BD Biosciences) and data were recovered using the CellQuest software (BD Biosciences). For BrdU incorporation measurements, cells were pulsed with 100 µM BrdU for 20 min, before harvesting. Cells were collected and fixed in 70% cold ethanol for 30 min on ice, then denatured in 4N HCl for 30 min at room temperature, washed in PBS containing 0.5% Tween 20 and incubated with FITC-conjugated BrdU antibody in PBS containing 0.1% BSA for 30 min. Cells were washed in PBS, stained with propidium iodide (PI) and analyzed by flow cytometry.
RT-PCR and RT-QPCR
Total cellular RNAs were isolated using Trizol reagent (Invitrogen). One µg of total RNA was reverse transcribed using oligo (dT) primer and MMLV reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. RT reaction (3 µl) was then amplified by PCR for 30 cycles using the following conditions: 94°C for 30 sec; 57°C for 45 sec; 72°C for 1 min. The specific primers used for mRNA amplification were as follows: SRSF2 forward: 5′-CCA-CTC-AGA-GCT-ATG-AGC-TAC-G-3′; SRSF2 reverse: 5′-ACT-CCT-TGG-TGT-AGC-GAT-CC-3′; p45SKP2 forward: 5′-TCAACTACCTCCAACACCTATCAC-3′; p45SKP2 reverse: 5′-GACAACTGGGCTTTTGCAGT-3′; cyclin E forward: 5′-AGTTCTCGGCTCGCTCCAGGAAGA-3′; cyclin E reverse: 5′-TCTTGTGTCGCCATATACCGGTCA-3′. Amplification of a fragment of the cDNA of gapdh (Invitrogen) was performed in the same PCR reaction as internal control. PCR products were run on a 2% agarose gel and visualized by ethidium bromide staining.
Quantitative real time RT-PCR was performed on Stratagene MX3005P apparatus (Agilent Technologies). In all conditions, 1 µg of total RNA were subjected to cDNA synthesis using Superscript III First-Strand Synthesis SuperMix for qPCR (Invitrogen) and subsequently amplified during 40 PCR cycles (10 min at 95°C, 15 sec at 95°C, 1 min at 60°C) using Power SYBR Green PCR Master Mix (Applied Biosystems). The specific primers used for mRNA amplification were as follows: cyclin E forward: 5′-GAA-ATG-GCC-AAA-ATC-GAC-AG-3′; cyclin E reverse: 5′-TCT-TTG-TCA-GGT-GTG-GGG-A-3′; SKP2 forward: 5′-GCT-GAA-GAG-CAA-AGG-GAG-TG-3′; SKP2 reverse: 5′-GAA-GGG-AGT-CCC-ATG-AAA-CA-3′; GAPDH forward: 5′-CGA-GAT-CCC-TCC-AAA-ATC-AA-3′; GAPDH reverse: 5′-ATC-CAC-AGT-CTT-CTG-GGT-GG-3′. In all experiments, RT-qPCR detection of the reference gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed for each sample. Relative gene expression was calculated for each sample, as the ratio of target gene copy number to GAPDH mRNA copy number multiplied by 100.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation experiments were performed in H1299 cells transfected for 72 h with either mismatch or SRSF2 siRNA. ChIP experiments were performed using the ChIP-ITR Express Magnetic Chromatin Immunoprecipitation kit from Active Motif (La Hulpe, Belgium) according to manufacturer’s instructions. Briefly, cells were formaldehyde cross-linked, and chromatin was isolated and sonicated using a Bioruptor apparatus. An equal amount of chromatin (30 µg) was precleared, immunoprecipitated with either a polyclonal antibody specific for E2F1 (C-20, Santa Cruz) or unrelated rabbit IgG, overnight at +4°C, washed and reverse cross-linked. One-twentieth of the immunoprecipitated chromatin was analyzed for the presence of cyclin E or SKP2 promoter DNA by Q-PCR using previoulsy described primers that flanked the E2F1 binding sites on both promoters.29,30 A sequence corresponding to the gapdh promoter was used as a negative control in E2F1 ChIP. Q-PCR studies were performed using the ChIP-qPCRTM assay from SuperArray (TEBU-Bio) according to the manufacturer’s instructions. Input DNA sample corresponding to 1% of immunoprecipitated chromatin was analyzed in parallel in order to normalized the results of each ChIP DNA sample to the corresponding input DNA sample. The primers used were as follow: cyclin E forward 5′-GCC-ATC-GGC-CAT-CTT-CCT-GGC-TC-3′ ; cyclin E reverse 5′-TCA-GGC-CGC-GGG-CCC-AGT-A-3′ ; SKP2-95 5′-CTCCCCGCCTACCCCGTGG-3′, SKP2-+135 5′-CAGACCCGCTAAGCCTAGCAACG-3′, gapdh forward 5′-AGCTCAGGCCTCAAGACCTT-3′ and gapdh reverse 5′-AAGAAGATGCGGCTGACTGT-3′.
GST pull-down assay
Beads coated with GST, GST-E2F1 or different GST-truncated E2F1 fusion proteins were prepared according to the manufacturer’s protocol (Bulk GST Purification module, Pharmacia Biotech). Beads were incubated for 45 min at room temperature with equivalent amounts of in vitro translated SRSF2 protein in a final volume of 150 µl binding buffer (25 mM HEPES pH 7.6, 8 mM MgCl2, 150 mM KCl, 0.1% NP40, 20% glycerol) containing 0.2 mg/ml BSA. Beads were washed three times with NETN buffer (100 mM NaCl, 1 mM EDTA, 0.5% NP40, 20 mM Tris pH 8.0), once with PBS and then analyzed by 10% SDS-PAGE.
Luciferase assays
Cells were lysed 48 h after transfection in 300 µl lysis buffer (Passive lysis buffer from Promega). The cell debris were removed by centrifugation at 13,200 rpm for 2 min, and luciferase activity was measured on a 10 µl aliquot in a luminometer using the luciferase kit from Promega. Each sample was normalized according to the protein amount. Results are the mean of three independent experiments performed in duplicate.
Analysis of E2F1 DNA-binding activity by ELISA assay
The DNA-binding activity of E2F1 was assessed using Transcription Factor (TF) ELISA Kit (Panomics) according the manufacturer’s instructions. Briefly, nuclear extracts obtained from Nuclear Extraction Kit (Panomics) were incubated with biotinylated oligonucleotides immobilized on a streptavidin-coated 96-well plate and containing an E2F1-consensus binding site (E2F1-Probe). E2F1 protein, bound to the oligonucleotides, was detected by an antibody directed against E2F1. An additional HRP-conjugated secondary antibody provides a sensitive colorimetric readout quantified by spectrometry.
Immunohistochemistry and immunohistochemical staining evaluation
Tissue samples were collected from lung resection of lung tumors and stored for scientific research in a biological resource repository (Centre de Ressources Biologiques, CHU Albert Michallon, Grenoble Hospital). National ethical guidelines were followed. All patients enrolled in this trial provided written informed consent. Tissue banking and research conduct was approved by the Ministry of Research (approval AC-2010-1129) and by the regional IRB (CPP 5 Sud Est). E2F1, cyclin E, SRSF2 and P-SRSF2 immunohistochemical stainings were performed as previously described12-15 in a series of 27 neuroendocrine lung carcinoma comprising 12 large cell neuroendocrine carcinoma (LCNEC) and 15 small cell lung carcinoma (SCLC). SRSF2, P-SRSF2, E2F1 and cyclin E immunostainings were evaluated by two independent observers in distinct areas of the slide sections for correlation and confirmation of tissues analysis and scored by taking into account the tumor heterogeneity. A final score (0–300) was established by multiplying the percentage of labeled cells (0–100%) with the intensity of staining (1–3). According to these final scores, tumor samples were divided in two classes for each staining, with either tumors overexpressing (+) or not (−) the protein of interest compared with normal lung tissues. The scores in normal lung tissues for SRSF2 and P-SRSF2 proteins were 100 and 50, respectively. Therefore, tumors displaying a score ≥ 200 or > 100 were considered as tumors overexpressing SRSF2 or P-SRSF2, respectively. As previously reported,12,13 tumors with a score ≥ 40 were considered as tumors overexpressing these proteins. Statistical analyses were perfomed using the Chi2-test with a p value ≤ 0.05 being considered significant.
Supplementary Material
Acknowledgments
We thank Sophie Michallet, Pascal Perron, Celine Barrial-Lampreia and Floriane Albert for technical assistance. This work was supported by the Comité Départemental Isère de la Ligue contre le Cancer, by the Fondation ARC pour la Recherche Contre Le Cancer and by the Conseil Scientifique National d’AGIR à dom. Valerie Edmond was supported by a grant from the Conseil Scientifique National d’AGIR à dom. Galina Merdzhanova was supported by fellowships from the Research French Ministry and the Fondation pour la Recherche Medicale (FRM). Stephanie Gout was supported by a post-doctoral fellowship from the Fondation ARC pour la Recherche Contre Le Cancer.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24363
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24363
References
- 1.Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–32. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–26. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 3.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DG, Degregori J. Putting the Oncogenic and Tumor Suppressive Activities of E2F into Context. Curr Mol Med. 2006;6:731–8. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- 5.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 6.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9:899–902. doi: 10.1016/S0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 7.Wang HY, Xu X, Ding JH, Bermingham JR, Jr., Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Mol Cell. 2001;7:331–42. doi: 10.1016/S1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 8.Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–96. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–25. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Rasheva VI, Knight D, Bozko P, Marsh K, Frolov MV. Specific role of the SR protein splicing factor B52 in cell cycle control in Drosophila. Mol Cell Biol. 2006;26:3468–77. doi: 10.1128/MCB.26.9.3468-3477.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, et al. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eymin B, Gazzeri S, Brambilla C, Brambilla E. Distinct pattern of E2F1 expression in human lung tumours: E2F1 is upregulated in small cell lung carcinoma. Oncogene. 2001;20:1678–87. doi: 10.1038/sj.onc.1204242. [DOI] [PubMed] [Google Scholar]
- 13.Salon C, Merdzhanova G, Brambilla C, Brambilla E, Gazzeri S, Eymin B. E2F-1, Skp2 and cyclin E oncoproteins are upregulated and directly correlated in high-grade neuroendocrine lung tumors. Oncogene. 2007;26:6927–36. doi: 10.1038/sj.onc.1210499. [DOI] [PubMed] [Google Scholar]
- 14.Merdzhanova G, Edmond V, De Seranno S, Van den Broeck A, Corcos L, Brambilla C, et al. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ. 2008;15:1815–23. doi: 10.1038/cdd.2008.135. [DOI] [PubMed] [Google Scholar]
- 15.Gout S, Brambilla E, Boudria A, Drissi R, Lantuejoul S, Gazzeri S, et al. Abnormal expression of the pre-mRNA splicing regulators SRSF1, SRSF2, SRPK1 and SRPK2 in non small cell lung carcinoma. PLoS One. 2012;7:e46539. doi: 10.1371/journal.pone.0046539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaustein M, Pelisch F, Tanos T, Muñoz MJ, Wengier D, Quadrana L, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–44. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 17.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, et al. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011;42:185–98. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–26. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cramer P, Cáceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, et al. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–8. doi: 10.1016/S1097-2765(00)80372-X. [DOI] [PubMed] [Google Scholar]
- 21.Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, et al. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc Natl Acad Sci U S A. 2004;101:2270–4. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auboeuf D, Hönig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–9. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 23.Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–9. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwek KY, Murphy S, Furger A, Thomas B, O’Gorman W, Kimura H, et al. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–5. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 25.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–8. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 26.Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, et al. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011;30:510–23. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salon C, Eymin B, Micheau O, Chaperot L, Plumas J, Brambilla C, et al. E2F1 induces apoptosis and sensitizes human lung adenocarcinoma cells to death-receptor-mediated apoptosis through specific downregulation of c-FLIP(short) Cell Death Differ. 2006;13:260–72. doi: 10.1038/sj.cdd.4401739. [DOI] [PubMed] [Google Scholar]
- 29.Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G. Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells. Cancer Res. 2007;67:4149–56. doi: 10.1158/0008-5472.CAN-06-4484. [DOI] [PubMed] [Google Scholar]
- 30.Cartier J, Berthelet J, Marivin A, Gemble S, Edmond V, Plenchette S, et al. Cellular inhibitor of apoptosis protein-1 (cIAP1) can regulate E2F1 transcription factor-mediated control of cyclin transcription. J Biol Chem. 2011;286:26406–17. doi: 10.1074/jbc.M110.191239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.