Abstract
The epidermal growth factor receptor (EGFR)-directed monoclonal antibody cetuximab is the only targeted therapy approved for the treatment of head and neck squamous cell carcinoma (HNSCC), but is only effective in a minority of patients. Epithelial-to-mesenchymal transition (EMT) has been implicated as a drug resistance mechanism in multiple cancers, and the EGFR and Hedgehog pathways (HhP) are relevant to this process, but the interplay between the two pathways has not been defined in HNSCC. Here we show that HNSCC cells that were naturally sensitive to EGFR inhibition over time developed increased expression of the HhP transcription factor GLI1 as they became resistant after long-term EGFR inhibitor exposure. This robustly correlated with an increase in Vimentin expression. Conversely, the HhP negatively regulated an EGFR-dependent, EMT-like state in HNSCC cells, and pharmacological or genetic inhibition of HhP signaling pushed cells further into an EGFR-dependent phenotype, increasing expression of ZEB1 and VIM. In vivo treatment with cetuximab resulted in tumor shrinkage in four out of six HNSCC patient-derived xenografts; however they eventually re-grew. Cetuximab in combination with the HhP inhibitor IPI-926 eliminated tumors in two cases and significantly delayed re-growth in the other two cases. Expression of EMT genes TWIST and ZEB2 was increased in sensitive xenografts suggesting a possible resistant mesenchymal population. In summary, we report that EGFR-dependent HNSCC cells can undergo both EGFR-dependent and -independent EMT and HhP signaling is a regulator in both processes. Cetuximab plus IPI-926 forces tumor cells into an EGFR-dependent state delaying or completely blocking tumor recurrence.
Keywords: Head and neck squamous cell cancer, hedgehog pathway, epidermal growth factor receptor pathway, epithelial to mesenchymal transition
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is a devastating disease afflicting >45,000 people in the United States each year (1). The epidermal growth factor receptor (EGFR)-directed inhibitor cetuximab is the only approved targeted therapy for the treatment of HNSCC (2, 3). However, efficacy is low and patients that initially demonstrate a promising response often become refractory (4, 5). The EGFR and hedgehog pathways (HhP) have been implicated as key drivers of proliferation and survival of cancer cells. HhP signaling may also be a crucial pathway in the survival and characteristics of cancer stem cells and is a likely candidate for drug resistance (6, 7).
EGFR and HhP signaling converge and/or synergize upstream of GLI1 through the MEK/ERK signaling pathway in cancer cells and during keratinocyte oncogenic transformation (8–10). Epidermal growth factor (EGF) stimulates expression of GLI1 and target genes BCL2 and PTCH1 in gastric cancer (11), and the HhP ligand sonic hedgehog (SHH) signals through MAPK and PI3K to increase expression of HhP specific targets in renal cancer (12).
Both pathways have been closely linked to epithelial-to mesenchymal-transition (EMT) (13, 14). In this process epithelial cells gain a more spindle or fibroblast-like phenotype and become more mobile and invasive, Molecularly, EMT is characterized by expression of the pro-EMT ZEB, SNAIL and TWIST transcription factors, loss of E-cadherin (E-CAD) and increased levels of Vimentin (Vim) (15). The ability of cells to alter their morphology is often associated with drug resistance, allowing tumor cells to escape from cytotoxic and pathway targeted therapies (16–18). Recently, reports have described an EGF-induced EMT-like state in EGFR-dependent HNSCC and prostate cancer cell lines (19, 20). On the other hand, chronic gefitinib treatment was found to generate a mesenchymal drug resistant population in HNSCC cells independent of EGFR activation (21). The dichotomy of these EGFR-dependent and resistant states and the role of HhP signaling have yet to be clarified in HNSCC.
The relationship between these pathways and their individual roles in EMT and drug resistance was previously investigated in immortalized keratinocytes or cancer cell lines (8, 11). We have generated and characterized a direct patient xenograft bank of HNSCC tumors implanted directly into mice with no time spent in culture. These in vivo tumor models may better mimic tumor heterogeneity and the relationship with the microenvironment (22).
We aimed to define the roles of EGFR and HhP signaling in early (EGFR-dependent) and late (EGFR-independent) EMT, migration/invasion, and anti-EGFR therapy susceptibility in HNSCC. We characterized the crosstalk between EGFR and HhP in HNSCC, and conducted combination studies targeting EGFR and HhP signaling in patient-derived xenografts.
MATERIALS AND METHODS
Cell lines and in vitro drugs
HN11, Tu-167, FaDu and 584 HNSCC cell lines were previously described (23–28) and grown in DMEM with 10% FBS, 200units/mL penicillin, and 200ug/mL streptomycin. Low serum media (LSM) contained 0.5% FBS. Erlotinib, AZD6244 and ZSTK474 were acquired commercially. IPI-926 was supplied by Infinity Pharmaceuticals Inc. To generated resistant cell lines, cells were continuously cultured in erlotinib (1, 5, 10 and 25μM) or DMSO (control). Erlotinib concentration was increased when cultures proliferated at >50% of controls. Final selection at 50μM erlotinib was completed 3× for 72h allowing regrowth in-between.
Gene silencing
siRNA experiments were completed in serum free media (SFM) using 1μl/ml Dharmafect1 and 100nM siRNA (Thermo). GLI1 silencing was completed using doxycycline (0.5μg/ml) inducible pTRIPZ lentiviral contructs (RHS4696-99636732, Open Biosystems) expressing small hairpin RNA (shRNA). Infection of cells with scramble or GLI1 sequences was conducted per the supplier's instructions.
In vitro Matrigel invasion assay and colony formation
Cells were added to 6-well Matrigel-coated 8μm pore inserts (BD Biosciences) and incubated for 24h. Invasion was quantified as cells/view, 6 fields/insert, repeated twice. Next, invading and non-invading cells were collected and seeded (300 cells/well). Cells were allowed to adhere (6–12h) prior to drug and incubated for 24–72h. Plates were incubated for 7days. Resulting colonies (>50 cells) were fixed with 4% formalin and stained using 0.1% crystal violet.
Sulforhodamine B colorimetric assay (SRB)
Cells (2,500–5,000) were plated in 96-well plates and incubated overnight. Drug was added and plates were incubated for 96h. Cells were fixed with 50μl of 10% TCA at 4°C (30min), washed 5× with dH20, 70μl/well SRB reagent was added, wells were washed 5μ with 1% acetic acid, 200μl/well 10mMTris base was added, and absorbance was measured using a Synergy 2 microplate reader (Bio-Tek).
Time-lapse imaging and cell tracking
Media on cells seeded on a 24-well plate was changed to LSM containing vehicle or drug for 24h before EGF treatment (100ng/mL). Images were taken on a Zeiss Axiovert equipped with an environmental chamber and a Hamamatsu CCD camera. Images were taken every 10–15min over the 48h time course. Acquisition and cell tracking were done using Volocity software (Perkin Elmer). Three points were chosen per well, and ten cells were tracked per point, for a total of 30 tracked cells/condition.
GLI1 promoter luciferase assay
A GLI1 promoter luciferase reporter construct was purchased (SwitchGear Genomics) containing a ~1000bp DNA fragment upstream of GLI1 overlapping the transcribed region by 50bp. Positive, negative and GAPDH promoter controls were used. Cells were seeded (1,500/well in 100μl medium) in white walled 96-well plates and transfected per the manufacturer's instructions. Wells were treated with 1μM erlotinib for 24h then plates were frozen at −80°C for >12h. Plates were thawed and 100μl of luciferase substrate was added. Luminescence was measured on a Synergy 2 micro-plate reader (Bio-Tek).
Human xenograft generation and in vivo studies
Briefly, fresh tumor tissue from HNSCC patients consented at the University of Colorado Hospital in accordance with the protocol approved by the Colorado Multiple Institutional Review Board (COMIRB # 08-0552) were collected. Prepared 3×3×3mm tumor pieces were dipped in Matrigel (BD Biosciences) and inserted into a “pocket” in both hind flanks of nude mice. Upon reaching 1500mm3, tumors were passed to a second colony of animals for therapeutic studies.
We tested cetuximab (acquired commercially), IPI-926, and the combination in six patient cases to generate efficacy data. For each case, we implanted 40 tumors in 20 mice. When tumors reached 200mm3 mice were distributed into 4 groups (at least n=8 tumors per group) and treated: control, cetuximab 40mg/kg 2/week IP, IPI-926 40mg/kg 5/week PO, or cetuximab 40mg/kg 2/week plus IPI-926 40mg/kg 5/week for 4 weeks. Tumor size was evaluated twice weekly using the formula: volume = [length × width2]/2. Six hours after the last drug administration tumors were extracted and portions were flash-frozen and embedded in paraffin. For shRNA studies, 100,000 cells transfected with scrambled or GLI1 constructs were injected SC in the flank of nude mice.
Immunohistochemistry (IHC)
IHC analyses were performed on tissue arrays constructed using a manual Tissue Puncher (Beecher Instruments, Silver Spring, MD). For IHC staining slides were de-paraffinized and re-hydrated in graded concentrations of alcohol by standard techniques before antigen retrieval in citrate buffer pH 6.0 (Dako Corp. Carpinteria, CA) at 105°C for 20min. All staining was done in a Dako Autostainer. Slides were incubated in 3% H2O2 for 10min, followed by primary pS6K antibody (2211, Cell Signaling) and incubated for 60min at RT. Staining was developed by: EnVision+ Dual Link System HRP (Dako) for 30min and substrate-chromogen (DAB+) Solution (Dako) for 7min. Slides were then counterstained with Automated Hematoxylin (Dako) for 5min. The intensity (0,1+,2+,3+) and the percentage (0–100%) of cells positive were interpreted blinded to the case and treatment.
RNA isolation and gene expression analysis
Samples (~30mg) were placed in 300μL of QIAzol, and homogenized using the MP Biomedicals Fast Prep 24. RNA was extracted using Qiagen kits according to the manufacturer's instructions. RNA concentration and quality was measured using the Nanodrop. RNA was reverse-transcribed to cDNA in 20μL reactions using the Verso cDNA Synthesis Kit (Fisher Scientific). Reverse transcription reactions followed the protocol recommendations and were performed using the Verti 96-Well Thermal Cycler (Applied Biosystems). TaqMan primer probes (Applied Biosystems), PCR amplification and probe detection were accomplished using the StepOnePlus Real-Time PCR System (Applied Biosystems). All data are representative of experiments performed at least two times in triplicate.
Western blotting
Cell pellets were lysed in 30–100μl RIPA lysis buffer containing 5μl/ml PMSF. Tissue sample (50mg) portions were thawed in a 4× volume of RIPA Buffer and homogenized using single-use plastic pestles. Protein was measured using the ELx800 absorbance microplate reader (BioTek) according to the manufacturer's instructions. 30ng of protein was loaded per well into NuPage Novex 4–12% Bis-Tris Midi Gel (Invitrogen), transferred using the iBlot Gel Transfer Stack System (Invitrogen) then processed. Primary antibodies were purchased from Cell Signaling Technologies: 2236 Phospho-EGF Receptor (Tyr1068) (1H12) Mouse mAb, 4405 EGF Receptor (15F8) Rabbit mAb, 4060 Phospho-Akt (Ser473) (D9E) XP(R) Rabbit mAb, 4821 Akt (pan) (40D4) Mouse mAb (Biotinylated), 4094 Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® Rabbit mAb (Biotinylated), 9102 p44/42 MAPK (Erk1/2) Antibody, 2534 GLI1 and 4968 Pan-Actin and used as recommended. Secondary anti-rabbit IgG (Immuno Research) was used at a 1:50,000 dilution. The signal was visualized using Immuobilon Western chemiluminescent HRP substrate (Millipore).
Immunocytochemistry (ICC)
Cells grown in chamber wells were fixed in 4% paraformaldehyde for 10min at RT then rinsed with PBST 3× for 5min before permeablizing in 0.2% Triton X-100 for10 min, before blocking in 1% BSA in PBS for 30min at RT with shaking. Cells were incubated with E-cadherin (Cell Signaling 4295) and/or Vimentin (Cell Signaling 9854) for 1.5h at RT at 1:50 and 1:200 dilutions, respectively. Alternatively, cells were incubated with isotype controls, including rabbit IgG conjugated with Alexa fluor 488 (Cell Signaling 2975) and rabbit IgG conjugated with Alexa Fluor 555 (Cell Signaling 3969). Cells were rinsed in PBST 3× for 5min, once with PBS and mounted with Prolong anti-fade with DAPI (Invitrogen). Images were taken on an Olympus IX81 microscope equipped with a Hamamatsu CCD camera. Each experiment (200 cells analyzed) was repeated twice.
Analysis of Vimentin-positive cells by flow cytometry
Cells (1×106) were fixed in 2ml 4% paraformaldehyde in PBS at 4°C for 15min and then 100% cold ethanol was added drop-wise to a final concentration of 80%. Cells were resuspended in permeabilization buffer (0.25% Triton X-100 in PBS) for 30min and washed 2× with cold PBS. Cells were suspended in anti-Vimentin-FITC antibody (Cell Signaling 9454) at 1:50 in PBS+2% FBS, incubated for 1h, and analyzed on a Cyan flow cytometer (Beckman Coulter).
Statistics
Data are represented graphically as the mean±SEM or SD for comparison between groups. P values less than 0.05 were considered significant.
RESULTS
EGFR inhibition up-regulates the HhP
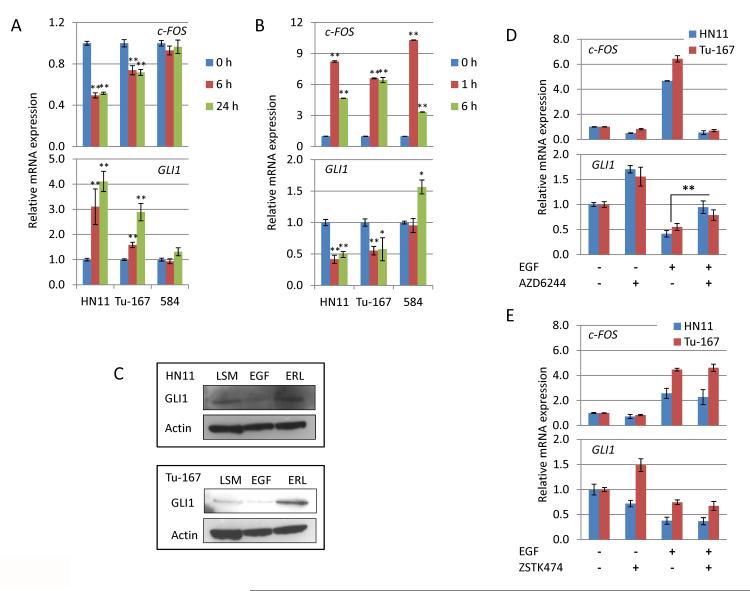
Acquired resistance to EGFR inhibition is common. Previous reports synergistically linked EGFR and HhP through MEK/ERK (9) and we hypothesized that this relationship may provide a mechanism for EGFR resistance in HNSCC. Sensitivity to anti-EGFR therapy was previously correlated with c-FOS suppression in HNSCC cell lines (24). Following 1μM erlotinib treatment, c-FOS levels decreased by 6h and remained suppressed at 24h for the epithelial HN11 and Tu-167 cell lines, while levels remained unchanged in the mesenchymal 584 cells. GLI1 mRNA increased in HN11 and Tu-167 cells at both time points but remained unchanged in 584 cells (Fig. 1A). The induction of GLI1 corresponded with reduced levels c-FOS in HN11 and Tu-167, suggesting that GLI1 expression is only induced after treatment in EGFR-dependent cell lines.
Figure 1. Inhibition of EGFR signaling increases expression of HhP gene in EGFR dependent cells.
A. 1μM erlotinib suppresses c-FOS expression in both HN11 and Tu-167 cells, but not 584 cells, at 6 and 24h. Expression of GLI1 is significantly increased in both HN11 and Tu-167 cells but is unchanged in 584 cells. B. EGFR activation by EGF (100ng/ml) induces c-FOS expression in all cell lines within 1h and significantly suppresses GLI1 levels in HN11 and Tu-167 cells at both 1 and 6h. However, GLI1 levels are increased in 584 cells at 6h after EGF treatment. C. 96h treatment with EGF and erlotinib suppresses and increase GLI1 levels respectively in both HN11 and Tu-167 cells. D and E. Suppression of GLI1 by EGFR activation occurs through the MEK/ERK pathway as the MEK inhibitor AZD6244 blocks suppression of Gli1 by EGF while the PI3K inhibitor ZSTK474 does not. *P<0.05, **P<0.01. All significance was calculated to control unless indicated by a bar between experimental groups.
Given that EGFR inhibition increased GLI1 mRNA in sensitive cells led us to indentify the effects of EGFR activation. Serum starved cells were treated with 100ng/ml EGF, leading to increased levels of c-FOS within 1h. Twenty-four hours after EGFR activation, expression of GLI1 was significantly reduced in HN11 and Tu-167 cells but not in 584 (Fig. 1B). Suppression of GLI1 by EGFR activation was confirmed in HN11 and Tu-167 cells using the EGFR ligands TGF-α and AR (Supplementary Fig. S1A). Using a GLI1 promoter-based luciferase assay we confirmed that the GLI1 promoter was activated by erlotinib while suppressed by EGF in both HN11 and Tu-167 cells with no change in 584 cells (Supplementary Fig. S1B). In contrast to previous reports (8), we found that neither erlotinib nor EGF treatment modulated GLI1 expression in human keratinocytes (Supplementary Fig. S1C and S1D). In HN11 and TU-167 cells, 72h EGF treatment suppressed GLI1 protein levels whereas erlotinib increased GLI1 levels (Fig. 1C). Combined, these results suggest that EGFR signaling down regulates the HhP transcription factor GLI1 in EGFR-dependent HNSCC cells.
EMT and anti-EGFR therapy resistance in HNSCC
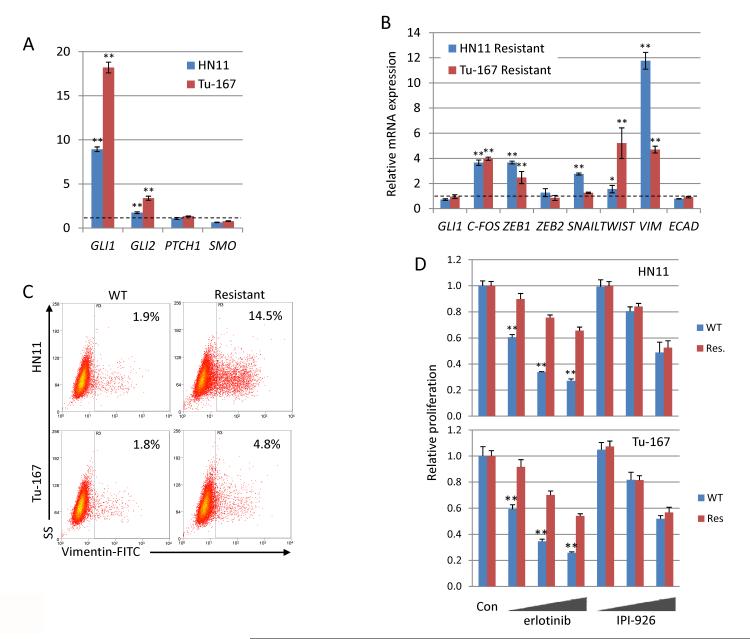
EMT and drug resistance are often related (18, 21), as observed in our HNSCC cell lines (Supplementary Fig. S2). We attempted to establish this connection by generating cell lines with acquired, high-level resistance to erlotinib (≥50μM). HN11 and Tu-167 cells continuously treated with 10μM erlotinib highly expressed the HhP transcription factor GLI1 and, to a lesser extent, GLI2 (Fig. 2A). Erlotinib-resistant (50μM) HN11 and Tu-167 cells released from erlotinib treatment had higher c-FOS, ZEB1, SNAIL, TWIST and VIM expression when compared to parental cells (Fig. 2B), consistent with a mesenchymal phenotype By flow cytometry erlotinib-resistant cells also had significant increases in Vim-positive cell populations (Fig. 2C). Interestingly, cellular morphology did not differ dramatically from controls. GLI1 and GLI2 were upregulated in cells chronically treated with erlotinib; therefore erlotinib-resistant cells were tested for sensitivity to HhP inhibition with the novel smoothened (SMO) inhibitor IPI-926 (29). Erlotinib-resistant cells demonstrated decreased sensitivity to erlotinib compared to wild-type; susceptibility to HhP inhibition was similar between the erlotinib-resistant and wild type cells (Supplementary Fig. S2E and Fig. 2D). Taken together these results suggest that chronic EGFR inhibition generates cells with EMT-like characteristics.
Figure 2. Chronic erlotinib treatment generates resistant cells with a mesenchymal gene expression pattern.
A. Expression of GLI transcription factors are significantly increased in EGFR dependent cells chronically treated with 10μM erlotinib. B. Erlotinib resistant cells highly express pro-EMT genes after removal from selection media. C. Percentage of Vim-positive cells is significantly increased in erlotinib resistant cell lines, measured by flow cytometry D. Erlotinib resistant cells are not sensitized to HhP inhibition by IPI-926. Cells were treated with erlotinib (1,5,10μM) or IPI-926 (1,5,10μM) and proliferation was measured by the SRB assay. Sensitivity to erlotonib decreased in resistant cells while sensitivity to IPI-926 was unchanged. *P<0.05, **P<0.01.
EGFR activation induces EMT and motility in HNSCC cells
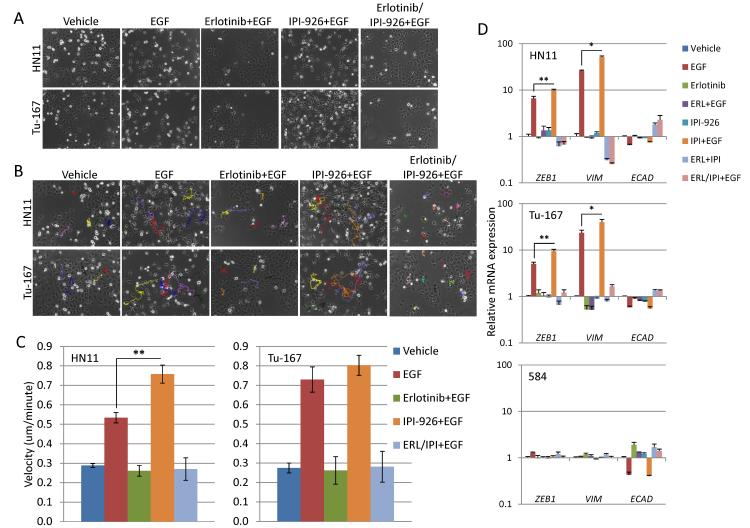
Increased GLI1 expression after EGFR inhibition, as well as the generation of erlotinib resistant cell lines, provided a rationale for studying of the relationship between the EGFR pathway, HhP, and EMT. HNSCC cell lines were treated with erlotinib and IPI-926 alone or in combination, with or without EGFR activation by addition of EGF ligand. As previously reported (19, 20), EGF (100ng/ml) induced an EMT-like state, defined by spindle-like cellular morphology, in all three epithelial cell lines (Fig. 3A) but not in EGFR-independent 584 cells. It is important to note that 500ng/ml SHH had no noticeable effect on the morphology of any cell line (not shown). As expected, erlotinib completely blocked the phenotypic transition but IPI-926 had no effect. EGF induces an EMT-like phenotype, with increased motility that was suppressed by erlotinib on time-lapse track analysis (Fig. 3B). EGF had no effect on motility in 584 cells (not shown). Interestingly, IPI-926 increased cellular velocity in both HN11 (p<0.01) and Tu-167 cells after EGF treatment but this effect was not observed in combination with erlotinib. These results suggest that EGFR activation generates an EMT-like state that is not prevented by HhP inhibition, but that co-inhibition of the EGFR and HhP suppresses EMT.
Figure 3. Inhibition of the hedgehog pathway may augment EGFR driven EMT-like state in EGFR dependent cell lines.
A. EGFR signaling activation by the EGF ligand (100ng/ml) generates an EMT-like phenotype in HN11 and Tu-167 cells that is completely blocked by erlotinib (1μM) but not IPI-926 (1μM). B and C. EGF increases cellular motility in EGFR dependent cells which was blocked by erlotinib but augmented by IPI-926. D. EGF increased expression of pro-EMT genes ZEB1 and VIM as well as suppressed E-cad generating an EMT-like gene expression profile in HN11 and Tu-167 cells but not 584 cells. However, E-CAD expression was suppressed by erlotinib treatment in 584 cells. *P<0.05, **P<0.01.
EGFR signaling increases EMT expression
Next, EMT-related gene expression was measured after EGF treatment with or without erlotinib, IPI-926, or in combination. Expression of the pro-EMT transcription factor ZEB1 was significantly increased by EGF treatment in HN11, Tu-167 and FaDu cells but not 584 cells (Fig. 3D and Supplementary Fig. S4C). In comparison, EGF did not substantially modulate the expression of ZEB2, SNAIL and TWIST1 (not shown). VIM expression followed that of ZEB1 while E-CAD was suppressed by EGFR activation. These effects were reversed by pretreatment with erlotinib while pretreatment with IPI-926 before addition of EGF increased expression of both ZEB1 and VIM when compared to EGF alone. These data suggest that the HhP inhibition might contribute to EGFR-driven EMT.
We used siRNAs to determine if ZEB1 is responsible for increased expression of VIM by EGFR activation. Both siRNAs suppressed expression of ZEB1 after EGF treatment that in turn led to significantly (p<0.01) lowered levels of VIM mRNA (Supplementary Fig. S3A and S3B). However, silencing of ZEB1 did not dramatically change the generation of spindle-like cells by EGF but both siRNAs significantly reduced EGF induced invasion (not shown), indicating that ZEB1 may be a key transcriptional driver of the EGF-induced EMT-like state.
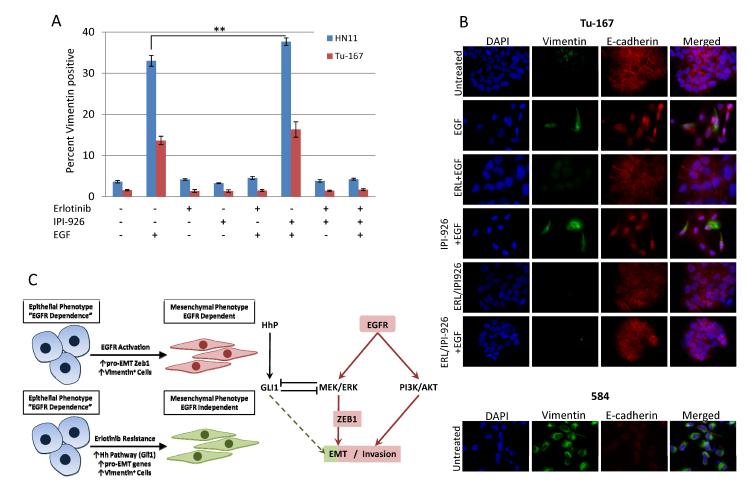
Flow cytometry and ICC confirmed modulation of Vim by EGF at the protein level. In normal culture ~1–3% of HN11 and Tu-167 cells were Vim-positive (Fig. 4 and Supplementary Fig. S4) while 100% of 584 cells are Vim-positive. EGF treatment increased the percentage of Vim-positive cells 7 and 10-fold in Tu-167 and HN11 cells, respectively. Erlotinib completely blocked the EGF-driven increase in Vim-positive cells while HhP inhibition slightly increased the positive population in Tu-167 cells and significantly increased the Vim-positive population (P<0.05) in HN11 cells. This further suggests that HhP signaling may promote EGF-induced EMT (Fig. 4A). ICC results demonstrate that Vim-positive and spindle-like cell populations overlap and HhP inhibition again increases the Vim-positive population after EGFR activation. Finally, 584 cells are 100% Vim-positive and express low levels of E-cad (Fig. 4B and Supplementary Table S1). Similarly, silencing of GLI1 (Supplementary Fig. S5D) slightly increased the Vim-positive population in Tu-167 cells while remaining unchanged in HN11 cells (Supplementary Fig. S6A).
Figure 4. EGFR driven EMT-like state is associated with Vimentin-positive cells.
A. EGF increased the percentage of Vim-positive cells in HN11 and Tu-167 cell lines measured by flow cytometry. Erlotinib blocked this effect while IPI-926 significantly increased the positive population in HN11 cells. B. EGF increased the number of Vim-positive cells and induced cellular dissociation and slight decreases in E-cad. 584 cells are all positive for Vim and express very low levels of E-cad. C. Schemata depicting (Left) EGFR-dependent and -independent EMT and (Right) the relevant pathways downstream of EGFR. *P<0.05, **P<0.01.
We next inquired if EGF could induce an EMT-like state in erlotinib-resistant cells. Unlike in parental cells, 1μM erlotinib did not increase GLI1 in resistant cells but instead appeared to suppress expression in HN11 and Tu-167 cells. However, EGF continued to suppress GLI1 expression slightly in Tu-167 cells (Supplementary Fig. S5A). Induction of ZEB1 and VIM by EGF was nearly completely suppressed in both HN11 and Tu-167 erlotinib resistant cells. However, ZEB1 and VIM continued to be down-regulated by erlotinib while EGF continued to suppress E-CAD.
Increased invasion associated with EGFR-driven EMT
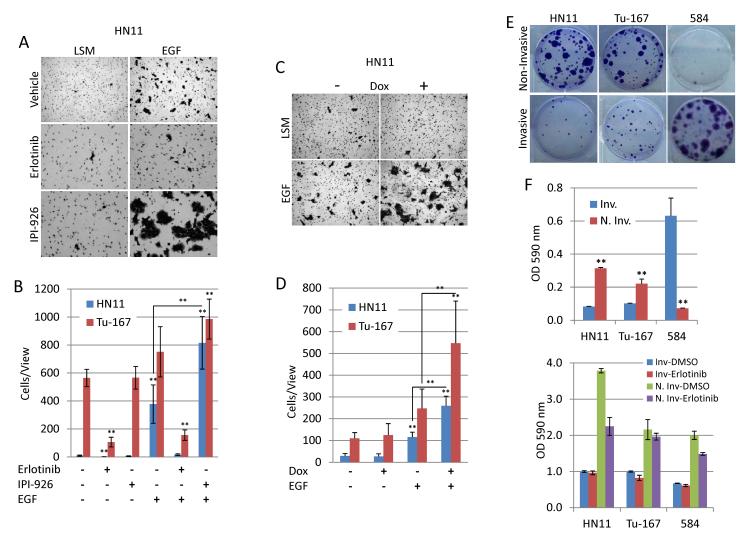
The EGFR induced EMT-like state increased invasion in EGFR-dependent cells. Increased invasion was blocked by erlotinib treatment (Fig. 5A). Consistent with our previous findings, IPI-926 augmented EGFR-driven invasion slightly in Tu-167 cells and significantly (p<0.05) in the HN11 cell line (Fig. 5A). To confirm this was not an unintended effect of IPI-926, independent of HhP inhibition, we tested the invasiveness of HN11 and Tu-167 cells expressing shRNA against GLI1. Similar to IPI-926, silencing of GLI1 significantly (P<0.05) increased the invasiveness of cells treated with saturating levels of EGF in both HN11 and Tu-167 cells. We next questioned whether invasiveness was indicative of highly proliferative state sensitizing cells to erlotinib. Non-invasive cells more readily formed colonies compared to invasive cells in HN11 and Tu-167 while the opposite was true for 584 (Fig. 5E). Finally, invasive EGFR-dependent cells did not demonstrate increased sensitivity to erlotinib but non-invasive HN11 cells had increased sensitivity when compared to their invasive counterparts (Fig. 5F). Combined, these findings indicate that though the EGFR driven EMT-like state increases motility and invasiveness, proliferation is suppressed and cells are not sensitized to EGFR inhibition.
Figure 5. EGF drives invasion in EGFR dependent cells and is augmented by inhibition of the HhP.
A and B. Erlotinib blocks EGF-induced invasion while IPI-926 significantly increases invasion by HN11 cells. C and D. Expression of shRNA against GLI1 increased EGF-induced invasion in HN11 and Tu-167 cells. E and F. Invasive cells are less proliferative than non-invasive cells from the EGFR dependent cell lines HN11 and Tu-167. The opposite is true for the mesenchymal 584 cells. *P<0.05, **P<0.01.
MEK/ERK and PI3K/AKT signaling are required for EGF- induced invasion
We next attempted to clarify what pathways downstream of EGFR regulate this EMT-like event. Prior to dosing with EGF HN11 and Tu-167 cells were treated with 1μM of the MEK/ERK inhibitor AZD6244 or 1μM of the PI3K/AKT inhibitor ZSTK474. Similar to erlotinib, AZD6244 completely blocked phenotypic change and alone induced tightly associated cells and suppressed expression of ZEB1 and VIM. ZSTK474 did not affect the EMT induction, and significantly increased (P<0.05) expression of ZEB1 and VIM while further suppressing E-CAD by EGF. However, unlike control cells that form spindle-like cells and dissociate, cells treated with ZSTK474 formed a distinct phenotype, elongated yet still tightly associated with adjacent cells (Supplementary Fig. S7A and S7B). Previous reports support that MEK/ERK is primarily responsible for generating the mesenchymal phenotype but PI3K/AKT may be crucial to cell-to-cell adhesion and expression of matrix metalloproteinases (MMPs) (19, 30). Inhibition of MEK/ERK or PI3K/AKT signaling effectively blocked EGF induced invasion (Supplementary Fig. S7C and S7D). Taken together these results suggest that EGFR activation in EGFR-dependent cells generates a highly motile and invasive EMT-like state that is augmented by inhibition of the HhP via MEK/ERK signaling.
Inhibition of EGFR and HhP signaling in HNSCC cell lines
The above findings lead us to hypothesize that by inhibiting the HhP we could achieve two objectives: 1) increase EGFR dependence and enhance anti-EGFR therapy efficacy, and 2) prevent the EGFR inhibitor-driven EMT switch. These results also suggested that only dual targeting would achieve this goal. To this end we conducted combination treatment experiments in vitro with erlotinib and IPI-926. IPI-926 has minimal activity in vitro at concentrations of ≤1μM and there was little additive effect with erlotinib. However, 250 μg/ml SHH ligand significantly reduced the anti-proliferative effects of erlotinib (Supplementary Fig. S8), suggesting again that activation of HhP signaling may be a mechanism of resistance for anti-EGFR therapy.
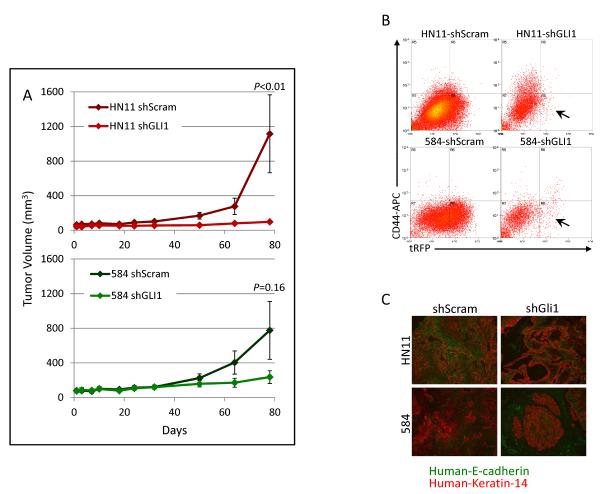
Using in vitro systems we had observed that suppression of the HhP increased EGFR driven motility and invasion. To explore this in a more complex system, tumorigenicity of HNSCC cell lines expressing shGLI1 in immune-compromised mice was measured. Tumor formation was inhibited in both 584-shGli1 (P=0.16) and HN11-shGli1 (P<0.01) (Fig. 6A). Resulting tumors were analyzed by flow cytometry for CD44 and turboRFP, expressed as part of the shRNA insert. While CD44 expression was not altered dramatically, turboRFP expression was all but lost in shGLI1 tumors, suggesting that silencing of GLI1 inhibited tumor formation and the small resulting tumors may have arisen from cells that had lost the viral insert (Fig. 6B). Resulting tumor tissue was stained for E-cad and K14 and tumors expressing scramble sequence consisted primarily of cancer cells with interspersed stromal cells. However, cancer cells in shGli1 tumors were found in smaller pockets within larger areas of mouse stromal cells possibly indicating that silencing GLI1 leads to proliferation inhibition in vivo (Fig. 6C).
Figure 6. Silencing of GLI1 inhibits tumor formation in nude mice.
A. Tumor formation of HN11 and 584 cells expressing shRNA against GLI1 was suppressed when compared to scrambled controls. B. shRNA expressing cells simultaneously express turboRFP, which was nearly completely lost in shGli1 expressing tumors compared to scrambled controls. C. shGli1 expressing tumors contained fewer tumors cells and higher levels of mouse stroma when compared to tumors expressing a scrambled sequence.
Combination cetuximab and IPI-926 treatment in HNSCC patient xenografts
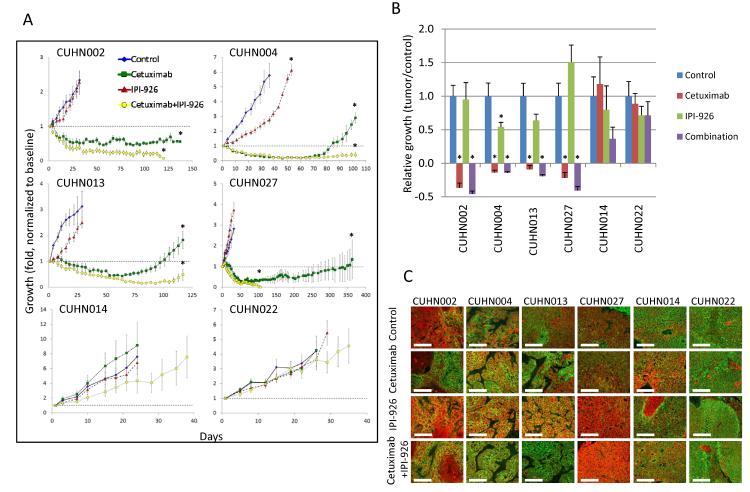
To better mimic the true tumor environment we next tested the efficacy of cetuximab, IPI-926, and the combination using patient-derived HNSCC xenografts. Four of six cases had tumor shrinkage after EGFR inhibition by cetuximab (Fig. 7A and 7B), but none regressed completely and 75% re-grew to baseline during the study. IPI-926 demonstrated modest single-agent efficacy with only moderate growth retardation in one xenograft (CUHN004). IPI-926 did not increase the rate of tumor regression in combination with cetuximab or dramatically decrease the T/C value at the end of treatment (Fig. 7B). However, after being released from therapy the re-growth kinetics for the cetuximab and combination arms of the study diverged dramatically for all four cetuximab sensitive cases. All tumors regressed completely and never re-grew in the combination arm of two cases (CUHN002 and CUHN027), while re-growth was delayed by ~1 month in the other two cases (Supplementary Table S2), suggesting IPI-926 increases tumor control in combination with EGFR inhibition.
Figure 7. Combination treatment of human HNSCC xenografts with cetuximab and IPI-926 blocks tumor re-growth.
A. Growth kinetics of 6 HNSCC xenograft cases treated with cetuximab, IPI-926 and cetuximab+IPI-926. B. Tumor/Control values for treated cases. C. Staining of E-cadherin (green) and Keratin-14 (red) in HNSCC xenografts after treatment. Treatment with IPI-926 decreased E-cadherin, pushing cells into an EGFR-driven EMT-like state. Scaling bars represent 50μm at 40×. *P<0.05.
To unravel the molecular events underlying this dramatic efficacy, gene and protein expression were quantified for genes relevant to the EGFR pathway, HhP and EMT. As expected, expression of c-FOS decreased significantly in sensitive xenografts (CUHN002, CUHN004, CUHN013, CUHN027) after cetuximab treatment while remaining unchanged in resistant xenografts (CUHN014, CUHN022) (Supplementary Fig. S9A and S9B). IPI-926, cetuximab or combination treatment decreased expression of mouse GLI1 in both sensitive and resistant xenografts while human GLI1 was relatively unchanged in sensitive xenografts. GLI1 increased in the combination arm of resistant cases, again confirming that the mouse stroma compartment may be target of HhP inhibition. Also, pro-EMT factors ZEB2 and TWIST1 and SNAIL were found to be up-regulated after cetuximab and combination treatment in sensitive cases while remaining unchanged in the resistant group. Increased expression of pro-EMT transcription factors was observed for the cetuximab sensitive xenografts and resembled the expression profile of erlotinib-resistant HNSCC cell lines. These findings suggest that after a one week PD study tumor cells may have undergone an EMT or selected for cells in an EMT state; these are likely the cells responsible for the re-population of the tumor and thus need to be specifically targeted in order to achieve a durable antitumor effect.
Protein analysis revealed that cetuximab reduced pEGFR in all cases except CUHN027, with a further reduction of total and pEGFR in combination therapy. Total EGFR was also reduced in CUHN004 and interestingly in the resistant CUHN014 and CUHN022 cases, suggesting that intrinsic dependence rather than the pharmacodynamic effect (that is necessary but not sufficient) determines susceptibility to EGFR inhibitors. Downstream assessment was more informative; pMAPK and pAKT were suppressed in all sensitive cases treated with cetuximab or in combination. IPI-926 increased pMAPK in all sensitive cases, pAKT in both CUHN004 and CUHN027 xenografts and pEGFR in CUHN004 (Supplementary Fig. S9C). Also, IPI-926 treatment increased pS6K in both CUHN004 and CUHN013 xenografts. Similarly, pS6K were suppressed in both cetuximab and combination treatment arms in sensitive xenografts while remaining unchanged in resistant cases by IHC (Supplementary Fig. S9D). Finally, IPI-926 decreased E-CAD in cetuximab sensitive cases following our findings in vitro suggesting that HhP inhibition allows cells to enter an EGFR-dependent EMT-like state (Fig. 7C).
These results suggest that cetuximab significantly suppresses MEK/ERK and PI3K/AKT signaling in turn reducing expression of the downstream factor c-FOS in sensitive xenografts. Also, the hedgehog and EGFR signaling pathways may negatively regulate each other indicated by increased levels ofpEGFR, pMAPK and pAKT and pS6K after IPI-926 treatment, pushing tumor cells into a pro-EGFR state. Again, this confirms that while HhP inhibition is ineffective as a single agent, it can push tumor cells further into an EGFR-dependent state, leading to increased killing by anti-EGFR therapy resulting in tumor obliteration.
DISCUSSION
Preventing intrinsic and acquired resistance to EGFR inhibition with cetuximab wouldbe of benefit to patients suffering from HNSCC (2, 3), so we attempted to define the basis of EGFR resistance. Contrary to previous studies using keratinocytes (8), we identified increased expression of the HhP transcription factor GLI1 after EGFR inhibition as a unique characteristic of EGFR-dependent HNSCC cells. GLI1 has been demonstrated to be a key driver of tumor growth and metastasis in multiple cancers (31, 32). As previously described (9) we found that “crosstalk” between the EGFR and hedgehog pathways occurs through the MEK/ERK cascade, but that HhP inhibition may make cells more EGFR dependent.
Chronic erlotinib treatment in HN11 and Tu-167 cells dramatically increased GLI1 expression compared to parental controls and may signify the upregulation of a resistance pathway. Erlotinib-resistant cells were next generated to determine whether the HhP was a key driver of survival and proliferation when EGFR signaling is suppressed. Both strains had characteristics indicating that they had undergone an EMT event including erlotinib resistance, increased expression of pro-EMT factors ZEB1, ZEB2, TWIST and SNAIL, and in Vim-positive cell populations. Similarly generated gefitinib resistant HNSCC cells lines had also undergone an EMT event (21). However, resistant cells no longer over-expressed GLI1, indicating upregulation only occurred during the transition to the mesenchymal state, as has previously been reported (13, 33). These results confirm that EMT is at least an attribute, if not a direct mechanism, of drug resistance. Combined, these results generated a strong rationale for combination therapy inhibiting the EGFR and hedgehog pathways in xenograft models.
Next, we defined how activation and inhibition of the EGFR and hedgehog pathways, alone, and in combination, regulate signaling and may increase sensitivity to EGFR inhibition in HNSCC cell lines. From these studies, and as observed above, we established that there are two distinct mechanisms of EMT in HNSCC cells. As previously described in epithelial cancer cell lines (19, 20), activation of the EGFR pathway generates an EMT-like state in EGFR dependent HNSCC cells. This “early” EMT-like event is defined by increased motility and mesenchymal morphology with increased expression of EMT signature genes and a Vim-positive population leading to invasiveness. Interestingly, this phenotype does not increase sensitivity or resistance to EGFR or HhP inhibition. Though this EMT-like state does not lead to drug resistance, it does grant cancer cells invasive potential, and is a potentially valuable in vitro model of tumor cell migration and invasion. Also, inhibition of the HhP, both pharmacologically and genetically, augmented the EGFR driven phenotype by increasing activation of both MEK/ERK and PI3K/AKT. On the contrary, HhP activation by SHH increased resistance to erlotinib, confirming that the HhP negatively regulates EGFR signaling, whereas inhibiting HhP signaling allows cells to enter a state of greater EGFR dependence. Again, these findings support the rationale for combined inhibition of EGFR and HhP signaling as a means of enhancing anti-EGFR efficacy. We characterized two distinct EMT-like events in HNSCC cells, both EGFR-dependent and -independent and that both states are mutually exclusive as seen in 584 and erlotinib resistant cells. Finally, HhP signaling may help initiate EGFR-independent EMT while suppressing the EGFR driven EMT-like state.
The use of combination therapy in vitro was relatively uninformative, and our experiments highlighted the risks of artificial, growth-factor saturated models. To clarify whether HhP signaling was a genuine target in HNSCC in vivo GLI1 was silenced by shRNA in two cell lines and found GLI1 suppression inhibited tumor formation. Subsequently, and in an optimal utilization of an advanced patient-derived animal model, direct HNSCC patient xenografts were treated with cetuximab and IPI-926 alone and in combination. Cetuximab treatment induced tumor shrinkage in four of six cases but IPI-926 demonstrated little response in this in vivo model. However, cetuximab treated xenografts re-grew to baseline after the end of treatment while combination therapy with IPI-926 delayed re-growth by a month in two cases and all tumors were ablated in the other two xenografts. As expected, cetuximab blocked signaling through MEK/ERK and/or PI3K/AKT and combination treatment enhanced this effect while IPI-926 increased pMAPK, pAKT and pS6K in EGFR dependent cases. It is important to note that IPI-926 also targets the stromal compartment as mouse GLI1 was suppressed after treatment and was previously observed in multiple carcinoma models (34, 35). However, our results do not rule out inhibition of the HhP in tumor cells, given that GLI1 shRNA abrogated tumor growth in vivo, an effect that cannot be explained by stromal effects.
These findings support the hypothesis that inhibition of HhP signaling pushes HNSCC cells into an even more EGFR-dependent state, sensitizing them to anti-EGFR therapy while blocking a relevant resistance mechanism. As for the efficacy of the novel therapeutic IPI-296, these findings support the use of a more complex animal model that better mimics the human environment. These provoking results provided the rationale for the clinical translation of this novel combinatory paradigm (NCT01255800).
Supplementary Material
Acknowledgements
The authors would like to thank Karen McGovern, Jeff Kutock and Veronica Campell of Infinity pharmaceuticals for constructive manuscript discussions and supplying the IPI-926 compound for this study.
Grant Support Supported by National Institutes of Health grants 1R01CA149456 (A.J.) and R21DE019712 (A.J.). The Vora Family Foundation provided support (to A.J.) for equipment purchasing and molecular testing. The Janet Mordecai Family Foundation gave support (to A.J.) for molecular testing. Infinity Pharmaceuticals provided financial support (to A.J.) for the animal and molecular studies related to the investigational compound
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. The New England journal of medicine. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 3.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. The New England journal of medicine. 2008;359:1116–27. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–56. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benavente S, Huang S, Armstrong EA, Chi A, Hsu KT, Wheeler DL, et al. Establishment and characterization of a model of acquired resistance to epidermal growth factor receptor targeting agents in human cancer cells. Clin Cancer Res. 2009;15:1585–92. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–98. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C, et al. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–92. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isohata N, Aoyagi K, Mabuchi T, Daiko H, Fukaya M, Ohta H, et al. Hedgehog and epithelial-mesenchymal transition signaling in normal and malignant epithelial cells of the esophagus. Int J Cancer. 2009;125:1212–21. doi: 10.1002/ijc.24400. [DOI] [PubMed] [Google Scholar]
- 11.Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada M, Miyabayashi K, et al. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol Carcinog. 2009;48:703–12. doi: 10.1002/mc.20516. [DOI] [PubMed] [Google Scholar]
- 12.Dormoy V, Danilin S, Lindner V, Thomas L, Rothhut S, Coquard C, et al. The sonic hedgehog signaling pathway is reactivated in human renal cell carcinoma and plays orchestral role in tumor growth. Mol Cancer. 2009;8:123. doi: 10.1186/1476-4598-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta H, Aoyagi K, Fukaya M, Danjoh I, Ohta A, Isohata N, et al. Cross talk between hedgehog and epithelial-mesenchymal transition pathways in gastric pit cells and in diffuse-type gastric cancers. Br J Cancer. 2009;100:389–98. doi: 10.1038/sj.bjc.6604846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, et al. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelialmesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–76. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr S, Thomson S, Buck E, Russo S, Petti F, Sujka-Kwok I, et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin Exp Metastasis. 2008;25:685–93. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 18.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan Y, Shi C, Inge L, Hibner M, Balducci J, Huang Y. Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene. 2010;29:4947–58. doi: 10.1038/onc.2010.240. [DOI] [PubMed] [Google Scholar]
- 20.Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng GQ, et al. Activation of EGFR promotes squamous carcinoma SCC10A cell migration and invasion via inducing EMT-like phenotype change and MMP-9-mediated degradation of E-cadherin. J Cell Biochem. 2011;112:2508–17. doi: 10.1002/jcb.23175. [DOI] [PubMed] [Google Scholar]
- 21.Maseki S, Ijichi K, Tanaka H, Fujii M, Hasegawa Y, Ogawa T, et al. Acquisition of EMT phenotype in the gefitinib-resistant cells of a head and neck squamous cell carcinoma cell line through Akt/GSK-3beta/snail signalling pathway. Br J Cancer. 2012;106:1196–204. doi: 10.1038/bjc.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yonesaka K, Zejnullahu K, Lindeman N, Homes AJ, Jackman DM, Zhao F, et al. Autocrine production of amphiregulin predicts sensitivity to both gefitinib and cetuximab in EGFR wild-type cancers. Clin Cancer Res. 2008;14:6963–73. doi: 10.1158/1078-0432.CCR-08-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimeno A, Kulesza P, Kincaid E, Bouaroud N, Chan A, Forastiere A, et al. C-fos assessment as a marker of anti-epidermal growth factor receptor effect. Cancer Res. 2006;66:2385–90. doi: 10.1158/0008-5472.CAN-05-2882. [DOI] [PubMed] [Google Scholar]
- 25.Amador ML, Oppenheimer D, Perea S, Maitra A, Cusatis G, Iacobuzio-Donahue C, et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004;64:9139–43. doi: 10.1158/0008-5472.CAN-04-1036. [DOI] [PubMed] [Google Scholar]
- 26.Frederick MJ, Holton PR, Hudson M, Wang M, Clayman GL. Expression of apoptosis-related genes in human head and neck squamous cell carcinomas undergoing p53-mediated programmed cell death. Clin Cancer Res. 1999;5:361–9. [PubMed] [Google Scholar]
- 27.Frederick BA, Helfrich BA, Coldren CD, Zheng D, Chan D, Bunn PA, Jr., et al. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol Cancer Ther. 2007;6:1683–91. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- 28.Rangan SR. A new human cell line (FaDu) from a hypopharyngeal carcinoma. Cancer. 1972;29:117–21. doi: 10.1002/1097-0142(197201)29:1<117::aid-cncr2820290119>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 29.Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, et al. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926) J Med Chem. 2009;52:4400–18. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- 30.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–33. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 31.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–6. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–72. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Deng W, Lobo-Ruppert SM, Ruppert JM. Gli1 acts through Snail and E-cadherin to promote nuclear signaling by beta-catenin. Oncogene. 2007;26:4489–98. doi: 10.1038/sj.onc.1210241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCann CK, Growdon WB, Kulkarni-Datar K, Curley MD, Friel AM, Proctor JL, et al. Inhibition of Hedgehog signaling antagonizes serous ovarian cancer growth in a primary xenograft model. PLoS One. 2011;6:e28077. doi: 10.1371/journal.pone.0028077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.