Abstract
MicroRNAs (miRNAs) are a class of short noncoding RNAs which are endogenously expressed in many organisms and regulate gene expression by binding to messenger RNA (mRNA). MiRNAs are either produced from their independent transcription units in intergenic regions or lie in intragenic regions. Intragenic miRNAs and their host mRNAs are produced from the same transcript by the microprocessor and the spliceosome protein complex respectively. The details and exact timing of the processing events have implications to downstream RNA interference (RNAi) efficiency and mRNA stability. Here we engineer and study in mammalian cells a range of synthetic intragenic miRNAs co-expressed with their host genes. Furthermore, we study transcripts which carry the target of the miRNA, thereby emulating a common regulation mechanism. We perform fluorescence microscopy and flow cytometry to characterize the engineered transcripts and investigate the properties of the underlying biological processes. Our results shed additional light on miRNA and pre-mRNA processing but importantly provide insight towards engineering transcripts customized for combined delivery and use in synthetic gene circuits.
Introduction
MicroRNAs function as a fundamental layer of post-transcriptional regulation of gene expression by binding to approximately 20 nucleotide long1, partially complementary, microRNA target sites typically present in the 3’ untranslated region (UTR) of a target mRNA2, 3. The RNA-induced silencing complex (RISC) facilitates miRNA binding and leads to translation inhibition or mRNA degradation and cleavage4, 5. Mapping of miRNA locations in the genome has revealed the presence of miRNA in either intergenic or intragenic regions6–8. Intergenic miRNAs are autonomous transcription units while intragenic miRNAs are transcribed together with protein coding genes7, 9, 10 and can be physically located in the intron, exon, or UTR of the host gene11. Intragenic miRNAs interface with the cellular environment by regulating several genes simultaneously. Notably, these genes are often important to the immediate interaction network of the host gene6. The regulation of the host mRNA by the microRNA can be indirect (e.g. by targeting a transcription factor of the host transcript12) or direct, forming an incoherent feedforward loop architecture13.
Today, there are open questions regarding the timing and efficiency of the splicing of the introns by the spliceosome, the processing of the miRNA by the microprocessor, and the associated implications for the miRNA function and mRNA stability 14, 15. Possible scenarios16 for the miRNA microprocessor and spliceosome activities include that the spliceosome and microprocessor work independent of each other, that the spliceosome and microprocessor work together and have a cooperative relationship, and finally that the spliceosome and microprocessor mutually inhibit with each other. Experimental results17–20 suggest a mutually cooperative, physical, and functional coupling of intronic microRNA biogenesis and splicing at the host intron, but importantly warrant additional synthetic biology-inspired experimentation towards rational engineering of intragenic transcripts. Here we use custom intragenic miRNAs as a method for combined gene and functional miRNA/RNA delivery. Specifically, we engineer and study in mammalian cells a range of synthetic intragenic miRNAs co-expressed with their host genes. Our long-term objective is to harness the properties of these transcripts for custom combined delivery21 and implementation of complex circuits13, 22.
Results and Discussion
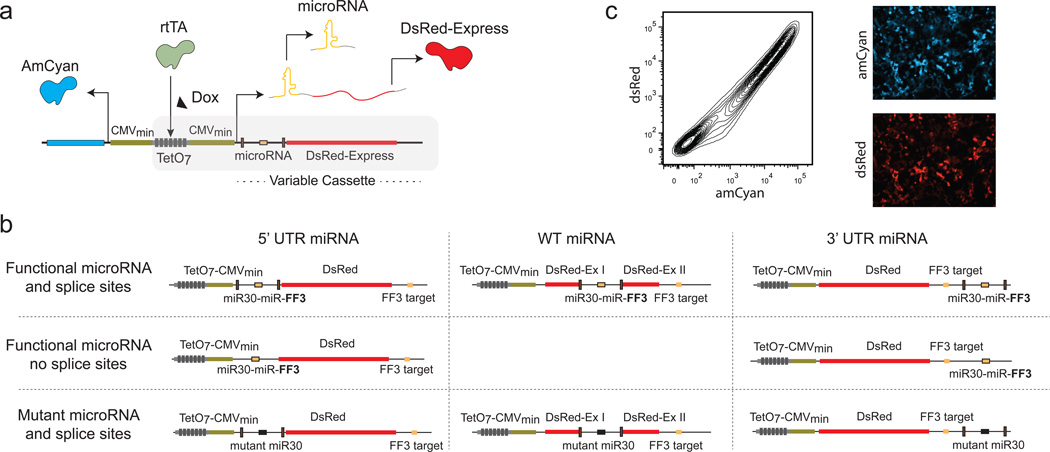
To probe the intragenic miRNA biogenesis and pre-mRNA splicing crosstalk, and towards combined delivery of miRNA and protein, we engineered a range of constructs that contain a synthetic miRNA (namely miRNA-FF313, 23, 24) and an mRNA (with and without the miRNA target) coding for the fluorescent protein dsRedExpress (we will refer to it as dsRed). The intragenic miRNA and host mRNA (dsRed) transcript is controlled by an inducible bidirectional promoter which divergently transcribes an amCyan fluorescent protein (Figure 1a). The amCyan serves as a control for transfection efficiency.
Figure 1. Architectures and representative results.
(a) A schematic representation of the backbone plasmid. The plasmid consists of a bidirectional promoter flanked by diverging minimal CMV promoters. We use rtTA-expressing HEK293 TET-On cells with saturating Dox concentration. (b) The miRNA is placed at the 3’UTR, 5’ UTR (with and without splice sites) and in an intron between two dsRed exons while keeping the microRNA target at the 3’UTR. Constructs are arranged according to the position of the miRNA (functional and mutant) in the dsRed mRNA. Eight additional plasmids were constructed without the miRNA target at the 3’UTR. (c) Representative microscopy and flow cytometry data of a control plasmid (without miRNA target).
We placed the miRNA-FF3 in three locations: a region flanking gene coding exons (defined as wild-type transcript) and in the UTR regions (defined as 3’ and 5’ transcripts) of the dsRed gene, with the FF3 target at the 3’UTR. Most introns have clear demarcations, which are detected by the spliceosome and trigger the splicing process. A typical intron includes the splice donor and acceptor sites, and a pyrimidine rich region. The splice donor site is usually a conserved GU sequence at the 5’ end of the intron. Intron terminates with an almost invariant AG sequence that is called the splice acceptor site. Upstream of the splice acceptor site, there is a pyrimidine rich region25.
We engineered synthetic splice sites to flank a synthetic miRNA. Furthermore, we built constructs with UTR miRNAs without splice sites to nullify the effect of the spliceosome and study the independent effect of the microprocessor on the mRNA transcript. Finally, we designed control transcripts with mutant miRNAs to abolish the effect of the microprocessor.
As illustrated in Figure 1b, we generated eight constructs: wild-type (miRNA intron between protein coding exons), 3’ UTR intron region with splice sites, 5’ UTR intron region with splice sites, 3’ UTR region without splice sites, wild type intronic region with mutant miRNA and splice sites, 3’UTR region with mutant miRNA and splice sites, 5’UTR region with mutant miRNA and splice sites, 5’ UTR region without splice sites, and their eight counterparts lacking the miRNA target (Supplementary Material, Constructs).
As a backbone for the transcripts, we used the plasmid pTRE-Tight-Bi (Clontech) which contains a bidirectional pTRE-Tight promoter consisting of seven rtTA binding sites flanked by diverging minimal CMV promoters (CMVMIN) and dsRed and amCyan on either side of the promoter. We use rtTA-expressing HEK293 TET-On cells with saturating Doxycycline (Dox) concentration, effectively turning the pTRE-Tight into a constitutive promoter (Supplementary Figure 1). We transiently transfected HEK293 TetOn cells using Lipofectamine LTX and assayed the output 48 hours post-transfection using microscopy and flow cytometry (FACS). A typical experiment using the fully induced promoter is illustrated in Figure 1c, where we present a flow cytometry scatter plot of amCyan versus dsRed fluoroscence and representative microscopy images of each channel.
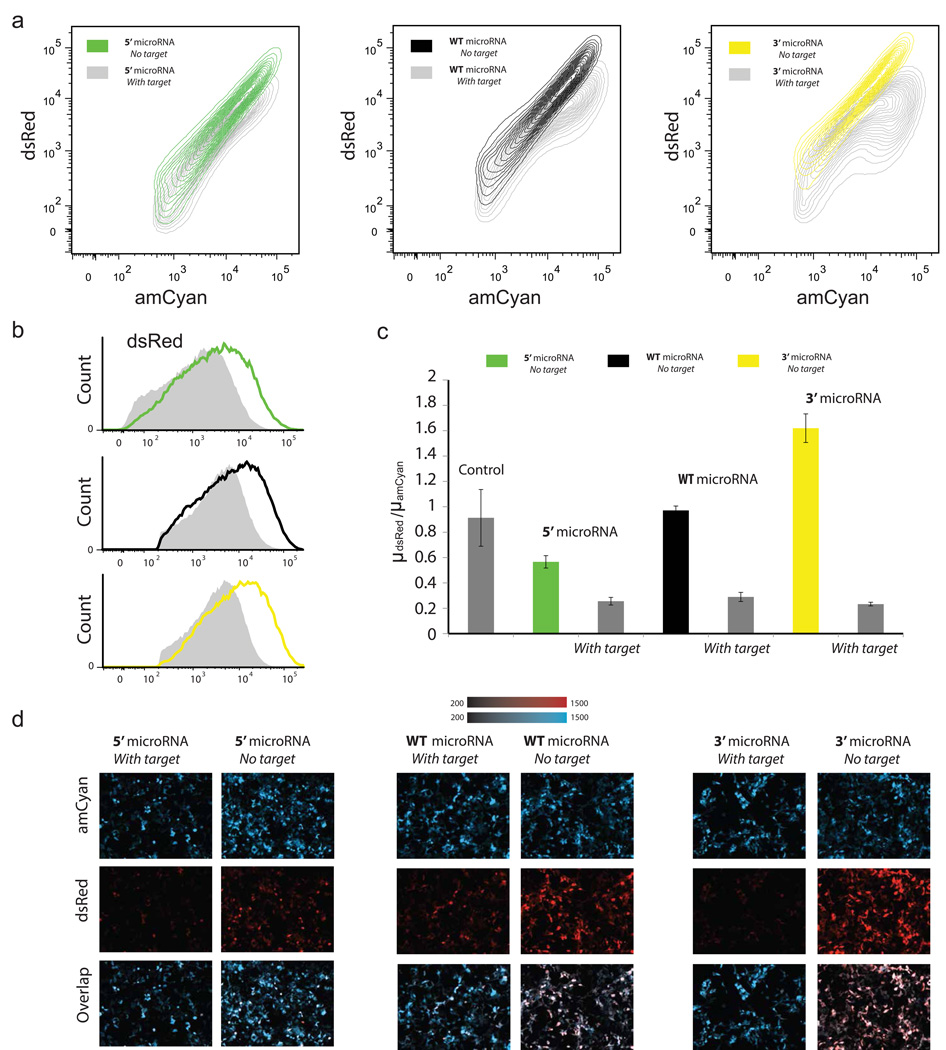
We first examined the expression of the host mRNA when we permutated the synthetic miRNA with the artificial splice sites (Figure 1b, functional miRNA and splice sites), with and without the miRNA target in the 3’UTR. The flow cytometry results (Figure 2a) for the three locations of the miRNA, show the dsRed protein is produced efficiently for all cases. Judging by the repression observed from the transcripts that carry the microRNA target (Figure 2a and 2b) we can conclude the microRNAs are also processed correctly for all cases. If we quantify the flow cytometry results and calculate the mean of the dsRed populations normalized by the amCyan mean (thereby correcting for the transfection efficiency) we observe an increase in expression correlated with the distance from transcription start site (Figure 2c). In particular, the normalized expression of dsRed for the 3’UTR case is higher than the intronic and both are higher than the 5’UTR case. When we examine the transcripts with miRNA target (Figure 2c), we observe in all cases the microRNAs reduce the dsRed level to approximately the same level. In Figure 2d, we provide representative microscopy snapshots of the amCyan and dsRed for all cases.
Figure 2. Functional miRNAs and splice sites.
(a) Density contour plots of the constructs with and without target. (b) Overlaid flow cytometry histogram of dsRed with (gray) and without (color) target for the three constructs. (c) Flow cytometry. Y axis is the ratio of the mean of dsRed over the mean of amCyan. Control is plasmid without miRNA. (d) Representative microscopy images of the 5’ UTR miRNA circuit, wild-type miRNA circuit and 3’UTR miRNA circuit (with and without miRNA target).
From this set of initial experiments we arrive to a number of observations. If the spliceosome and microprocessor work exclusively together as a complex and assuming it takes the same time for the complex to splice and produce the functional miRNA for all miRNA locations, then one would expect (approximately) the same expression level for all three constructs—a hypothesis refuted by the experiments. The increase in the yield (or efficiency) as the location of the miRNA insert changes from the 5’ to the 3’ UTR is possibly the result of a combined processing of the transcript by the spliceosome and microprocessor complex and stand-alone microprocessor units. Compared to spliceosome which is a mega complex26, the microprocessor is essentially a two protein complex 14, 27; therefore it is conceivable that the spliceosome may inhibit the microprocessor function but not vice versa. We can also assume that the spliceosome-microprocessor complex requires more time to process an intron carrying miRNA versus processing a non-miRNA coding intron. Considering the above, one potential explanation for the observed differential expression is that single microprocessor units remove the microRNA prior to processing by the spliceosome-microprocessor complex.
To shed additional light to the process, we introduced introns with mutated miRNA into the untranslated regions and between the dsRed exons. To mutate the miRNA, the 22nt sequence involved in the stem loop formation was deleted and a random sequence was inserted which does not form a stem loop (Supplement Material). Comparing the expression of dsRed (Figure 3a and Figure 3b), we observe similar expression for the wild-type and 3’UTR transcripts. Interestingly the 5’UTR miRNA mutant results in low dsRed expression compared with 5’UTR miRNA without mutations (Figure 2c and Figure 3c), which suggests the function of the microprocessor assists the spliceosome in the 5’ modifications. Again, in Figure 3d, we provide representative microscopy snapshots of the amCyan and dsRed for all cases.
Figure 3. Mutant microRNAs and splice sites.
(a) Density contour plots of the constructs with and without target. (b) Overlaid flow cytometry histogram of dsRed with (gray) and without (color) target for the three constructs. (c) Flow cytometry data. Y axis is the ratio of the mean of dsRed over the mean of amCyan. Control is plasmid without miRNA. (d) Representative microscopy images of the 5’ UTR miRNA circuit, wild-type miRNA circuit and 3’UTR miRNA circuit (with and without miRNA target).
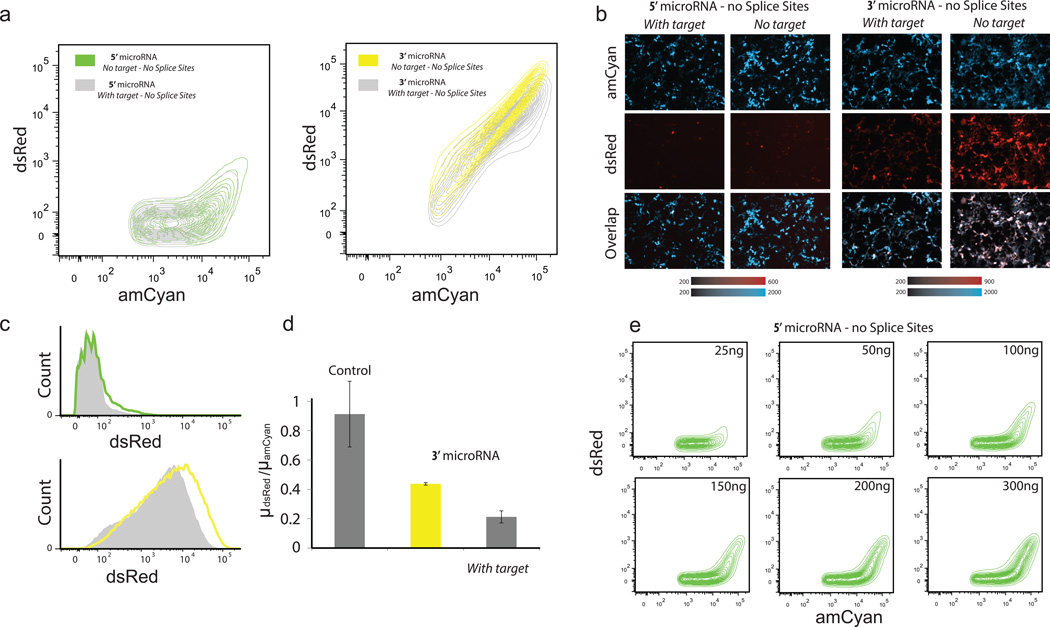
For all cases with splice sites there is strong expression of dsRed protein and the miRNA is always functional (Figure 2c), which suggests that when the spliceosome operates on splice sites there is always microprocessing of the miRNAs. To establish that the microprocessor can work independently of the spliceosome, we removed the splice signals from the microRNA intron of the 3’ and 5’ UTR with splice sites constructs. The flow cytometry data (Figure 4a) and representative microscopy snapshots (Figure 4b) show that for both cases there is production of dsRed but the behavior of the two transcripts is considerably different. More specifically, the dsRed expression is weak for the 5’UTR transcript (Figure 4c). Furthermore, we observe that microRNA is processed correctly and efficiently in both transcripts, as judged by the transcripts with the miRNA target (Figure 4a and 4c), and control experiments where we cotransfected the 5’UTR transcript with a reporter protein carrying the miRNA target (Supplementary Figure 2).
Figure 4. Functional microRNAs without splice sites.
(a) Density contour plots of the constructs (with and without miRNA target). (b) Representative microscopy images of the 5’ UTR miRNA circuit and 3’UTR miRNA circuit (with and without miRNA target). (c) Overlaid flow cytometry histogram of dsRed with (gray) and without (color) target for the two constructs. (d) Flow cytometry data. Y axis is the ratio of the mean of dsRed over the mean of amCyan. Control is plasmid without miRNA. (e) Titration of the concentration of the plasmid carrying the 5’UTR miRNA.
For both 5’ and 3’ UTR cases, the microprocessor efficiently crops the miRNA. This confirms that the microprocessor can work independently from the spliceosome. We observe low level of dsRed for the 5'UTR transcript without splice sites. Our hypothesis is that the microprocessor acts on the transcript and cleaves the miRNA before the end of transcription and the majority of the transcripts are consequently degraded. In the case of 3’ UTR without splice sites, the microprocessor cleaves the microRNA shortly before the end of the transcription and the transcripts are transferred immediately for further processing (3’ polyadenylation) salvaging the dsRed expression. Furthermore, we observe (Figure 4a) that in the 5’UTR case there is dsRed expression only for high amCyan levels (which indicates high copy-number transfected cells). We hypothesize that when the transcribed mRNA units exceed the free microprocessor copies then the transcripts can escape the microprocessor processing and are transferred to the cytoplasm with the microRNA as part of their UTR. In order to further examine the microprocessor saturation hypothesis we transfected variable amounts of the 5’UTR transcript, and indeed we observe (Figure 4e) that for low concentrations the vast majority of the cells show negligible dsRed expression levels which gradually increases as we increase the concentration of the transfected plasmid.
To confirm that the low expression of dsRed is due to cropping of the microRNA by the microprocessor we performed a control experiment, where we inserted the mutant microRNA without splice sites at the 5’ UTR. The expression of this construct is significantly higher than the 5’ microRNA (Supplement Figure 3) which suggests that the stability of the mRNA is not compromised exclusively by the sequence insertion. Therefore, the above experimental evidence suggests that the 5’UTR transcript without splice sites operates as a thresholding device, which can be used to efficiently delivery functional miRNAs but restrict the protein production only to the high-copy transfected cells.
Conclusion
We use synthetic biology to study intragenic miRNAs as a means for combined gene and functional miRNA delivery. We engineered a range of transcripts carrying synthetic miRNAs co-expressed with their host genes. Our experiments confirm a synergistic relationship between the spliceosome and microprocessor complex and provide proof that the microprocessor units operate independently from the spliceosome. From a synthetic biology perspective, our results show that the transcripts with the microRNA intron between protein coding exons result to intermediate expression levels while the 3’UTR with splice sites transcript yields the most efficient combined microRNA and protein delivery. Furthermore, the insertion of microRNAs in the 5’UTR without splice sites can be used to restrict protein delivery only to high-copy transfected cells while the intragenic miRNA remains functional for all transfected cells. In summary, our results highlight the complex and dynamic interplay between the RNA processing events and point to opportunities for rational engineering of these transcripts for custom combined delivery and use in sophisticated circuits.
Methods
Recombinant DNA cloning
The restriction enzymes, Taq polymerase, and T4 Ligase enzyme used for cloning and ligation were obtained from NEB. QIAGEN Plasmid isolation, gel extraction and PCR purification kits were used. For transformation, competent DH5α cells (originally obtained from Life Technologies) were prepared using the standard CaCl2 method of competent cell preparation. Bacterial culture media and agar (BD Biosciences) and prepared according to manufacturer’s instructions. Primers for all the experiments were designed using A Plasmid Editor (Ape – version 1.17) and synthesized from IDT and Sigma. The primers received were diluted into stocks of 100pmol/µl. Sequencing was performed by Genewiz.
Cell Culture
HEK293 TET-On Advanced cell line (Clontech, Cat-No#630931) was used in all the experiments. These cells are derived from human embryonic kidney cells. The cells were grown at 37°C, 100% humidity and 5% CO2, in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Cat-No#11965-118) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Cat-No#26140-079), 0.1mM non-essential amino acids (Invitrogen, Cat-No#11140-050), 0.045ug/mL of Penicillin and 0.045ug/mL of Streptomycin antibiotics (Penicillin-Streptomycin liquid, Invitrogen, Cat-No#15140-122) and sterilized using a filter (VWR, Cat-No#28199-778). The adherent culture was harvested from this medium by trypsinizing with Trypsin-EDTA (0.25% Trypsin with EDTA, Invitrogen Cat-No#25200-114) and diluting in a fresh medium upon reaching 50–90% confluence. In the Tetracycline inducible expression (HEK293 TET-ON) system, the gene expression is turned on when Doxycycline (Dox, a derivative of tetracycline) is added to the culture medium. The critical component of the TET-ON system is the regulatory protein rtTA (reverse tetracycline controlled transactivator).
Transfection and data collection
140,000 cells in 1mL of complete medium were plated into each well of 12 plastic plates (Grenier Bio-one, Cat-No# 665180) and grown for 24 hours. Lipofectamine LTX (Invitrogen, Cat-No# 15338-100) was used for transient transfection according to the manufacturer’s protocol. In short, the transfection mix was prepared by adding 200ng of plasmids in 200µL of DMEM for each well. 1.5µL of Lipofectamine LTX was added to the mix and incubated at room temperature for 30mins. The transfection mix was then applied to the plates and mixed by gentle shaking. Dox induction was performed after 2 hours. The cells were incubated for about 48 hours before analysis.
Measurements
All the microscopy data was collected using an Olympus motorized inverted research microscope (IX81). All microscope images were taken from live cells grown in 12 well plates (CellStar, Cat.-No. 665180) in the transfection medium supplemented with 10% FBS. The microscope is equipped with an environmental chamber (Weather station) held at 37°C during measurements. The images were collected by a Hamamatsu camera at 10X objective. The filter sets are as follows: 430/25× (excitation) and 470/30m (emission) for amCyan, and 565/55× (excitation) and 650/70 (emission) for dsRed. The software used to analyze the microscopy data is Slidebook version 5.0. All the flow cytometry data was collected from a BD LSRFortessa cell analyzer. The cells were prepared for FACS analysis by trypsinizing each well with 0.1 mL 0.25% trypsin-EDTA, adding 0.5ml DMEM media with FBS to neutralize the trypsin, collecting the cell suspension and centrifuging at 4000 rpm for 2 min. Media was removed and the pellet re-suspended by short vortexing in 0.5 mL PBS buffer (Cellgro,Cat-No#45000-432). AmCyan was measured with a 200nm laser and a 450/50 emission filter, dsRed-monomer with a 300nm laser and a 575/26 emission filter. The data from the FACS is analyzed using a software package called FlowJo.
Supplementary Material
Acknowledgements
This work was funded by the US National Institutes of Health (NIH) grant GM096271, the US National Science Foundation (NSF) grant CBNET-1105524, and the University of Texas at Dallas. We would like to thank the members of the lab for discussions and feedback.
Footnotes
Competing financial interests
The author declares no competing financial interests.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13(8):1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′ UTR evolution. Cell. 2005;123(6):1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Hinske L, Galante P, Kuo W, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts' interactome. BMC Genomics. 2010;11(1):533. doi: 10.1186/1471-2164-11-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ying Shao-Yao, Lin Shi-Lung. Current perspectives in intronic micro RNAs (miRNAs) J Biomed Sci. 2006;13(1):5–15. doi: 10.1007/s11373-005-9036-8. 01. [DOI] [PubMed] [Google Scholar]
- 8.He C, Li Z, Chen P, Huang H, Hurst LD, Chen J. Young intragenic miRNAs are less coexpressed with host genes than old ones: Implications of miRNA–host gene coevolution. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nature Structural & Molecular Biology. 2008;15(9):902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R. Deep annotation of drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21(2):203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SC, Tang P, Lin WC. Intronic microRNA: Discovery and biological implications. DNA Cell Biol. 2007;26(4):195–207. doi: 10.1089/dna.2006.0558. [DOI] [PubMed] [Google Scholar]
- 13.Bleris L, Xie Z, Glass D, Adadey A, Sontag E, Benenson Y. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Molecular Systems Biology. 2011;7(1) doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory RI, Yan K, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 15.Volk N, Shomron N. Versatility of MicroRNA biogenesis. PloS One. 2011;6(5):e19391. doi: 10.1371/journal.pone.0019391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shomron N, Levy C. MicroRNA-biogenesis and pre-mRNA splicing crosstalk. Journal of Biomedicine and Biotechnology. 2009;2009 doi: 10.1155/2009/594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataoka N, Fujita M, Ohno M. Functional association of the microprocessor complex with the spliceosome. Mol Cell Biol. 2009;29(12):3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA, Fisher DE, Shomron N, Levy C, Novina CD. Feed-forward microprocessing and splicing activities at a MicroRNA–Containing intron. PLoS Genetics. 2011;7(10):e1002330. doi: 10.1371/journal.pgen.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26(3):775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182(1):61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130(2):363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 22.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science Signalling. 2011;333(6047):1307. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 23.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nature Nanotechnology. 2010;5(9):666–670. doi: 10.1038/nnano.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. MicroRNA circuits for transcriptional logic. Methods in Molecular Biology (Clifton, NJ) 2012;813:169. doi: 10.1007/978-1-61779-412-4_10. [DOI] [PubMed] [Google Scholar]
- 25.Clancy S. RNA splicing: Introns, exons and spliceosome. Nature Education. 2008;1(1) [Google Scholar]
- 26.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419(6903):182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 27.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.