Abstract
Purpose
The identification of gene mutations and structural genomic aberrations that are critically involved in CLL pathogenesis is still evolving. One may postulate that genomic driver lesions with effects on CLL cell proliferation, apoptosis thresholds or chemotherapy resistance should increase in frequency over time when measured sequentially in a large CLL cohort.
Experimental Design
We sequentially sampled a large well-characterized CLL cohort at a mean of 4 years between samplings and measured acquired copy number aberrations (aCNA) and LOH using SNP 6.0 array profiling and the mutational state of TP53, NOTCH1 and SF3B1 using Sanger sequencing. The paired analysis included 156 patients, of whom 114 remained untreated and 42 received intercurrent therapies, predominantly potent chemo-immunotherapy, during the sampling interval.
Results
We identify a strong effect of intercurrent therapies on the frequency of acquisition of aCNAs in CLL. Importantly, the spectrum of acquired genomic changes was largely similar in patients that did or did not receive intercurrent therapies; therefore, various genomic changes that become part of the dominant clones are often already present in CLL cell populations prior to therapy. Further, we provide evidence that therapy of CLL with pre-existing TP53 mutations results in outgrowth of genomically very complex clones which dominate at relapse.
Conclusions
Using complementary technologies directed at the detection of genomic events that are present in substantial proportions of the clinically relevant CLL disease bulk, we capture aspects of genomic evolution in CLL over time, including increases in the frequency of genomic complexity, specific recurrent aCNAs and TP53 mutations.
Keywords: CLL, genomic copy number aberrations, mutations
INTRODUCTION
Chronic lymphocytic leukemia is a heterogeneous disease caused by differences in biology and manifested in varied clinical presentations(1, 2). Given the important clinical consequences of CLL disease heterogeneity, including implications for patient counseling, therapy selections and therapy development, substantial efforts have been directed at deriving a molecular and cellular understanding of this disease variety. Such efforts have identified molecular traits that associate with initial disease stability or progression, the response durations to standard up-front therapies and ultimately, overall survival. Most of these efforts have been directed at the characterization of CLL at diagnosis(3–11).
However, CLL is phenotypically an adaptive neoplasm that originates in monoclonal B-cell lymphocytosis and ultimately degenerates in a substantial fraction of patients into drug-resistant or intervention-resistant disease(12). Understanding the determinants or drivers of CLL cell accumulation before and after therapy, the molecular characteristics of CLL in relapse, the frequency and drivers of CLL clonal evolution and devolution and therapy resistance is of importance and will facilitate development of better risk-adapted counseling and therapy approaches.
With the goal of identifying drivers of CLL pathogenesis, investigators have compared the frequencies of selected molecular characteristics in relapsed or refractory CLL patient cohorts with frequencies in unrelated CLL cohorts analyzed at diagnosis, providing hypothesis-generating findings about mechanisms of CLL disease progression and aggressive disease behavior. Such efforts have implicated TP53 mutations/del17p in acquired therapy resistance and have generated a multitude of other hypothesis-generating findings about the involvement of specific factors in CLL disease aggressiveness (13).
However, the comparative analysis of the incidence of molecular aberrations in unrelated CLL cohorts assayed at different disease stages alone is insufficient to implicate specific events in CLL progression or aggressiveness, as many molecular abnormalities are enriched in relapsed CLL and as various biases are unaccounted for. To counteract some of these hidden biases, longitudinal analysis of CLL cohorts assayed repeatedly over substantial time spans is needed. Such paired longitudinal analysis has uncovered the acquisition of selected genomic aberrations as detected through clinical FISH testing in CLL patients over time as well as acquisition of novel aCNA and LOH in CLL when assayed longitudinally on high-resolution SNP array platforms(14–19). However, despite the importance of these studies for our understanding of CLL genomic evolution, FISH testing substantially underestimates genomic aberration loads in CLL, and SNP array-based analysis would benefit from simultaneous measurements of gene mutations to allow for more complete estimates of genomic clonal evolution in CLL.
Here, we report a multi-dimensional genomic analysis of 156 paired CLL specimens procured a mean of approximately 4 years apart, including a SNP 6.0 array-based genome-wide analysis of aCNA, LOH, copy-neutral LOH (cn-LOH) and exon resequencing of TP53, NOTCH1 and SF3B1. Through these analyses, we identify a strong effect of intercurrent therapy and of TP53 mutations on clonal evolution and genomic degeneration in CLL. Further, we identify recurrent genomic changes that increased in frequency during CLL progression and exclude others, like mutations in NOTCH1 or SF3B1 that had previously been implicated in CLL chemotherapy resistance or relapse. We demonstrate that the type and spectrum of longitudinally acquired genomic changes in CLL is similar in treated and untreated patients, providing strong evidence for a therapy-independent origin. Finally, we provide evidence that the dominant CLL clones in individual patients that had undergone genomic evolution at longitudinal analysis can be related back in all informative cases to the dominant antecedent clone. In aggregate, these data have implications for our biological understanding of CLL as a disease, the evolution of the CLL genome, and the timing and selection of therapies in CLL.
METHODS
Patients
Between January 2005 and January 2010, 271 patients evaluated at the University of Michigan Comprehensive Cancer Center were enrolled onto this study. As specified in the protocol, patients were resampled following initial enrollment. The trial was approved by the University of Michigan Institutional Review Board (IRBMED #2004-0962) and written informed consent was obtained from all patients prior to enrollment. Data from 259 of these 271 patients are reported here (5 patients enrolled on the study were excluded due to a diagnosis that was not CLL, and 7 patients had insufficient cryopreserved cells available for the analyses described). Data from 156 paired samples (enrollment and one longitudinal sample) are included in this report; of these, 27 samples were assayed at two separate longitudinal time points. Of the 156 samples analyzed longitudinally, 134 were from previously untreated patients at trial enrollment while 22 samples were procured from patients relapsed at trial enrollment.
Regardless of whether the subjects were originally diagnosed at our institution or another, we used the same CLL diagnostic criteria, based on the National Cancer Institute-Working Group Guidelines for CLL(20). Eligible patients needed to have an absolute lymphocytosis (greater than 5000 mature lymphocytes per μl), and lymphocytes needed to express CD19, CD23, sIg (weak) and CD5 in the absence of other pan-T-cell markers.
CLL treatment was defined as cytotoxic chemotherapy and/or monoclonal antibody therapy for CLL. Clinical information, including Rai stage and all treatments given, was collected on all patients. Patient samples were characterized for selected CLL-associated chromosomal aberrations on the day of trial enrollment as a routine clinical test at the Mayo Clinic (Rochester, MN) using FISH (CLL-FISH).
Cell Isolation
Flow cytometry sorting of CLL specimens
Cryopreserved PBMCs (frozen after Ficoll-gradient purification) from CLL blood specimens were prepared for FACS sorting into CD19+ and CD3+ cells as previously described(6).
Preparation of Sample DNA
DNA used for SNP 6.0 profiling was extracted from FACS-sorted CD19+ and CD3+ cells as described. For eleven cases, paired buccal DNA was used instead of CD3+ cell-derived DNA(7).
Array Data Analysis
The DNA was prepared for hybridization to SNP 6.0 arrays according to the manufacturer's recommendations. Purification of PCR products was performed using the precipitation/centrifugation method. Affymetrix CEL files for each sample were analyzed using Genotyping Console software for initial quality control, followed by use of the Affymetrix “Birdseed” algorithm to generate tab-delimited SNP call files in text format. Call rates for the longitudinal group of samples included in this report were between 95.33% and 99.68%, with a mean and median call rate of 98.67% and 98.92%, respectively; none of the tumor DNA samples gave out-of-bounds results.
Sample copy number heatmap displays were obtained from CEL files through use of the freely available software dChip(21), adapted to run on a 64-bit computer environment. To generate displays of LOH, a two-step, internally developed, Java-based software analysis system was employed. The Pre-LOH Unification Tool (PLUT) served to align all individual patient SNP calls to their respective dbSNP rsID numbers and genomic physical positions prior to incorporation into the LOH tool version 2, an updated version of the LOH tool able to accommodate Affy SNP 6.0 array data(22).
For genomic copy number analysis, two observers visually inspected parallel heatmap copy number images of CD19+ cell-derived and paired normal CD3+ cell-derived DNA samples generated through dChip and using the median smoothening functionality. Only those copy number changes detected in CD19+ cell-derived DNA that were not found at the same position in paired normal CD3+ cell-derived DNA were called somatic. Lesions had to be at least 30 SNP positions in length to be scored positive (see Supplementary Table 1).
For LOH analysis between paired samples, a filter setting within the LOH tool version 2 was employed, allowing visualization of individual paired SNP calls as LOH only if present within 3000 base pairs of another such call. This step filtered out many false, sporadically distributed single LOH calls due to platform noise. Further, LOH calls for at least three closely spaced SNPs were required to make an LOH call in any particular genomic region. SNP 6.0 array data files for all patient samples analyzed will be deposited in the GEO public database (accession number GSEXXX).
Genomic losses and gains were also independently nominated using a published algorithmic lesion calling approach(23). This algorithm was developed to be highly specific but slightly less sensitive than visual approaches, thus avoiding lesion overcalling that is common with unsupervised algorithmic approaches. Overall, visual and algorithmic approaches demonstrated excellent agreement in lesions called. The discordant calls between visual and algorithmic calling approaches were operationally resolved according to the following rules: i) if visual loss positive, reconfirmed positive, and algorithmic negative, and the ratio of the mean copy number estimates as determined through dChip for each individual tumor DNA lesion divided by the mean copy number estimates in paired CD3+ cell-derived DNA is less than 0.8, the call is positive; ii) if visual gain positive, reconfirmed positive, and algorithmic negative, and the ratio of the mean copy number estimates for each lesion in tumor DNA divided by the mean copy number estimates in paired CD3+ cell-derived DNA is greater than 1.33, the call is positive; and iii) if visual gain negative, reconfirmed negative, and algorithmic positive, and no overt array artifacts were present in either CD3+ or CD19+ samples, and the ratio of the mean copy number estimates for each lesion in tumor DNA divided by the mean copy number estimates in paired CD3+ cell-derived DNA is greater than 1.33, the call is positive.
The methods for algorithmic lesion calling
The method for algorithmic lesion calling of paired SNP 6.0 array data has been published(23).
Exon resequencing of TP53, NOTCH1 and SF3B1
Primers to amplify and sequence exons 2–10 of human TP53, exon 34 of NOTCH1 and exons 13–17 of SF3B1 and adjacent intronic sequences were designed using the primer 3 program (http://frodo.wi.mit.edu/primer3/) and sequence information was generated using direct sequencing as described(24). Mutations were confirmed to be somatic using unamplified FACS-sorted CD19+ cell-derived DNA and paired patient CD3+ cell-derived DNA as templates.
RESULTS
Patient Characteristics
Characteristics of the 156 CLL patients that were subjected to longitudinal genomic analysis in this study are summarized in Table 1. Characteristics of the 103 CLL patients that were not subjected to longitudinal genomic analysis are summarized in Supplementary Table 2. As is evident from review of Table S2, most patients had died prior to repeat sampling, had either low ALC following therapy or were not followed at this center after enrollment. Also evident is a higher-risk profile (biomarker-based) of these patients, suggesting that estimates of clonal genomic evolution events in this report are underestimates of the true incidence of this phenomenon in CLL. Both observations are intrinsic to longitudinal cohort analysis.
Table 1.
Characteristics of 156 CLL patients analyzed in longitudinal genomic analysis.
| Patient Characteristics | No intercurrent therapy no. (%) | Intercurrent therapy no. (%) |
|---|---|---|
| Sample size N= 156 patients | 114 (73%) | 42 (27%) |
| Age at enrollment, years | ||
| Median | 62 | 63 |
| Range | 40 – 90 | 39 – 84 |
| Gender | ||
| Female | 43 (38%) | 12 (29%) |
| Male | 71 (62%) | 30 (71%) |
| Rai stage at enrollment | ||
| Low, 0 | 64 (56%) | 8 (19%) |
| Intermediate, I–II | 50 (44%) | 29 (69%) |
| High, III–IV | 0 (0%) | 5 (12%) |
| NOTCH1 exon 34 mutations at enrollment | ||
| Wild-type | 106 (93%) | 35 (83%) |
| Mutated | 8 (7%) | 7 (17%) |
| SF3B1 exons 13–17 mutations at enrollment | ||
| Wild-type | 111 (97%) | 36 (86%) |
| Mutated | 2 (2%) | 6 (14%) |
| SF3B1 data not available | 1 (1%) | 0 (0%) |
| P53 exons 2–10 mutations at enrollment | ||
| Wild-type | 104 (91%) | 36 (86%) |
| Mutations within coding exons | 10 (9%) | 6 (14%) |
| Prioritized interphase FISH-25* | ||
| 17p deletion | 7 (6%) | 4 (10%) |
| 11q deletion | 5 (4%) | 5 (12%) |
| Trisomy 12 | 15 (13%) | 6 (14%) |
| Normal karyotype | 29 (25%) | 9 (21%) |
| 13q deletion (sole abnormality) | 52 (46%) | 17 (40%) |
| Ig translocation | 2 (2%) | 0 (0%) |
| FISH data not available | 4 (4%) | 1 (2%) |
| IgVH mutational status | ||
| Unmutated (≥ 98% homology to germline) | 40 (35%) | 25 (60%) |
| Mutated (< 98% homology to germline) | 68 (60%) | 16 (38%) |
| Not Evaluable | 6 (5%) | 1 (2%) |
| ZAP-70 expression | ||
| Positive (> 20%) | 35 (31%) | 30 (71%) |
| Negative (≤ 20%) | 77 (68%) | 11 (26%) |
| ZAP-70 data not available | 2 (2%) | 1 (2%) |
| Treatment status at enrollment | ||
| Not treated | 105 (92%) | 29 (69%) |
| Treated | 9 (8%) | 13 (31%) |
| Number of prior therapies | ||
| 0 | 105 (92%) | 29 (69%) |
| 1 | 7 (6%) | 9 (21%) |
| 2 | 2 (2%) | 2 (5%) |
| >2 | 0 (0%) | 2 (5%) |
FISH findings in ≥25% of nuclei. Order of prioritization: 17p > 11q > trisomy 12 > 13q > Ig translocations > normal FISH
Of the 156 CLL patients subjected to longitudinal genomic analysis, 105 (67%) were untreated at first sampling and remained untreated at longitudinal sampling. Twenty-nine patients (19%) untreated at first sampling required therapy and had relapsed at longitudinal sampling. Finally, of 22 patients already in relapse at first sampling, 9 (6%) did not receive additional therapy prior to longitudinal sampling, while 13 (8%) did receive additional therapy and were again in relapse at longitudinal sampling. Therefore, 42/156 (27%) of paired samples were from patients that received therapy between sampling events. The median and mean times between samplings were 1403 and 1392 days, respectively (range 240 – 2741 days). The types of therapies received between samplings are listed in Table 2, with many of the patients receiving potent chemoimmunotherapies.
Table 2.
Listing of therapies received by patients between sample procurements.
| CLL ID | Intercurrent therapy | Clonal evolution (CE) or stability? |
|---|---|---|
| CLL002 | R → FCR | CE (+aCNA) |
| CLL004 | FR | stable |
| CLL007 | FR | CE (+aCNA, −aCNA) |
| CLL008 | FR | CE (+aCNA) |
| CLL010 | FR | CE (+aCNA) |
| CLL011 | FR | CE (+aCNA) |
| CLL013 | R-CVP | stable |
| CLL017 | FR → FCR | stable |
| CLL021 | R-CVP | stable |
| CLL024 | FR | CE (+aCNA) |
| CLL027 | FR | stable |
| CLL029 | FR | stable |
| CLL033 | FR → PCR | CE (+aCNA) |
| CLL036 | FR | CE (+cnLOH) |
| CLL056 | R | stable |
| CLL060 | PCR | stable |
| CLL066 | R → PCR | stable |
| CLL069 | R → R | stable |
| CLL074 | FR | CE (+aCNA) |
| CLL078 | FR | stable |
| CLL084 | R → R+MP→O | stable |
| CLL085 | FCR | stable |
| CLL094 | A → BR → O | stable |
| CLL104 | F → A | CE (+aCNA, −aCNA) |
| CLL109 | FR | stable |
| CLL112 | FR | stable |
| CLL116 | FR | CE (+cnLOH) |
| CLL117 | FCR → BR | CE (+aCNA, +TP53 mut, +cnLOH, −NOTCH1 mut) |
| CLL120 | FR | stable |
| CLL121 | FCR | CE (+aCNA) |
| CLL125 | F → B | CE(+TP53 mut) |
| CLL135 | FR | stable |
| CLL155 | FCR | stable |
| CLL170 | PCR → flavopiridol → RICE | stable |
| CLL176 | FCR | stable |
| CLL207 | R → R | stable |
| CLL209 | PCR | CE (+aCNA) |
| CLL212 | BR | stable |
| CLL219 | R-CVP → B | CE (+TP53 mut) |
| CLL239 | R | CE (+aCNA) |
| CLL242 | F → FC → BR | stable |
| CLL271 | B | stable |
F: fludarabine; C: cyclophosphamide; R: rituximab; A: alemtuzumab; O: ofatumumab; V: vincristine; P: prednisone; P: pentostatin; M: methylprednisolone; I: ifosphamide; E: etoposide.
Recurrent acquisition of del13q14 type II, del11q, copy-neutral LOH at chromosome 13, del17p and TP53 mutations during clonal evolution in CLL
We proceeded with a detailed review of all dominant clonal changes in paired samples in this large CLL cohort. Of the 156 CLL patients subjected to longitudinal genomic analysis, 29 (19%) developed changes in aCNA, cn-LOH, TP53 mutations or (rarely) NOTCH1 mutations, while 127 (81%) did not demonstrate changes.
Overall, clonal evolution and acquisition involving aCNAs was detected in twenty-one of these cases, novel cn-LOH developed in six cases, TP53 mutations were acquired in five cases, clonal devolution of individual aCNAs occurred in two cases and a NOTCH1 mutation was gained or lost once. No changes in the status of SF3B1 were detected. These acquired aCNAs and gene mutations and the genomic aberrations present at enrollment together with the treatment status have been systematically tabulated in Supplementary Table 1.
Within the group of 21 CLL that underwent longitudinal changes in aCNA in paired samples, we identified 79 instances of acquisitions of novel aCNAs (see Supplementary Table 1), overall indicating a substantial increase in genomic complexity. Of these novel aCNAs, many were non-recurrent and therefore individually of unclear pathobiological relevance.
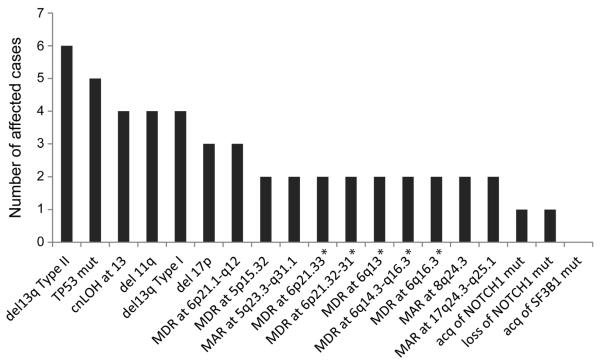
The most frequent recurrently acquired aCNAs were: del13q14 type II, which include RB1 (4%; 6/156), classical 11q deletions spanning ATM (2.5%; 4/156), cn-LOH at chromosome 13 (2.5%; 4/156), del17p spanning TP53 (2%; 3/156 representing 2 classical and 1 micro-deletion), and TP53 mutations (3%; 5/156) (25–28). Therefore, aCNAs/cn-LOH or gene mutations previously implicated through either CLL-FISH or CLL SNP array profiling in chemotherapy resistance, or, genomic complexity, or shorter survival are preferentially acquired during CLL disease progression, therefore providing evidence for their direct involvement as drivers of CLL evolution. The frequency of these changes have been graphically displayed together with other recurrent but infrequent minimally altered regions (MARs) in Figure 1 and have been tabulated in Supplementary Table 3.
Figure 1.
Frequency of individual acquired CNA/LOH, cn-LOH or selected gene mutations in 29 CLL patients with clonal changes in paired analysis: Indicated is the absolute number of events per genomic lesions. Asterisks indicate that the same two CLL cases (CLL # 74 and 104) carried these lesions. MDR: Minimally deleted region. MAR: Minimally altered region (gain and loss).
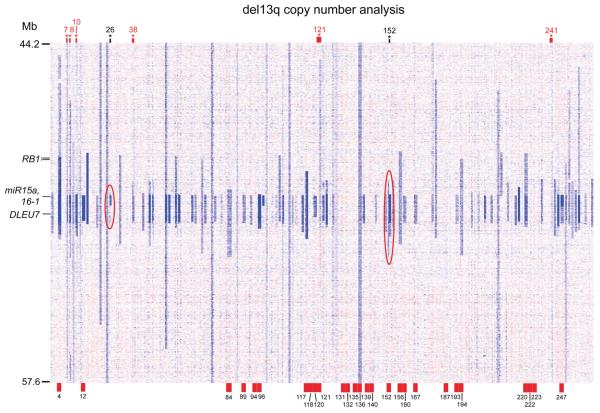
Finally, a heatmap of the genomic copy number analysis of 13q14 deletions in paired samples is presented in Figure 2, in summary, demonstrating stability for most patients and acquisition or change of 13q14 deletions in some patients as indicated.
Figure 2.
Longitudinal copy number analysis of chromosome 13 in 156 CLL patients: Copy number heatmap display generated from dChip software. Patients are oriented vertically. Each patient is ordered by first normal DNA, then study enrollment date tumor DNA, then longitudinal tumor DNA. Multiple longitudinal dates were analyzed for many patients. Relevant genes within the commonly deleted 13q14 region are marked. The RB1 locus delineates type II (loss lesions interrupt RB1) from type I (loss regions do not include RB1) lesions. Red case numbers at top of display indicate patients who acquired a del13q14 type II at longitudinal analysis. Black case numbers at top indicate patients whose del13q14 loss was smaller and homozygous at longitudinal measurement than at enrollment. These are highlighted by the red ovals. Red tick marks at bottom, with accompanying case numbers, indicate patients with more than one longitudinal measurement.
Acquisition of cn-LOH at chromosome 13 during disease evolution in CLL
Previous work had identified cn-LOH as an infrequent genomic event in CLL(25, 28). The identified cn-LOH were largely restricted to chromosome 13 and 17p, with the former associated with short (~0.8 – 1 Mb), homozygous 13q14 deletions centered at approximately 50 Mb, and the latter associated with homozygous TP53 mutations(29). The peculiar anatomy of cn-LOH at chromosome 13 suggested that attainment of homozygous genomic loss in the center of these cn-LOH-chr.13 events, possibly coupled with gene dosage effects within regions of chromosomal reduplication, was a pathological driver in CLL.
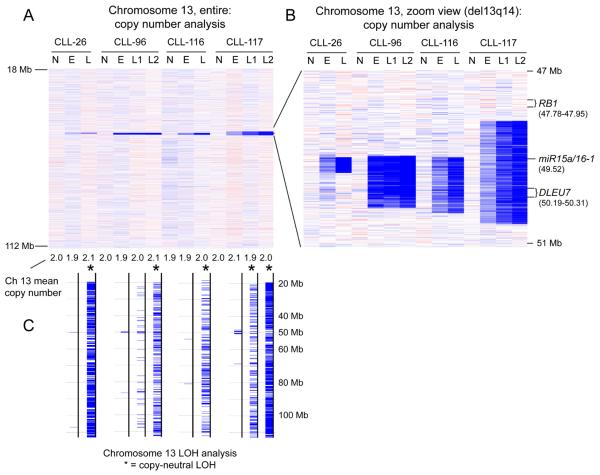
In this cohort we identified acquisition of four novel cn-LOH events on chromosome 13 (see Figure 3). These were not invariably related to intercurrent therapy, as two of these four events occurred in patients (CLL # 026 and 96) that did not receive therapy between samplings (although in CLL # 026, the emergence of cn-LOH-chr.13 co-incided with acquisition of a del17p, which likely is a co-dominant driver). Therefore, cn-LOH-chr.13 constitutes a novel candidate genomic lesion type implicated in CLL progression.
Figure 3.
The acquisition of copy neutral LOH (cn-LOH) at 13q14 in CLL in longitiudiunal analysis. Panels A and B: copy number heatmap displays generated using dChip software. Each column represents one patient. N: normal DNA. E: enrollment date tumor DNA. L: longitudinal date tumor DNA. Panel A: chromosome 13 copy number heatmap overview. Panel B: zoom view of the 13q14 region commonly deleted in CLL. Panel C: LOH analysis. cn-LOH across the entire length of chromosome 13 is indicated by the presence of many hash marks and by the black asterisks.
Lack of association of mutations in NOTCH1 or SF3B1 with clonal dominance in CLL
As part of this study we ascertained mutation frequencies in the three most commonly mutated genes in CLL: these are TP53, NOTCH1 and SF3B1. In the entire cohort of 259 CLL patients, of which 78% were untreated and 22% were relapsed at trial enrollment, the mutation frequencies of NOTCH1 and SF3B1 at trial enrollment were 9% and 6%, respectively. Interestingly, we detected only one acquired NOTCH1 mutation in longitudinal follow-up (CLL # 195—the mutation was also subclonal). We also identified the loss of a NOTCH1 mutation in the setting of acquisition o f a TP53 mutation in another patient (CLL # 117). We detected no change in SF3B1 mutation status in any of the patients analyzed. Therefore, this data fully support the important conclusion that NOTCH1 and SF3B1 mutations do not confer strong selective growth advantages on CLL cells and are not preferentially associated with CLL relapse or CLL clonal dominance.
The emergence of novel dominant CLL clones is facilitated by CLL-directed therapies but the spectrum of genomic changes is independent of therapy
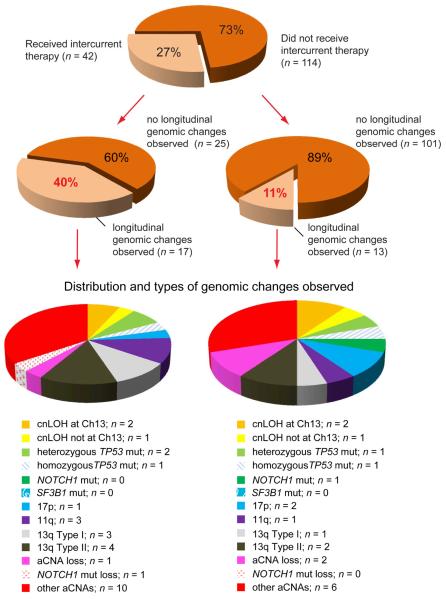
Categorizing the incidence of dominant clonal changes by intercurrent treatment status (yes/no) uncovered substantial differences. Of 105 patients that never received therapy, only 10% (11/105) demonstrated a genomic clonal change, and if all patients that did not receive therapy between samplings were assessed (N=114), the frequency was 11% (12/114). In contrast, the frequency of dominant genomic clonal changes in the twenty-nine CLL patients that were untreated at enrollment and received therapies between samplings was 48% (14/29), and if all patients that received therapies between samplings were included, the frequency was 40% (17/42). Therefore, one major conclusion from this data is that therapy frequently facilitates the emergence of dominant CLL clones that are linearly related to (based on shared identical aCNA or gene mutations) but partially genomically distinct from the dominant presentation clone and that subsequently constitute the clinically relevant disease at relapse.
Next, we compared the types of genomic changes detected in patients after receiving intercurrent therapy with the types detected in patients that did not receive therapy between samplings. The surprising finding was that the spectrum of genomic changes was largely similar (see Figure 4). To account for this, it should first be postulated that a CLL clone which rises to dominance, given time and the absence of any therapy, must therefore be out-competing the clones observed at presentation. If treated patients experience a similar spectrum of genomic changes, then in aggregate, these data support the conclusion that such therapies are facilitating the outgrowth of clones that are destined to become dominant by removing their competition.
Figure 4.
The distribution and types of genomic changes in longitudinal analysis organized by intercurrent therapy status (schema). Left: cases which received intercurrent therapy. Right: cases which did not receive intercurrent therapy.
TP53 mutations are never lost, often gained and associated with substantially increased genomic complexity in CLL following therapy
TP53 mutations were detected in 16 patients out of 156 patients at enrollment. TP53 mutations were acquired longitudinally in 5 out of 156 patients, one of which carried a pre-existing but different TP53 mutation at enrollment that was retained, resulting in two distinct TP53 mutations(30–33). TP53 mutations were never lost and identical mutations were detected at enrollment and longitudinally.
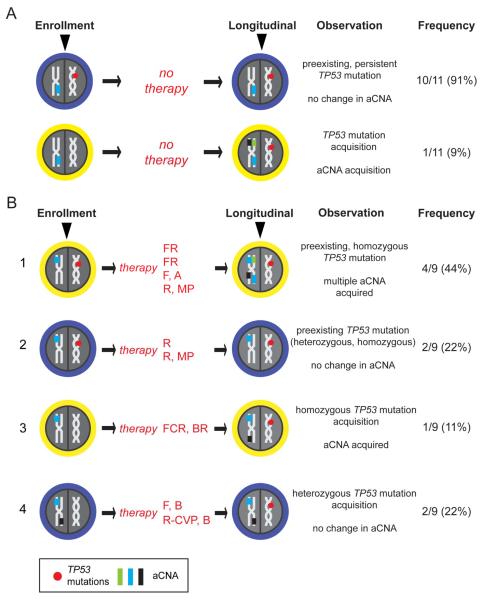
Of the 20 patients with either persistent or longitudinally acquired TP53 mutations, 11 remained untreated during the sampling interval. Of these 11 patients, 10 carried a completely stable aCNA pattern when comparing enrollment and longitudinal samples. Only one untreated CLL case (#140) without a preexisting TP53 mutation acquired a TP53 mutation, and with that acquisition gained 4 aCNAs. Conversely, 9 patients with either persistent or longitudinally acquired TP53 mutations received intercurrent therapy; of these, four patients with pre-existing TP53 mutations demonstrated strong numerical gains in aCNAs in their CLL genomes at relapse. Specifically, CLL #33 (treated with FR and PCR), CLL #74 (FR), CLL #104 (alemtuzumab) and CLL #239 (rituximab/Solumedrol) with baseline aCNA counts of 4, 7, 2 and 3 lesions, respectively, carried 11, 19, 21 and 7 aCNAs at relapse. Two CLL with pre-existing TP53 mutations, CLL #56 (rituximab) and CLL #84 (rituximab/Solumedrol), did not change aCNA counts. Furthermore, of the three CLL cases without preexisting TP53 mutations (CLL #117, treated with FCR and BR; CLL #125, treated with F and B sequentially; and CLL #219, treated with R-CVP and B sequentially) that acquired a TP53 mutation at relapse, one gained three aCNAs. These findings have been graphically summarized in Figure 5. Finally, the mean aCNA gain for TP53-mutated CLL after therapy was 5.1, while the mean aCNA gain for CLL with wild type TP53 after therapy was 0.6 (p=0.12).
Figure 5.
The relationship of aCNA acquisition and TP53 mutation and effects of therapy (schema). A: Frequencies of TP53 mutation events and aCNA acquisition or lack thereof in patients without intercurrent therapy. B: Frequencies of TP53 mutation events and aCNA acquisition in the context of therapies. FR: fludarabine and rituximab. F, A: fludarabine, alemtuzumab. R/MP: rituximab and methylprednisolone. FCR: fludarabine, cyclophosphamide, and rituximab. BR: bendamustine and rituximab. F, B: fludarabine, bendamustine. R-CVP: rituximab, cyclophosphamide, vincristine, and prednisone.
Summarizing the novel conclusions from the longitudinal TP53 mutation analysis, we provide evidence that: i) therapy of CLL with pre-existing TP53 mutations often facilitates outgrowth of CLL with markedly elevated genomic aCNA counts, a trait previously linked to very aggressive disease(7); and ii) that the genomic complexity of TP53 mutated CLL remains stable over time when left untreated, suggesting that treatment initiation at a clinically justified late time point in the course of these patients may be beneficial.
All novel longitudinally dominant CLL clones are clonally related to the dominant presentation clone in CLL
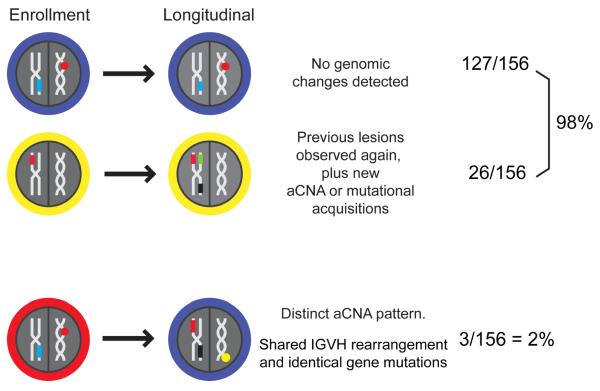
Comparing the genomic aberrations present at enrollment and at longitudinal analysis in the informative subset of CLL cases with detectable genomic aberrations at enrollment, we noted that 99% (130/132) of longitudinally analyzed cases shared genomic aberrations (aCNA/LOH or gene mutations) with the CLL at presentation. Further, when comparing the genomic aberrations present at enrollment and at longitudinal analysis in the 29 CLL cases with clonal changes, we noted that 90% (26/29) of the dominant longitudinal CLL clones were linearly related to the dominant CLL presentation clone (see Supplementary Table 1).
In contrast, the emergence of dominant CLL clones demonstrating branched evolution was rare. Specifically, CLL # 026 (untreated between samplings) lost a del13q14 of 1.225 Mb in length but gained a homozygous and shorter (0.41Mb) del13q14 in the setting of novel cn-LOH across chromosome 13. This case also gained a micro-del17p. CLL # 152 (untreated between samplings) lost a del13q14 of 3.124 Mb length but gained 5 unrelated aCNAs, including a homozygous but shorter del13q14 of 1.4 Mb in length (see Figure 1, red ovals). One additional informative case, CLL # 007 (treated with FR), lost three aCNA located on chromosomes 18 and 20, while gaining two aCNA at 13q14 at relapse. However, CLL # 007 carried an identical SF3B1 mutation at presentation and relapse. Finally, analysis of the IgVH status of all three CLL pairs disclosed identical IgVH rearrangements for the individual pairs demonstrating a shared clonal origin. These data have been schematically summarized in Figure 6.
Figure 6.
The frequency of genomic changes in CLL in longitudinal analysis and clonal relatedness (schema).
Therefore in summary, incomplete eradication of founder clones or the facilitated outgrowth of descendent subclones, which possess a greater degree of fitness and eventually become dominant underlied CLL evolution and relapse in all studied cases.
DISCUSSION
In this study, we have employed ultra-high-density SNP 6.0 arrays to interrogate the genomes of 156 CLL patients for acquired chromosomal copy number changes and LOH using paired longitudinally procured samples. Complementing this genome-wide analysis, we have performed exon resequencing of TP53, NOTCH1 and SF3B1, the most frequently mutated genes in CLL(34–38). Through paired genomic analysis of samples procured an average of 4 years apart, we are able to support the following principal conclusions: i) CLL-directed therapy facilitated outgrowth of more genomically complex CLL clones in ~40–50% of cases at relapse(16); ii) in CLL untreated between samplings, there was infrequent outgrowth or emergence of genomically evolved CLL clones that emerged over time when compared with the dominant presentation clone (11% incidence); iii) the types and spectrum of acquired genomic changes was similar in untreated and treated CLL, providing strong evidence for a therapy-independent origin; iv) TP53 mutations were acquired before and after therapy (but never lost), although the acquisition frequency of TP53 mutations even in relapsed patients was relatively low (~10% incidence)—this frequency is substantially lower than previously reported in highly selected drug-resistant CLL patient populations(13); v) importantly, the treatment of CLL harboring pre-existing TP53 mutations resulted in the dominance of very genomically complex CLL clones at relapse, and these clones displayed highly elevated aCNA counts previously measured in only a few percent of aggressive CLL cases(7); vi) the specific aCNAs and regions of LOH that are recurrently acquired in CLL at longitudinal time points of disease progression (del13q14 type II, which include RB1; del11q; del17p; and cn-LOH-chr.13) have been previously implicated in or suspected to contribute to either CLL proliferation or chemotherapy resistance, genomic complexity and shortened survival, therefore providing complementary evidence for their role as drivers of CLL evolution; and vii) the dominant CLL clone at longitudinal analysis could be traced back to the dominant presentation clone in all cases.
Importantly and inherent to longitudinal genomic analysis, some patients demonstrating clinically aggressive or genomically high-risk CLL (mutated TP53/del17p or elevated SNP 6.0 array-based genomic complexity) could not be repeatedly sampled; and therefore estimates of clonal evolution as reported here are very likely underestimates of the true frequency of this phenomenon in CLL.
This study is characterized by various methodological strengths that support the overall conclusions drawn, including: i) a very large patient cohort that was sampled longitudinally at time intervals long enough to provide meaningful estimates of clonal genomic evolution and devolution events; ii) genomic aCNA/LOH and gene resequencing analyses that were based on paired DNA samples (tumor and paired normal) for all CLL samples; iii) the use of high-purity, flow-sorted CD19+ and CD3+ cells (or rarely buccal DNA) as a source of DNA; and iv) a rigorous and conservative genomic copy number data analysis schema that had previously been externally validated using FISH(23).
In addition to aCNAs described previously as acquired in small fractions of patients over time (del11q, del17p), we identify multiple additional recurrent aCNA/cn-LOH as candidate CLL drivers(14, 16, 17, 19). However, despite analysis of 156 paired CLL samples in this study, an even larger number of analyzed samples procured before and after therapy is needed to provide better estimates of individual aCNA/cn-LOH or novel gene mutations as potential drivers of relapse.
The conclusions reported here are based on technologies that can measure genomic events at high sensitivity and specificity if they are represented in approximately 25% or more of the input DNA analyzed; therefore, minor subclones were not measured, and instead we focused on the characterization of CLL clones that actually constituted and contributed the bulk of the clinically overt disease. Therefore, this approach directly allows for clinically meaningful conclusions to be drawn. Recently, data have emerged quantifying cancer cell fractions of altered alleles in a limited number of paired CLL samples over time. Similar to findings reported here and previously by others cited above, enrichments for TP53 mutations and del11q were measured(39). In contrast however, we could not detect a role for SF3B1 - or NOTCH1 mutations (similar to findings very recently reported) (40) in evolving clonal dominance or as drivers of relapse in CLL, thus providing a stimulus for research into the role served by these recurrently mutated genes in CLL(41).
Interpreting the findings from an applied clinical perspective, we note that inappropriate early use of therapy in CLL could compromise patient prognoses through selection of genomically more aggressive CLL. On a more speculative note, the emergence of aggressive dominant CLL clones may in fact be constrained by the coexistence of less aggressive and quantitatively dominant CLL clones that compete for various resources. Inappropriate early use of therapy may reduce the potential beneficial effect of containment of aggressive clones by the more indolent clones and ultimately accelerates the disease. This concept is deserving of further in-depth study.
As one side observation of this study, we note that the frequency of acquisition of individual high-risk CLL genomic lesions like del17p, del11q, or TP53 mutations over time in an unselected CLL cohort was relatively low, suggesting that routine serial testing (unless guided by clinical concerns) is probably not indicated for most patients and that mutation testing for NOTCH1 and SF3B1 be restricted to research settings.
In summary, this data, based on the largest reported longitudinal multi-dimensional genomic analysis in CLL to date, demonstrate that emergence of novel dominant clones is frequent in relapsed CLL and relatively infrequent in untreated CLL, that the origin of most genomic changes however is likely therapy-independent and due to random mutagenesis, and that the acquisition of specific aCNAs, cn-LOH and TP53 mutations are the major recurrent clonal evolution events.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Postulating that genomic driver lesions with effects on CLL cell proliferation, apoptosis thresholds or chemotherapy resistance should increase in frequency over time, we measured aCNA/LOH and cnLOH and the mutational state of TP53, NOTCH1 and SF3B1 in 156 paired longitudinal CLL samples. The most frequently measured changes were acquisitions of del13q14 type II, which include RB1; del11q; del17p; copy neutral-LOH at chromosome 13; and TP53 mutations. Interestingly, while the frequency of acquisition of changes in treated as opposed to untreated CLL was approximately fourfold higher, the spectrum of genomic and mutational changes was largely similar, suggesting that the above cited and other recurrent genomic aberrations facilitate clonal dominance independent of - but aided by - therapies and further, that their origin is largely therapy-independent. Finally, the emergence of genomically hypercomplex CLL following the treatment of CLL with pre-existing TP53 mutations argues for careful justification of therapy initiation in this patient population.
Acknowledgement
Supported by the National Institutes of Health through CA136537 (SM), a Clinical Scholars Award of the Leukemia and Lymphoma Society of America (SM), the Translational Research Program of the Leukemia and Lymphoma Society of America (SM) and a collaborative research grant by the Lymphoma Research Foundation. This research is supported (in part) by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592). We are grateful for services provided by the microarray core of the University of Michigan Comprehensive Cancer Center.
Footnotes
Individual contributions: Peter Ouillette, Kamlai Saiya-Cork and Sami Malek performed the laboratory research. Kerby Shedden and Cheng Li assisted with statistical analysis and software development for data analysis.
Erlene Seymour assisted with database management.
Sami Malek conceived the study and supervised the work.
Peter Ouillette and Sami Malek wrote the paper.
Conflict of Interest: None to declare
REFERENCES
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Malek SN. The biology and clinical significance of acquired genomic copy number aberrations and recurrent gene mutations in chronic lymphocytic leukemia. Oncogene. 2012 doi: 10.1038/onc.2012.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 4.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 5.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Kujawski L, Ouillette P, Erba H, Saddler C, Jakubowiak A, Kaminski M, et al. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood. 2008;112:1993–2003. doi: 10.1182/blood-2007-07-099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouillette P, Collins R, Shakhan S, Li J, Peres E, Kujawski L, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118:3051–61. doi: 10.1182/blood-2010-12-327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malek S. Clinical utility of prognostic markers in chronic lymphocytic leukemia. ASCO Education Book 2010. 2010:263–7. [Google Scholar]
- 9.Van Bockstaele F, Verhasselt B, Philippe J. Prognostic markers in chronic lymphocytic leukemia: a comprehensive review. Blood Rev. 2009;23:25–47. doi: 10.1016/j.blre.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Rassenti LZ, Jain S, Keating MJ, Wierda WG, Grever MR, Byrd JC, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–30. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–84. [PubMed] [Google Scholar]
- 12.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–67. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Buhler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53–p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–97. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 14.Shanafelt TD, Hanson C, Dewald GW, Witzig TE, LaPlant B, Abrahamzon J, et al. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26:e5–6. doi: 10.1200/JCO.2008.16.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavazzini F, Rizzotto L, Sofritti O, Daghia G, Cibien F, Martinelli S, et al. Clonal evolution including 14q32/IGH translocations in chronic lymphocytic leukemia: analysis of clinicobiologic correlations in 105 patients. Leuk Lymphoma. 2012;53:83–8. doi: 10.3109/10428194.2011.606384. [DOI] [PubMed] [Google Scholar]
- 16.Knight SJ, Yau C, Clifford R, Timbs AT, Sadighi Akha E, Dreau HM, et al. Quantification of subclonal distributions of recurrent genomic aberrations in paired pre-treatment and relapse samples from patients with B-cell chronic lymphocytic leukemia. Leukemia. 2012;26:1564–75. doi: 10.1038/leu.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stilgenbauer S, Sander S, Bullinger L, Benner A, Leupolt E, Winkler D, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–5. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 18.Braggio E, Kay NE, VanWier S, Tschumper RC, Smoley S, Eckel-Passow JE, et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia. 2012;26:1698–701. doi: 10.1038/leu.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunnarsson R, Mansouri L, Isaksson A, Goransson H, Cahill N, Jansson M, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica. 2011;96:1161–9. doi: 10.3324/haematol.2010.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O'Brien S, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–7. [PubMed] [Google Scholar]
- 21.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–40. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 22.Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13:4777–85. doi: 10.1158/1078-0432.CCR-07-0456. [DOI] [PubMed] [Google Scholar]
- 23.Parkin B, Erba H, Ouillette P, Roulston D, Purkayastha A, Karp J, et al. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood. 2010;116:4958–67. doi: 10.1182/blood-2010-01-266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010;116:71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–21. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 26.Parker H, Rose-Zerilli MJ, Parker A, Chaplin T, Wade R, Gardiner A, et al. 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia. 2011;25:489–97. doi: 10.1038/leu.2010.288. [DOI] [PubMed] [Google Scholar]
- 27.Ouillette P, Collins R, Shakhan S, Li J, Li C, Shedden K, et al. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:6778–90. doi: 10.1158/1078-0432.CCR-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 29.Saddler C, Ouillette P, Kujawski L, Shangary S, Talpaz M, Kaminski M, et al. Comprehensive biomarker and genomic analysis identifies p53 status as the major determinant of response to MDM2 inhibitors in chronic lymphocytic leukemia. Blood. 2008;111:1584–93. doi: 10.1182/blood-2007-09-112698. [DOI] [PubMed] [Google Scholar]
- 30.Malcikova J, Smardova J, Rocnova L, Tichy B, Kuglik P, Vranova V, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114:5307–14. doi: 10.1182/blood-2009-07-234708. [DOI] [PubMed] [Google Scholar]
- 31.Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–57. [PubMed] [Google Scholar]
- 32.el Rouby S, Thomas A, Costin D, Rosenberg CR, Potmesil M, Silber R, et al. p53 gene mutation in B-cell chronic lymphocytic leukemia is associated with drug resistance and is independent of MDR1/MDR3 gene expression. Blood. 1993;82:3452–9. [PubMed] [Google Scholar]
- 33.Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 1991;88:5413–7. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 36.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–8. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villamor N, Conde L, Martinez-Trillos A, Cazorla M, Navarro A, Bea S, et al. NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia. 2012 doi: 10.1038/leu.2012.357. [DOI] [PubMed] [Google Scholar]
- 41.Shedden K, Li Y, Ouillette P, Malek SN. Characteristics of chronic lymphocytic leukemia with somatically acquired mutations in NOTCH1 exon 34. Leukemia. 2012;26:1108–10. doi: 10.1038/leu.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.